Abstract
Background
Head and neck squamous cell carcinoma (HNSCC) is characterized by high proliferation and limited differentiation. The altered expression of the p53 family members, and specifically of p63, represents a pivotal event in the pathogenesis of HNSCC. Physiologically, p63 affects metabolism through the direct transactivation of the enzyme hexokinase 2, and subsequently controls the proliferation of epithelial cells; nonetheless, its role in cancer metabolism is still largely unclear. The high energetic demand of cancer and the consequent needs of a metabolic reshape, also involve the serine and glycine catabolic and anabolic pathways, including the one carbon metabolism (OCM), to produce energetic compounds (purines) and to maintain cellular homeostasis (glutathione and S-adenosylmethionine).
Results
The involvement in serine/glycine starvation by other p53 family members has been reported, including HNSCC. Here, we show that in HNSCC p63 controls the expression of the enzymes regulating the serine biosynthesis and one carbon metabolism. p63 binds the promoter region of genes involved in the serine biosynthesis as well as in the one carbon metabolism. p63 silencing in a HNSCC cell line affects the mRNA and protein levels of these selected enzymes. Moreover, the higher expression of TP63 and its target enzymes, negatively impacts on the overall survival of HNSCC patients.
Conclusion
These data indicate a direct role of p63 in the metabolic regulation of HNSCC with significant clinical effects.
Similar content being viewed by others
Background
Head and neck squamous cell carcinoma (HNSCC) is the sixth malignancy in the world by incidence [1, 2] with a significant mortality rate [3]. HNSCC represents a heterogeneous group of tumors with a communal origin from the epithelial cells lining in the oral cavity, the oropharynx, the larynx, or the hypopharynx [4, 5]. Among known risk factors, tobacco and alcohol have a synergistic effect in HNSCC onset, but also human papilloma virus and/or Epstein–Barr virus infection can play a recognized role in its occurrence [6, 7]. The viral or environmental link has a direct effect on the clinical outcome. Surgery and chemo-radiotherapy, primary treatments for HNSCC, critically impact on patients’ quality of life and tumor relapse is invariably very common [8,9,10]. Surgical removal and the combination of radiotherapy with chemotherapy are the first line therapeutic approaches against this type of cancer as well as in other squamous malignancies such as the esophageal carcinoma [11,12,13,14]. Although advances in therapy based on a better understanding of HNSCC pathogenesis, as the association of Cetuximab (anti EGFR monoclonal antibody) to standard chemotherapy, treatments remain, at date, not very effective and only 40–50% of HNSCC patients will survive 5 years after the diagnosis [15, 16]. From a molecular point of view, the amplification or deregulated expression of the genomic locus encoding for one of the members of the p53 family [17, 18], p63, is a frequent event in HNSCC [19], beyond the contribution of p53 itself [20,21,22,23], whose genomic alteration is present in 70% of HNSCC cases [24]. As well described elsewhere, the involvement of p53 in tumour suppression [25,26,27] is widely recognized [28], impinging on translation [29, 30], cell death [31, 32], epigenetic regulation [33,34,35] or metabolism [36,37,38,39]. p63 is specifically expressed in epithelia and regulates cellular proliferation and stem cells capacity [40,41,42]. An alteration of the expression of a amino-shorter isoform of p63 (ΔΝp63), codified by the second promoter, seems to play a pivotal role in HNSCC; indeed, recent studies indicate its involvement in aberrated cell survival, renewal, and senescence suppression [43,44,45]. Moreover, in normal epithelia ΔΝp63 is involved, together with the catalytic subunits of SWI/SNF complex (Brg1 or BRM), in maintaining a cell-type specific open chromatin to control the enhancer landscape for normal cell differentiation [45,46,47]. By contrast, the association between ΔΝp63 and another component of the SWI/SNF complex, ACTL6A, is responsible of the reduction of chromatin accessibility and of the alteration of gene transcription profiling typical of HNSCC [48]. In normal epithelial cells, i.d. keratinocytes, ΔΝp63 controls cellular metabolism through direct transactivation of the first enzyme of glycolysis hexokinase 2 (HK2) [49]. This evidence suggests that ΔΝp63 might be able to control cellular energy production to sustain cell proliferation [42, 49]. Cancer cells undergo a metabolic adaptation to maintain a high rate of cellular proliferation, where glucose and the amino acid glutamine are the major metabolic sources to maintain glycolysis and Krebs cycle [50,51,52]. The high energetic demand of cancer cells involves the anabolic pathway of serine biosynthesis where the enzyme phosphoglycerate dehydrogenase (PHGDH) converts 3-phospho- glycerate generated from glycolysis into the precursor 3-phosphohydroxypyruvate; subsequently the enzymes phosphoserine aminotransferase 1 (PSAT1) and phosphoserine phosphatase (PSPH) convert the 3-phosphohydroxypyruvate into serine [53]. Distinct studies demonstrate that PHGDH upregulation and serine biosynthesis can play a role in sustaining cancer cell growth and transformation [54, 55]. This biosynthetic pathway, known as “serine de novo biosynthesis”, refuels Krebs cycle through PSAT1 converting glutamate to α-ketoglutarate, an anaplerotic intermediate of the cycle. In general, serine synthesis represents the major source of intermediate of Krebs cycle (approximately half of the anaplerotic flux). The amino acid serine, produced by this reaction, becomes the substrate of other two enzymes, the hydroxymethyltransferases SHMT1 and SHMT2, localized in the cytosol and mitochondria, respectively [53]. The reversible conversion of tetrahydrofolate (THF) and serine to methylenetetrahydrofolate (me-THF) and glycine catalyzed by SHMTs enzymes, represents the major source of hydroxy-methyl groups (–CH3) required for glutathione (GSH), purines and S-adenosylmethionine biosynthesis (methyl groups donor for DNA/histone methylation) [56]. All together these metabolic reactions are known as one carbon metabolism (OCM), fundamental for cell energy production and homeostasis maintenance [57, 58]. In human keratinocytes the extracellular availability of serine and glycine is essential for normal cell proliferation and the intracellular inhibition of SHMTs enzymes, responsible for serine and THF conversion into glycine and 5,10 methylene-THF ameliorates psoriatic skin hyperproliferation and inflammation [59]. In addition, the modulation of the activity of SHMT2 and methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2 (MTHFD2) can affect cutaneous squamous cell carcinoma proliferation [60]. In the recent years the study of cancer metabolism, that comprises tumor and immune cell reprogramming, extracellular and intracellular metabolites availabilities for cellular transformation, migration and invasion, and the metabolic response to chemotherapy, represents a fundamental point to completely understand tumor cell transformation and/ or resistance to therapy [61,62,63,64] and to better evaluate some of the molecular alterations that are linked to HNSCC pathogenesis [65, 66].
Despite the alteration of the serine biosynthetic pathway and OCM has been shown in many human cancers, its role in HNSCC has not been yet described.
Results
p63, serine and OCM enzymes are altered and differentially expressed in HNSCC
The p53 family members (p53, p63 and p73) are strongly altered at the genomic level in a cohort of HNSCC human samples analyzed with the OncoPrint profiling from the TCGA Firehose Legacy dataset through the cBioportal platform (71%, 26% and 7% genomic and/or transcriptomic alterations, respectively). Additionally, the serine de novo biosynthesis and OCM enzymes show various alterations at a genomic and transcriptional level. Among all the enzymes analyzed in the same cohort of patients from the TCGA, PSPH represents the most altered gene (Fig. 1a), frequently amplified. Moreover, patients’ overlap analysis of expression alterations in the OCM and serine de novo genes with TP63 alterations in the same HNSCC dataset analyzed on cBioportal, shows a greater number of patients altered in the TP63-PSPH group compared to the other overlaps (Fig. 1b). The expression analysis of TP63 and serine de novo and OCM enzymes using a publicly available dataset of HNSCC (GSE12452) shows that these genes are differentially up regulated in HNSCC samples compared to the normal control (Fig. 1c). In addition, single cell RNA sequencing revels that there is an expression gradient starting from non-lesional samples (NL), leukoplakia samples (LP) to positive lymph nodes samples (LN) and primary cancer samples (CA) (Additional file 1: Figure S1a). Altogether, this evidence suggests that OCM and serine de novo enzymes, together with p63, could be involved in HNSCC pathogenesis.
Serine and one carbon metabolism enzymes are overexpressed and deregulated in HNSCC patients. a OncoPrint profiling of TP53, TP63, TP73, serine de novo and one carbon metabolism (OCM) enzymes alterations in HNSCC, analyzed from the TCGA Firehose Legacy dataset on the cBioportal platform. b Patients’ overlap analysis of expression alterations in the OCM and serine de novo genes with TP63 in HNSCC dataset analyzed on cBioportal. c Expression analysis of TP63 and serine de novo and OCM enzymes using a publicly available dataset of HNSCC (GSE12452). The p-value has obtained using Student’s t test with Welch’s correction. Values were considered significant with p value < 0.05 (n.s. = not significant)
p63 directly binds serine de novo metabolism and OCM genes promoter regions
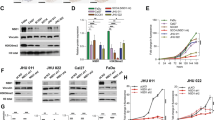
To better understand if there is a mechanistic link between the epithelial master regulator p63 and serine de novo biosynthesis enzymes, publicly available chromatin immunoprecipitation sequencing (ChIP seq) of p63 performed on human primary keratinocytes and HaCaT cells (GSE126390, GSE33571, GSE56674, GSE60814, GSE67382, GSE59827) were visualized and analyzed on Genome Browser (https://genome.ucsc.edu/) (Fig. 2a). The direct binding of p63 to the promoter regions of these genes is indicated as black bands and/or peaks corresponding to H3K27ac area of open chromatin; these areas are highlighted in light blue and represent the chromatin regions where p63 can bind PHGDH, PSAT1, PSPH and MTHFD2 promoters. To confirm this bioinformatic evidence, a chromatin Immunoprecipitation for p63 in the same genomic regions was performed on FaDu cells (Fig. 2b). As shown by p63 fold enrichment respect to the IgG control, we demonstrate the physical binding of this transcription factor on PHGDH, PSAT1, PSPH and MTHFD2 promoters. Additionally, these enzymes result to be more expressed in single cell RNA seq of primary and metastatic cancer compared to the normal cells (Additional file 1: Figure S2a–c).
p63 directly binds promoter regions of genes involved in serine metabolism and OCM. a Chromatin Immunoprecipitation sequencing (ChIP seq) of p63 performed on human primary keratinocytes and HaCaT cells, available on Geo Dataset (GSE126390, GSE33571, GSE56674, GSE60814, GSE67382, GSE59827), visualized on Genome Browser (https://genome.ucsc.edu/); b ChIP experiment on FaDu using anti-p63 antibody to evaluate p63 binding on the promoters of PHGDH, PSAT1, PSPH and MTHFD2 genes. One representative experiment is shown
Taken together these results show that there is a direct transcriptional link between p63 and serine de novo biosynthesis enzymes and OCM enzymes.
p63 modulates serine de novo metabolism and OCM expression levels and their molecular signature impacts on patients’ overall survival
After demonstrating that p63 can directly bind the promoter regions of serine de novo and OCM enzymes, we performed a correlation analysis between p63 protein expression and PHGDH, PSAT1, PSPH and MTHFD2 mRNA expression levels in HNSCC (Additional file 1: Figure S3a). To this end, we analyzed the mRNA expression level of PHGDH, PSAT1, PSPH and MTHFD2 after p63 depletion in FaDu and HaCaT cell lines and we found a significant reduction of their expression under silencing conditions (Additional file 1: Figure S3b–c). Moreover, we performed p63 silencing with 2 different siRNA in FaDu cells showing that there is a significant reduction of PHGDH, PSAT1, PSPH and MTHFD2 mRNA level at 48 h (Fig. 3a) and a significant reduction of PHGDH, PSPH and MTHFD2 and a tendence to decrease for PSAT1 at 72 h (Fig. 3b). In addition, after 72 h of silencing we also observe a significant reduction of PSPH protein level (Fig. 3c, d).
p63 silencing decreases the expression of phosposerine phosphatase enzyme (PSPH) in FaDu. a Relative mRNA levels of genes involved in de novo serine biosynthesis (PHGDH, PSAT1 and PSPH) and OCM (MTHFD2) after p63 silencing for 48 h; b relative mRNA levels of PHGDH, PSAT1 and PSPH and MTHFD2 after p63 silencing for 72 h; c Western blot analysis of PHGDH, PSAT1, PSPH and MTHFD2 after 72 h of p63 silencing. d Relative ratio of PHGDH, PSAT1, PSPH and MTHFD2 after p63 silencing for 72 h, normalized protein levels. One representative experiment of 3 is shown. The p-value was obtained using ordinary one-way analysis of variance (ANOVA). Values were considered significant when p value < 0.05 (n.s. = not significant)
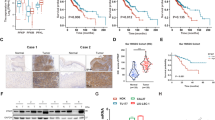
Despite the strong number of alterations in HNSCC, the high expression of TP63 alone and the high expression of PHGDH, PSAT1, PSPH and MTHFD2 (four-genes signature) are not sufficient to impact on HNSCC patients’ overall survival, but the high expression of TP63, PHGDH, PSAT1, PSPH and MTHFD2 (five-genes signatures) seems to be more impacting on patients’ overall survival (Fig. 4a–c). All together these data confirm the importance of p63 direct control of serine biosynthesis and OCM enzymes expression in HNSCC pathogenesis which might be summarized in Fig. 5.
p63 and OCM mRNA molecular signature impacts on HNSCC overall survival. a Overall Survival analysis of HNSCC based on mRNA expression; in the first panel, patients are divided between high and low TP63 expression, in the second panel between high and low expression of the OCM gene signature (PHGDH, PSAT1, PSPH and MTHFD2), in the last panel, between high and low expression of the TP63&OCM gene signature (TP63, PHGDH, PSAT1, PSPH and MTHFD2); the p value was obtained using the log-rank test. Values were considered significant when the p value < 0.05 (n.s. = not significant)
p63 controls serine de novo biosynthesis by transcriptionally regulation of PSPH. Schematic representation summarizing the biochemical pathways in which serine and OCM enzymes are involved, highlighting the transcriptional regulation by the p53 family members and, specifically, p63 regulation on PSPH gene
Discussion
p63-driven metabolic changes in HNSCC
Various cancers form distinct anatomical origin fall into the category of HNSCC. All these neoplasms have a common feature that is the origin from a squamous epithelium form the upper respiratory tract. HNSCC are largely caused by exogenous insults to the epithelium such as cigarette smoke and alcohol abuse. A distinct subset of HNSCC show a clear link with viral infections, specifically from human papilloma virus or Epstein-Barr virus. Cancers belonging to this group are often diagnosed at a local stage, which can be quite advanced [67], and are primarily treated with two approaches: surgery, if the mass is accessible (which is not the case of a few sinuses cancer) and non-locally advanced, or with a combination of daily radiotherapy with chemotherapy (usually with platinum-based compounds) with neoadjuvant or even curative intents [10, 68]. Even if radio-chemotherapy can achieve complete responses also in cases of locally advanced disease, local and/or metastatic recurrence is inevitable in almost half of the patients [69]. The association with targeted therapies, such as the case of Cetuximab, has shown incremented activity, but still failing in prolonging overall survival.
HNSCC shows an extremely high percentage of mutations in the p53 gene, up to the 70% of cases [24]. Indeed, like in several other cancers [70,71,72,73], p53 and its family members seems to be of high relevance. Anyhow, p63, another p53 family member, has been recognized as playing a pathogenetic role in this cancer since its gene locus is often amplified. TP63 gene deregulation is a frequent event in squamous cancers (it can be seen also in esophageal, lung and cervix cancer) since p63 is involved in the maintenance of epithelial homeostasis, especially in those cells in which p53 is somehow inactivated [74]. Being a transcription factor, p63 activity has been described in various circumstances in different cancers, sustaining cell survival, cell renewal and reducing senescence [45]. Moreover, since epithelial tissues require a high metabolic energetic flux due to constant proliferation, p63 has been described in regulating cellular metabolism in normal and pathological conditions such as cancer [40].
Cancer cells have a continuous demand for energy production, therefore their metabolism is reshaped to produce a greater number of Krebs cycle intermediates to refuel the respiratory chain with electrons [75, 76]. Numerous catalytic and anabolic metabolisms, which can provide such intermediates, are altered in cancer. Among them the serine de novo biosynthesis and one carbon metabolism are often deregulated since they can provide Krebs cycle intermediates from glycolysis derived pyruvate (in the usually glycolytic environment of the cancer cell), providing also the hydroxy-methyl groups which are essential for chromatin remodeling (through the synthesis of s-adenosylmethionine), glutathione and purine synthesis, which are also fundamental for cell proliferation [57, 77].
In HNSCC the expression of genes coding for enzymes involved either in the serine de novo biosynthesis and the OCM have been found deregulated at a genomic and transcriptomic level in a cohort of patients form the TCGA. In this group of patients, most of the enzymes of the two metabolisms show altered RNA expression, but the most striking altered expressed gene is PSPH, altered in up to the 11% of patients, most invariably amplified. The alteration of the expression levels of these genes seems to be specific for cancer cells since their expression levels in HNSCC tissues are invariably higher if compared to their normal matching controls.
Conclusion
P63, like its family members, has already been demonstrated to control specific cell metabolism in various contexts either through its transcriptional activity or through direct transactivation of key enzymes such as hexokinase 2 [40, 42, 49]. In keratinocytes, p63 has a direct interaction with the promoter regions of many of the genes involved in serine de novo biosynthesis and OCM (PHGDH, PSAT1, PSPH and MTHFD2), which is also true in FaDu cells, a well-established HNSCC cell line model, where p63 shows the same binding to the promoter region of the abovementioned genes. In FaDu cells, the silencing of p63 reduces the expression levels of the genes where its binding on the promoter has been shown and decreases the protein levels of the enzyme PSPH, the last enzyme of the serine biosynthesis.
Since the alteration of p63 in HNSCC is a documented event, the serine de novo biosynthesis and the OCM alteration, which can be under the regulation of p63 itself in HNSCC should be better evaluated, representing a potential therapeutical target since many compounds are rapidly being made available for modulating the activity of the enzymes of these two metabolisms (e.g. PHGDH inhibitors, SHMT2 inhibitors) and the influence of metabolism on sensitivity to therapy has been demonstrated in various tumoral contexts [63, 78,79,80]. Moreover, apart from therapeutical purposes, the alteration of the genes/enzymes of the two metabolisms, should also be thoroughly evaluated as predictive biomarkers, since a 5-genes signature (TP63, PHGDH, PSAT1, PSPH) seems to correlate with HNSCC overall survival and considering the lack of effective predictive tools [81,82,83].
Altogether our findings show a possible involvement of serine de novo synthesis and OCM in HNSCC pathogenesis, potentially under the control of p63, representing the first step towards the understanding of the metabolic reshape of this heterogeneous group of malignancies.
Material and methods
In vitro cell culture and transfection
FaDu cells were seeded, in MEM (Corning, Cat. No. 10-109-CV) enriched of 10% FBS, 2 mM L-glutamine, 10 mM HEPES, 1 mM sodium pyruvate, 100 u/ml penicillin, and 100 mg/mL streptomycin. The cells were cultured in the medium at 37 °C and 5% CO2. FaDu cell line was transfected for siRNA-mediated knockdown experiments for 48 and 72 h. The cells (6 × 105 cells) were seeded in 10 mm plates and transfected with specific siRNAs for p63 (Table 1) using Lipofectamine RNAiMAX transfection reagent (Invitrogen).
RNA extraction and RT-qPCR analysis
Total RNA was isolated from FaDu cells by using RLT lysis buffer (Qiagen) and it was purified using RNeasy Mini Kit (Qiagen). RNA quantification was performed by NanoDrop spectrophotometer (Thermo Scientific). cDNA was synthesized using SensiFAST™ cDNA synthesis kit (Bioline). The qPCR was performed by GoTaq Real-Time PCR System (Promega) in an Applied Biosystems 7500 Real-Time 15 PCR System (Applied Biosystems). Appropriate qPCR primers (Table 2) were used for quantitative PCR; The housekeeping gene TBP is used as internal control for data normalization. The expression of each gene was defined by the threshold cycle (Ct), and relative expression levels were calculated by using the 2−ΔΔCt method.
Immunoblotting analysis
FaDu cells were lysed with RIPA lysis buffer (50 mM Tris–cl pH 7.4; 150 mM NaCl; 1% NP40; 0.25% Na-deoxycholate; 1 mM AEBSF; and 1 mM DTT). The total protein extracts (20 µg) were separated using SDS polyacrylamide gels and transferred on PVDF membrane. Then, the membranes were incubated overnight at 4 °C with the primary antibody. The membranes were incubated with secondary antibody, later PBS-T washes, for 1 h at room temperature. Immunoblotting signals were captured using Uvitec Alliance Q9 Advanced (PMID: 9531492; PMID: 9794797; PMID: 15698515). The following primary antibodies were implied for western blot experiments: anti-p63 (Cell Signalling, Cat. No. 13109, 1:1000), anti-PHGDH (Sigma, Cat. No. HPA021241, 1:1000), anti-PSAT1 (Abcam ab154055, 1:500), anti-PSPH (Sigma, Cat. No. HPA020376, 1:300), anti-MTHFD2 (Abcam Cat. No. ab151447, 1:1000) and anti-vinculin (Sigma, Cat. No. V9131 1:10000). Uncropped images of the western blots are shown in additional file 1.
Statistical analysis
Data, which represent three independent experiments, are expressed as means ± SDs. GraphPad Prism 8.0 software (San Diego, CA, USA) was performed for statistical analysis, ordinary one-way analysis of variance (ANOVA) and parametric Student’s t test was applied for comparing different samples. Values of P < 0.05 was considered statistically significant.
Chromatin immunoprecipitation (ChIP) analysis
FaDu cells were cross-linked with 1% formaldehyde (Sigma, Cat. No F1635) for 10 min at room temperature to perform ChIP assay. The cross-linking reaction was blocked by addition of glycine (Sigma, Cat. No G-7126) 125 mM. After washing three times with PBS, chromatin was collected in cell lysis buffer (Pipes 5 mM pH 8.0, KCl 85 mM, NP40 0,5%) for 5 min at 4 °C. Later, chromatin was transferred in nuclei lysis buffer (Tris–Cl 50 mM pH8.0, EDTA 10 mM, SDS1%) and fragmented by sonication. To verify the amount of the chromatin in a range of 250–500 bp, after sonication, it was applied gel electrophoresis. Immunoprecipitation assay was performed overnight using Dynalbeads Protein G (Invitrogen, Cat. No 10004D) and antibodies (5–10 μg) anti-p63 (Cell Signaling, Cat. No. ab13109) or IgG (Invitrogen, Cat. No. 10500C), as control. Immunoprecipitates were washed in IP dilution buffer (SDS 0,01%, Triton X-100 1,1%, EDTA 1,2 mM, Tris–Cl 16,7 mM pH8, NaCl 167 mM) for 3 times, in dialysis buffer (EDTA 2 mM, Tris–Cl 50 mM pH8,0, Sarkosyl 0,2%) for 3 times, in TSE buffer (SDS 0,1%, Triton X-100 1%, Tris 20 mM pH8.1, EDTA 2 mM, NaCl 50 mM) for 3 times, in IP Wash buffer (Tris–Cl 100 mM pH9.0, LiCl 500 mM, NP40 1%, Deoxycholic acid 1%) for 3 times and TE (Tris–Cl pH7.0, EDTA 2 mM) for 1 time. Immunoprecipitation was eluted in elution buffer (NaHCO3 50 mM, SDS 1%) at 65 °C overnight and at least purificated by QIAquick PCR purification kit (Qiagen, Cat. No. 28104). ChIP-qPCR was performed using primers (Table 3) in an Applied Biosystems 7500 Real-Time 15 PCR System (Applied Biosystems, Carlsbad, CA). Real-Time PCR data were normalized to the amount of input chromatin.
Bioinformatics analysis
The Oncoprint profiling and Patients’ overlap analysis of expression alterations of TP63, serine metabolism and OCM enzymes in HNSCC (TCGA firehose legacy) was performed using cBioPortal for Cancer Genomic (http://www.cbioportal.org).
For TP63, serine metabolism and OCM enzymes expression at mRNA level in FaDu and in HaCaT cells two publicly databases available through GEO datasets (https://www.ncbi.nlm.nih.gov/gds) were analyzed (GSE88833 and GSE88832 respectively). ChIP sequencing publicly data (GSE126390, GSE33571, GSE56674, GSE60814, GSE67382, GSE59827) was visualized using Genome Browser (https://genome.ucsc.edu/). The correlation analysis between p63 (protein) and PHGDH, PSAT1, PSPH and MTHFD2 (mRNA) in HNSCC (TCGA firehose legacy) was performed using cBioPortal for Cancer Genomic (http://www.cbioportal.org).
Availability of data and materials
For Serine metabolism and OCM enzymes expression at mRNA level in HNSCC samples a publicly datasets available through GEO datasets (https://www.ncbi.nlm.nih.gov/gds) was analyzed (GSE12452). The expression of serine and OCM metabolism enzymes in NL: non lesional samples; LP: Leukoplakia samples; LN: Lymph nodes + samples; CA: primary cancer samples of HNSCC was analyzed using through Single Cell RNA sequencing publicly datasets GEO datasets (GSE181919). For p63 binding on serine and OCM enzymes promoter regions different Chromatin Immunoprecipitation sequencings (ChIP seq) (GSE126390, GSE33571, GSE56674, GSE60814, GSE67382, GSE59827) were visualized and analyzed using UCSC genome Browser (https://genome.ucsc.edu/).
Abbreviations
- HNSCC:
-
Head and neck squamous cell carcinoma
- HK2:
-
Hexokinase 2
- PHGDH:
-
Phosphoglycerate dehydrogenase
- PSAT1:
-
Phosphoserine aminotransferase 1
- PSPH:
-
Phosphoserine phosphatase
- SHMT1:
-
Serine hydroxymethyltransferases 1
- SHMT2:
-
Serine hydroxymethyltransferases 2
- THF:
-
Tetrahydrofolate
- Me-THF:
-
Methylenetetrahydrofolate
- GSH:
-
Glutathione
- OCM:
-
One carbon metabolism
- MTHFD2:
-
Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2
References
Guo D, Yang M, Li S, Zhu W, Chen M, Pan J, et al. Expression and molecular regulation of non-coding RNAs in HPV-positive head and neck squamous cell carcinoma. Front Oncol. 2023;13:1122982.
Leemans CR, Braakhuis BJM, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22.
Xue Y, Jiang X, Wang J, Zong Y, Yuan Z, Miao S, et al. Effect of regulatory cell death on the occurrence and development of head and neck squamous cell carcinoma. Biomark Res. 2023;11:2.
Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer. 2018;18:269–82.
Zhou W-H, Wang Y, Yan C, Du W-D, Al-Aroomi MA, Zheng L, et al. CC chemokine receptor 7 promotes macrophage recruitment and induces M2-polarization through CC chemokine ligand 19&21 in oral squamous cell carcinoma. Discov Oncol. 2022;13:67.
Gatti V, Fierro C, Annicchiarico-Petruzzelli M, Melino G, Peschiaroli A. ΔNp63 in squamous cell carcinoma: defining the oncogenic routes affecting epigenetic landscape and tumour microenvironment. Mol Oncol. 2019;13:981–1001.
Schindele A, Holm A, Nylander K, Allard A, Olofsson K. Mapping human papillomavirus, Epstein–Barr virus, cytomegalovirus, adenovirus, and p16 in laryngeal cancer. Discov Oncol. 2022;13:18.
Ionna F, Bossi P, Guida A, Alberti A, Muto P, Salzano G, et al. Recurrent/metastatic squamous cell carcinoma of the head and neck: a big and intriguing challenge which may be resolved by integrated treatments combining locoregional and systemic therapies. Cancers (Basel). 2021;13:2371.
Schaaij-Visser TBM, Graveland AP, Gauci S, Braakhuis BJM, Buijze M, Heck AJR, et al. Differential proteomics identifies protein biomarkers that predict local relapse of head and neck squamous cell carcinomas. Clin Cancer Res. 2009;15:7666–75.
Nomori H, Shiraishi A, Honma K, Shoji K, Otsuki A, Cong Y, et al. Differences between lung adenocarcinoma and squamous cell carcinoma in histological distribution of residual tumor after induction chemoradiotherapy. Discov Oncol. 2021;12:36.
Ho AL. Immunotherapy, chemotherapy, or both: options for first-line therapy for patients with recurrent or metastatic head and neck squamous cell carcinoma. J Clin Oncol. 2023;41:736–41.
Li C-C, Chen C-Y, Chou Y-H, Huang C-J, Ku H-Y, Lin Y-C, et al. High vs. low radiotherapy dose in locally advanced esophageal squamous cell carcinoma patients treated with neoadjuvant concurrent chemoradiotherapy: an endemic area population-based study. Discov Oncol. 2022;13:130.
Pierik AS, Leemans CR, Brakenhoff RH. Resection margins in head and neck cancer surgery: an update of residual disease and field cancerization. Cancers (Basel). 2021;13:2635.
Qin Q, Jun T, Wang B, Patel VG, Mellgard G, Zhong X, et al. Clinical factors associated with outcome in solid tumor patients treated with immune-checkpoint inhibitors: a single institution retrospective analysis. Discov Oncol. 2022;13:73.
Taberna M, Oliva M, Mesía R. Cetuximab-containing combinations in locally advanced and recurrent or metastatic head and neck squamous cell carcinoma. Front Oncol. 2019;9:383.
Fasano M, Della Corte CM, Viscardi G, Di Liello R, Paragliola F, Sparano F, et al. Head and neck cancer: the role of anti-EGFR agents in the era of immunotherapy. Ther Adv Med Oncol. 2021;13:175883592094941.
Vitale I, Pietrocola F, Guilbaud E, Aaronson SA, Abrams JM, Adam D, et al. Apoptotic cell death in disease-current understanding of the NCCD 2023. Cell Death Differ. 2023;30:1097–154.
Ganini C, Amelio I, Bertolo R, Bove P, Buonomo OC, Candi E, et al. Global mapping of cancers: The Cancer Genome Atlas and beyond. Mol Oncol. 2021;15:2823–40.
Compagnone M, Gatti V, Presutti D, Ruberti G, Fierro C, Markert EK, et al. ΔNp63-mediated regulation of hyaluronic acid metabolism and signaling supports HNSCC tumorigenesis. Proc Natl Acad Sci U S A. 2017;114:13254–9.
Kennedy MC, Lowe SW. Mutant p53: it’s not all one and the same. Cell Death Differ. 2022;29:983–7.
Rozenberg JM, Zvereva S, Dalina A, Blatov I, Zubarev I, Luppov D, et al. The p53 family member p73 in the regulation of cell stress response. Biol Direct. 2021;16:23.
Levine AJ. Exploring the future of research in the Tp53 field. Cell Death Differ. 2022;29:893–4.
Mammarella E, Zampieri C, Panatta E, Melino G, Amelio I. NUAK2 and RCan2 participate in the p53 mutant pro-tumorigenic network. Biol Direct. 2021;16:11.
Klinakis A, Rampias T. TP53 mutational landscape of metastatic head and neck cancer reveals patterns of mutation selection. EBioMedicine. 2020;58:102905.
Hoyos D, Greenbaum B, Levine AJ. The genotypes and phenotypes of missense mutations in the proline domain of the p53 protein. Cell Death Differ. 2022;29:938–45.
Boutelle AM, Attardi LD. p53 and tumor suppression: it takes a network. Trends Cell Biol. 2021;31:298–310.
Thomas AF, Kelly GL, Strasser A. Of the many cellular responses activated by TP53, which ones are critical for tumour suppression? Cell Death Differ. 2022;29:961–71.
Panatta E, Zampieri C, Melino G, Amelio I. Understanding p53 tumour suppressor network. Biol Direct. 2021;16:14.
Lindström MS, Bartek J, Maya-Mendoza A. p53 at the crossroad of DNA replication and ribosome biogenesis stress pathways. Cell Death Differ. 2022;29:972–82.
Kasteri J, Das D, Zhong X, Persaud L, Francis A, Muharam H, et al. Translation control by p53. Cancers (Basel). 2018;10:133.
El-Saafin F, Bergamasco MI, Chen Y, May RE, Esakky P, Hediyeh-Zadeh S, et al. Loss of TAF8 causes TFIID dysfunction and p53-mediated apoptotic neuronal cell death. Cell Death Differ. 2022;29:1013–27.
Engeland K. Cell cycle regulation: p53–p21-RB signaling. Cell Death Differ. 2022;29:946–60.
Levine AJ, Berger SL. The interplay between epigenetic changes and the p53 protein in stem cells. Genes Dev. 2017;31:1195–201.
Vrba L, Junk DJ, Novak P, Futscher BW. p53 induces distinct epigenetic states at its direct target promoters. BMC Genom. 2008;9:486.
Peng T, Liu M, Hu L, Guo D, Wang D, Qi B, et al. LncRNA Airn alleviates diabetic cardiac fibrosis by inhibiting activation of cardiac fibroblasts via a m6A-IMP2-p53 axis. Biol Direct. 2022;17:32.
Agostini M, Mancini M, Candi E. Long non-coding RNAs affecting cell metabolism in cancer. Biol Direct. 2022;17:26.
Panatta E, Butera A, Celardo I, Leist M, Melino G, Amelio I. p53 regulates expression of nuclear envelope components in cancer cells. Biol Direct. 2022;17:38.
Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700.
Priami C, Montariello D, De Michele G, Ruscitto F, Polazzi A, Ronzoni S, et al. Aberrant activation of p53/p66Shc-mInsc axis increases asymmetric divisions and attenuates proliferation of aged mammary stem cells. Cell Death Differ. 2022;29:2429–44.
Di Daniele N, Viticchiè G, Melino G, Lena AM, Novelli F, Piro MC, et al. Metabolic pathways regulated by p63. Biochem Biophys Res Commun. 2017;482:440–4.
Smirnov A, Lena AM, Cappello A, Panatta E, Anemona L, Bischetti S, et al. ZNF185 is a p63 target gene critical for epidermal differentiation and squamous cell carcinoma development. Oncogene. 2019;38:1625–38.
Latina A, Viticchiè G, Lena AM, Piro MC, Annicchiarico-Petruzzelli M, Melino G, et al. ΔNp63 targets cytoglobin to inhibit oxidative stress-induced apoptosis in keratinocytes and lung cancer. Oncogene. 2016;35:1493–503.
Rocco JW, Leong C-O, Kuperwasser N, DeYoung MP, Ellisen LW. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9:45–56.
Frezza V, Fierro C, Gatti E, Peschiaroli A, Lena AM, Petruzzelli MA, et al. ΔNp63 promotes IGF1 signalling through IRS1 in squamous cell carcinoma. Aging. 2018;10:4224–40.
Moses MA, George AL, Sakakibara N, Mahmood K, Ponnamperuma RM, King KE, et al. Molecular mechanisms of p63-mediated squamous cancer pathogenesis. Int J Mol Sci. 2019;20:3590.
Bao X, Rubin AJ, Qu K, Zhang J, Giresi PG, Chang HY, et al. A novel ATAC-seq approach reveals lineage-specific reinforcement of the open chromatin landscape via cooperation between BAF and p63. Genome Biol. 2015;16:284.
Mancini M, Cappello A, Pecorari R, Lena AM, Montanaro M, Fania L, et al. Involvement of transcribed lncRNA uc.291 and SWI/SNF complex in cutaneous squamous cell carcinoma. Discov Oncol. 2021;12:14.
Saladi SV, Ross K, Karaayvaz M, Tata PR, Mou H, Rajagopal J, et al. ACTL6A is co-amplified with p63 in squamous cell carcinoma to drive YAP activation, regenerative proliferation, and poor prognosis. Cancer Cell. 2017;31:35–49.
Viticchiè G, Agostini M, Lena AM, Mancini M, Zhou H, Zolla L, et al. P63 supports aerobic respiration through hexokinase II. Proc Natl Acad Sci U S A. 2015;112:11577–82.
Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21:297–308.
Melino S, Nepravishta R, Bellomaria A, Di Marco S, Paci M. Nucleic acid binding of the RTN1-C C-terminal region: toward the functional role of a reticulon protein. Biochemistry. 2009;48:242–53.
Melino S, Leo S, Toska Papajani V. Natural hydrogen sulfide donors from allium sp. as a nutraceutical approach in type 2 diabetes prevention and therapy. Nutrients. 2019;11:1581.
Amelio I, Cutruzzolá F, Antonov A, Agostini M, Melino G. Serine and glycine metabolism in cancer. Trends Biochem Sci. 2014;39:191–8.
Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–50.
Rathore R, Schutt CR, Van Tine BA. PHGDH as a mechanism for resistance in metabolically-driven cancers. Cancer Drug Resist. 2020;3:762–74.
Labuschagne CF, van den Broek NJF, Mackay GM, Vousden KH, Maddocks ODK. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014;7:1248–58.
Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metab. 2017;25:27–42.
Ganini C, Amelio I, Bertolo R, Candi E, Cappello A, Cipriani C, et al. Serine and one-carbon metabolisms bring new therapeutic venues in prostate cancer. Discover Oncology. 2021;12:45.
Cappello A, Mancini M, Madonna S, Rinaldo S, Paone A, Scarponi C, et al. Extracellular serine empowers epidermal proliferation and psoriasis-like symptoms. Sci Adv. 2022;8:7902.
Cappello A, Zuccotti A, Mancini M, Tosetti G, Fania L, Ricci F, et al. Serine and one-carbon metabolism sustain non-melanoma skin cancer progression. Cell Death Discov. 2023;9:102.
Manthalkar L, Ajazuddin, Bhattacharya S. Evidence-based capacity of natural cytochrome enzyme inhibitors to increase the effectivity of antineoplastic drugs. Discov Oncol. 2022;13:142.
Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47.
Zipinotti dos Santos D, dos Santos Guimaraes I, Hakeem-Sanni MF, Cochran BJ, Rye K-A, Grewal T, et al. Atorvastatin improves cisplatin sensitivity through modulation of cholesteryl ester homeostasis in breast cancer cells. Discov Oncol. 2022;13:135.
Navarro C, Ortega Á, Santeliz R, Garrido B, Chacín M, Galban N, et al. Metabolic reprogramming in cancer cells: emerging molecular mechanisms and novel therapeutic approaches. Pharmaceutics. 2022;14:1303.
Yang L, Qiao P, Zhang J, Chen X, Hu A, Huang S. Crosstalk between ROCK1 and PYROXD1 regulates CAFs activation and promotes laryngeal squamous cell carcinoma metastasis. Discov Oncol. 2022;13:120.
Yang L, Qiao P, Zhang J, Huang S, Hu A. Rho-associated kinase1 promotes laryngeal squamous cell carcinoma tumorigenesis and progression via the FAK signaling pathway. Discov Oncol. 2022;13:100.
Johnson DE, Burtness B, Leemans CR, Lui VWY, Bauman JE, Grandis JR. Head and neck squamous cell carcinoma. Nat Rev Dis Primers. 2020;6:92.
Möckelmann N, Kriegs M, Lörincz BB, Busch C, Knecht R. Molecular targeting in combination with platinum-based chemoradiotherapy in head and neck cancer treatment. Head Neck. 2016;38:E2173–81.
Rettig EM, D’Souza G. Epidemiology of head and neck cancer. Surg Oncol Clin N Am. 2015;24:379–96.
Ilacqua N, Anastasia I, Aloshyn D, Ghandehari-Alavijeh R, Peluso EA, Brearley-Sholto MC, et al. Expression of Synj2bp in mouse liver regulates the extent of wrappER-mitochondria contact to maintain hepatic lipid homeostasis. Biol Direct. 2022;17:37.
Wu S, Gong Y, Chen J, Zhao X, Qing H, Dong Y, et al. Identification of fatty acid metabolism-related lncRNAs in the prognosis and immune microenvironment of colon adenocarcinoma. Biol Direct. 2022;17:19.
Zawacka-Pankau JE. The role of p53 family in cancer. Cancers (Basel). 2022;14:823.
Butera A, Roy M, Zampieri C, Mammarella E, Panatta E, Melino G, et al. p53-driven lipidome influences non-cell-autonomous lysophospholipids in pancreatic cancer. Biol Direct. 2022;17:6.
Bergholz J, Xiao Z-X. Role of p63 in development, tumorigenesis and cancer progression. Cancer Microenviron. 2012;5:311–22.
Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–37.
Ling R, Chen G, Tang X, Liu N, Zhou Y, Chen D. Acetyl-CoA synthetase 2(ACSS2): a review with a focus on metabolism and tumor development. Discov Oncol. 2022;13:58.
Shuvalov O, Petukhov A, Daks A, Fedorova O, Vasileva E, Barlev NA. One-carbon metabolism and nucleotide biosynthesis as attractive targets for anticancer therapy. Oncotarget. 2017;8:23955–77.
Ducker GS, Ghergurovich JM, Mainolfi N, Suri V, Jeong SK, Li SHJ, et al. Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2017;114:11404–9.
Bonagas N, Gustafsson NMS, Henriksson M, Marttila P, Gustafsson R, Wiita E, et al. Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress. Nat Cancer. 2022;3:156–72.
Nguyen MQ, Teh JLF, Purwin TJ, Chervoneva I, Davies MA, Nathanson KL, et al. Targeting PHGDH Upregulation reduces glutathione levels and resensitizes resistant NRAS-mutant melanoma to MAPK kinase inhibition. J Investig Dermatol. 2020;140:2242-2252.e7.
Song D, Wei Y, Hu Y, Chen X, Zheng Y, Liu M, et al. Identification of prognostic biomarkers associated with tumor microenvironment in ceRNA network for esophageal squamous cell carcinoma: a bioinformatics study based on TCGA database. Discov Oncol. 2021;12:46.
Zhang L, Chen Z, Xue D, Zhang Q, Liu X, Luh F, et al. Prognostic and therapeutic value of mitochondrial serine hydroxyl-methyltransferase 2 as a breast cancer biomarker. Oncol Rep. 2016;36:2489–500.
Wang B, Wang W, Zhu Z, Zhang X, Tang F, Wang D, et al. Mitochondrial serine hydroxymethyltransferase 2 is a potential diagnostic and prognostic biomarker for human glioma. Clin Neurol Neurosurg. 2017;154:28–33.
Acknowledgements
We thank A. M. Lena, M. Mancini and for discussion and scientific support. We also thank Professor Camillo Porta for the support.
Funding
This work was mainly supported by Ministry of Health and IDI-IRCCS, Grant RF-2019-12368888 and RicercaCorrente 2022 (to EC). Partially supported by LazioInnova, Grant A0375-2020-36568 UTV-IDI (to EC) and PNRR-M4C2-I1.3 Project PE_00000019 "HEAL ITALIA" (to FB, EC, GM). Funding from the Associazione Italiana Ricerca sul Cancro (AIRC) under IG Grant 22206 to EC and IG-2022 ID-27366; 2023-2027 to GM is also gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
EC designed research; AC performed most of the experiments and bioinformatical analysis, GT performed the ChIP experiments and qPCR; AS and XY performed single cell RNA seq analysis; CG performed analysis on cBioportal and critically revised the manuscript; YS, YW, FB, GM, EC and AC discussed and analyzed the data; AC and EC wrote the paper.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
GM is Editor-in-Chief of Biology Direct.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Fig S1.
Serine and OCM enzymes are differentially expressed in single-cell RNAseq of HNSCC and leukoplakia samples. (a) Average expression of serine and one carbon metabolism enzymes in single-cell RNA sequencing of normal and HNSCC samples (GSE181919); NL: non lesional samples; LP: Leukoplakia samples; LN: Lymph nodes + samples; CA: primary cancer samples (b) PHGDH, PSAT1, PSPH and MTHFD2 expression in all cell types. (c) PHGDH, PSAT1, PSPH and MTHFD2 expression in leukoplakia samples cell types. Fig S2. Serine and OCM enzymes are differentially expressed in single-cell RNAseq of normal and HNSCC samples. (a) PHGDH, PSAT1, PSPH and MTHFD2 expression in single-cell RNA seq of normal cell types (GSE181919) (b) PHGDH, PSAT1, PSPH and MTHFD2 expression in single-cell RNA seq of primary tumor samples cell types. (GSE181919) (c) PHGDH, PSAT1, PSPH and MTHFD2 expression in single-cell RNA seq of metastatic samples cell types, (GSE181919). Fig. S3. p63 controls PHGDH, PSAT1 and MTHFD2 expression in FaDu and HaCaT cells. (a) correlation analysis of the mRNA expression of p63 (protein) and PHGDH, PSAT1, PSPH and MTHFD2 (mRNA) in HNSCC (TCGA firehose legacy). (b) Expression analysis of PHGDH, PSAT1, PSPH and MTHFD2 in a publicly available dataset of FaDu cells (ctr vs sh-p63) (GSE88833). (c) Analysis of PHGDH, PSAT1, PSPH and MTHFD2 mRNA expression in a publicly available dataset of HaCaT cells (ctr vs sh-p63) (GSE88832); the p-value was obtained using ordinary one-way analysis of variance (ANOVA). Values were considered significant when the p-value < 0.05 (n.s. = not significant). Fig. S4. Western blots uncropped images. Corresponding panels used in the main figures are indicated.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Cappello, A., Tosetti, G., Smirnov, A. et al. p63 orchestrates serine and one carbon metabolism enzymes expression in head and neck cancer. Biol Direct 18, 73 (2023). https://doi.org/10.1186/s13062-023-00426-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13062-023-00426-1