Abstract
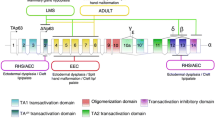
The p53-related protein p63 has pleiotropic functions, including cell proliferation, survival, apoptosis, differentiation, senescence, and aging. The p63 gene is expressed as multiple isoforms that either contain an N-terminal p53-homologous transactivation domain (TAp63) or that lack this domain (ΔNp63). Multiple studies have demonstrated that p63 plays a crucial role in stratified epithelial development, and have shown the importance of p63 for maintaining proliferation potential, inducing differentiation, and preventing senescence. Additionally, much research focuses on the role of p63 in cancer progression. Clinical evidence suggests that p63 may play a role in inhibiting metastasis. Similarly, genetic mice models together with cell culture data strongly indicate that p63 deficiency may be a causative factor for metastatic spread. Moreover, the role of p63 in cancer metastasis has been shown to be greatly related to the ability of mutant p53 to promote cancer malignancy. However, there is still much confusion as to what the role of each specific isoform is. In this review, we highlight some of the major findings in the current literature regarding the role of specific p63 isoforms in development, tumorigenesis, and particularly in cancer metastasis.



Similar content being viewed by others
References
Levine AJ, Oren M (2009) The first 30 years of p53: growing ever more complex. Nat Rev Cancer 9(10):749–758
Vousden KH, Prives C (2009) Blinded by the light: the growing complexity of p53. Cell 137(3):413–431
Yang A et al (1998) p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell 2(3):305–316
Kaghad M et al (1997) Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 90(4):809–819
Melino G (2011) p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ
Vanbokhoven H et al (2011) p63, a story of mice and men. J Invest Dermatol
Yang A et al (1999) p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398(6729):714–718
May P, May E (1999) Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene 18(53):7621–7636
Khoury MP, Bourdon J-C (2011) p53 isoforms: an intracellular microprocessor? Genes Cancer 2(4):453–465
Marcel V, Hainaut P (2009) p53 isoforms - a conspiracy to kidnap p53 tumor suppressor activity? Cell Mol Life Sci 66(3):391–406
Yang A et al (2000) p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature 404(6773):99–103
Weber A et al (2002) Expression of p53 and its homologues in primary and recurrent squamous cell carcinomas of the head and neck. Int J Cancer 99(1):22–28
Zawacka-Pankau J et al (2010) p73 tumor suppressor protein: a close relative of p53 not only in structure but also in anti-cancer approach? Cell Cycle 9(4)
Tomasini R et al (2008) TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev 22(19):2677–2691
Deyoung MP, Ellisen LW (2007) p63 and p73 in human cancer: defining the network. Oncogene 26(36):5169–5183
Mills AA et al (1999) p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398(6729):708–713
Westfall MD et al (2003) The Delta Np63 alpha phosphoprotein binds the p21 and 14-3-3 sigma promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol Cell Biol 23(7):2264–2276
Dohn M, Zhang S, Chen X (2001) p63alpha and DeltaNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene 20(25):3193–3205
Ghioni P et al (2002) Complex transcriptional effects of p63 isoforms: identification of novel activation and repression domains. Mol Cell Biol 22(24):8659–8668
Helton ES, Zhu J, Chen X (2006) The unique NH2-terminally deleted (DeltaN) residues, the PXXP motif, and the PPXY motif are required for the transcriptional activity of the DeltaN variant of p63. J Biol Chem 281(5):2533–2542
Mangiulli M et al (2009) Identification and functional characterization of two new transcriptional variants of the human p63 gene. Nucleic Acids Res 37(18):6092–6104
Thanos CD, Bowie JU (1999) p53 Family members p63 and p73 are SAM domain-containing proteins. Protein Sci 8(8):1708–1710
Serber Z et al (2002) A C-terminal inhibitory domain controls the activity of p63 by an intramolecular mechanism. Mol Cell Biol 22(24):8601–8611
Coutandin D et al (2009) Conformational stability and activity of p73 require a second helix in the tetramerization domain. Cell Death Differ 16(12):1582–1589
Joerger A et al (2009) Structural evolution of p53, p63, and p73: implication for heterotetramer formation. Proc Natl Acad Sci USA
Gaiddon C et al (2001) A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol 21(5):1874–1887
Rocco JW et al (2006) p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell 9(1):45–56
Davison TS et al (1999) p73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53. J Biol Chem 274(26):18709–18714
Ying H et al (2005) DNA-binding and transactivation activities are essential for TAp63 protein degradation. Mol Cell Biol 25(14):6154–6164
Barbareschi M et al (2001) p63, a p53 homologue, is a selective nuclear marker of myoepithelial cells of the human breast. Am J Surg Pathol 25(8):1054–1060
Signoretti S et al (2000) p63 is a prostate basal cell marker and is required for prostate development. Am J Pathol 157(6):1769–1775
Candi E et al (2007) DeltaNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc Natl Acad Sci USA 104(29):11999–12004
Laurikkala J et al (2006) p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development 133(8):1553–1563
Gonfloni S et al (2009) Inhibition of the c-Abl-TAp63 pathway protects mouse oocytes from chemotherapy-induced death. Nat Med 15(10):1179–1185
Suh E-K et al (2006) p63 protects the female germ line during meiotic arrest. Nature 444(7119):624–628
Barbieri CE et al (2006) Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res 66(15):7589–7597
Carroll DK et al (2006) p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol 8(6):551–561
Romano R-A et al (2009) An active role of the DeltaN isoform of p63 in regulating basal keratin genes K5 and K14 and directing epidermal cell fate. PLoS One 4(5):e5623
Ferretti E et al (2011) A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev Cell 21(4):627–641
Aberdam D et al (2007) Key role of p63 in BMP-4-induced epidermal commitment of embryonic stem cells. Cell Cycle 6(3):291–294
Mikkola ML (2007) p63 in skin appendage development. Cell Cycle 6(3):285–290
Herfs M et al (2010) Regulation of p63 isoforms by snail and slug transcription factors in human squamous cell carcinoma. Am J Pathol
Higashikawa K et al (2007) Snail-induced down-regulation of DeltaNp63alpha acquires invasive phenotype of human squamous cell carcinoma. Cancer Res 67(19):9207–9213
Petitjean A et al (2005) The expression of TA and DeltaNp63 are regulated by different mechanisms in liver cells. Oncogene 24(3):512–519
Yao J-Y, Pao C-C, Chen J-K (2010) Transcriptional activity of TAp63 promoter is regulated by c-jun. J Cell Physiol 225(3):898–904
Wu J et al (2010) TAp63 is a transcriptional target of NF-kappaB. J Cell Biochem 109(4):702–710
Lena A. et al (2008) miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ
Yi R et al (2008) A skin microRNA promotes differentiation by repressing ‘stemness’. Nature 452(7184):225–229
Manni I et al (2009) The microRNA miR-92 increases proliferation of myeloid cells and by targeting p63 modulates the abundance of its isoforms. FASEB J
Papagiannakopoulos T, Shapiro A, Kosik KS (2008) MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res 68(19):8164–8172
Liefer KM et al (2000) Down-regulation of p63 is required for epidermal UV-B-induced apoptosis. Cancer Res 60(15):4016–4020
Westfall MD et al (2005) Ultraviolet radiation induces phosphorylation and ubiquitin-mediated degradation of DeltaNp63alpha. Cell Cycle 4(5):710–716
Papoutsaki M et al (2005) A p38-dependent pathway regulates DeltaNp63 DNA binding to p53-dependent promoters in UV-induced apoptosis of keratinocytes. Oncogene 24(46):6970–6975
Chatterjee A et al (2010) Regulation of p53 Family Member Isoform Delta}Np63{alpha by the Nuclear Factor-{kappa}B Targeting Kinase I{kappa}B Kinase {beta}. Cancer Res 70(4):1419–1429
Fomenkov A et al (2004) RACK1 and stratifin target DeltaNp63alpha for a proteasome degradation in head and neck squamous cell carcinoma cells upon DNA damage. Cell Cycle 3(10):1285–1295
Lazzari C et al (2011) HIPK2 phosphorylates ΔNp63α and promotes its degradation in response to DNA damage. Oncogene 30(48):4802–4813
Rossi M et al (2006) The E3 ubiquitin ligase Itch controls the protein stability of p63. Proc Natl Acad Sci USA 103(34):12753–12758
Rossi M et al (2006) Itch/AIP4 associates with and promotes p63 protein degradation. Cell Cycle 5(16):1816–1822
Deutsch GB et al (2011) DNA damage in oocytes induces a switch of the quality control factor TAp63α from dimer to tetramer. Cell 144(4):566–576
Celli J et al (1999) Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell 99(2):143–153
McGrath JA et al (2001) Hay-Wells syndrome is caused by heterozygous missense mutations in the SAM domain of p63. Hum Mol Genet 10(3):221–229
Brunner HG, Hamel BCJ, van Bokhoven H (2002) P63 gene mutations and human developmental syndromes. Am J Med Genet 112(3):284–290
van Bokhoven H, McKeon F (2002) Mutations in the p53 homolog p63: allele-specific developmental syndromes in humans. Trends Mol Med 8(3):133–139
Rinne T et al (2006) Delineation of the ADULT syndrome phenotype due to arginine 298 mutations of the p63 gene. Eur J Hum Genet 14(8):904–910
Senoo M et al (2007) p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129(3):523–536
Shalom-Feuerstein R et al (2010) ΔNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ
Candi E et al (2006) Differential roles of p63 isoforms in epidermal development: selective genetic complementation in p63 null mice. Cell Death Differ 13(6):1037–1047
Nguyen B-C et al (2006) Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev 20(8):1028–1042
Wu G et al. ΔNp63α and tap63α regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res
Su X et al (2009) TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell 5(1):64–75
Katoh I et al (2000) p51A (TAp63gamma), a p53 homolog, accumulates in response to DNA damage for cell regulation. Oncogene 19(27):3126–3130
DeYoung MP et al (2006) Tumor-specific p73 up-regulation mediates p63 dependence in squamous cell carcinoma. Cancer Res 66(19):9362–9368
Truong AB et al (2006) p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev 20(22):3185–3197
Antonini D et al (2010) Transcriptional repression of miR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J Invest Dermatol 130(5):1249–1257
Keyes WM et al (2005) p63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev 19(17):1986–1999
Guo X et al (2009) TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol 11(12):1451–1457
Flores ER et al (2005) Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell 7(4):363–373
Keyes WM et al (2006) p63 heterozygous mutant mice are not prone to spontaneous or chemically induced tumors. Proc Natl Acad Sci USA 103(22):8435–8440
Hagiwara K et al (1999) Mutational analysis of the p63/p73L/p51/p40/CUSP/KET gene in human cancer cell lines using intronic primers. Cancer Res 59(17):4165–4169
Sunahara M et al (1999) Mutational analysis of p51A/TAp63gamma, a p53 homolog, in non-small cell lung cancer and breast cancer. Oncogene 18(25):3761–3765
Björkqvist AM et al (1998) DNA gains in 3q occur frequently in squamous cell carcinoma of the lung, but not in adenocarcinoma. Genes Chromosomes Canc 22(1):79–82
Hibi K et al (2000) AIS is an oncogene amplified in squamous cell carcinoma. Proc Natl Acad Sci USA 97(10):5462–5467
Massion PP et al (2003) Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res 63(21):7113–7121
Sniezek JC et al (2004) Dominant negative p63 isoform expression in head and neck squamous cell carcinoma. Laryngoscope 114(12):2063–2072
Hu H et al (2002) Elevated expression of p63 protein in human esophageal squamous cell carcinomas. Int J Cancer 102(6):580–583
Wang TY et al (2001) Histologic and immunophenotypic classification of cervical carcinomas by expression of the p53 homologue p63: a study of 250 cases. Hum Pathol 32(5):479–486
Matos I et al (2005) p63, cytokeratin 5, and P-cadherin: three molecular markers to distinguish basal phenotype in breast carcinomas. Virchows Arch 447(4):688–694
Perou CM et al (2000) Molecular portraits of human breast tumours. Nature 406(6797):747–752
Leong C-O et al (2007) The p63/p73 network mediates chemosensitivity to cisplatin in a biologically defined subset of primary breast cancers. J Clin Invest 117(5):1370–1380
Ramsey MR et al (2011) Physical Association of HDAC1 and HDAC2 with p63 mediates transcriptional repression and tumor maintenance in squamous cell carcinoma. Cancer Res 71(13):4373–4379
Wu G et al (2005) DeltaNp63alpha up-regulates the Hsp70 gene in human cancer. Cancer Res 65(3):758–766
Keyes WM et al (2011) ΔNp63α is an oncogene that targets chromatin remodeler Lsh to drive skin stem cell proliferation and tumorigenesis. Cell Stem Cell 8(2):164–176
Pruneri G et al (2005) The transactivating isoforms of p63 are overexpressed in high-grade follicular lymphomas independent of the occurrence of p63 gene amplification. J Pathol 206(3):337–345
Quade BJ et al (2001) Expression of the p53 homologue p63 in early cervical neoplasia. Gynecol Oncol 80(1):24–29
Stefanou D et al (2004) p63 expression in benign and malignant breast lesions. Histol Histopathol 19(2):465–471
Wang X et al (2002) p63 expression in normal, hyperplastic and malignant breast tissues. Breast Cancer 9(3):216–219
Koga F et al (2003) Impaired p63 expression associates with poor prognosis and uroplakin III expression in invasive urothelial carcinoma of the bladder. Clin Cancer Res 9(15):5501–5507
Koga F et al (2003) Impaired Delta Np63 expression associates with reduced beta-catenin and aggressive phenotypes of urothelial neoplasms. Br J Cancer 88(5):740–747
Urist MJ et al (2002) Loss of p63 expression is associated with tumor progression in bladder cancer. Am J Pathol 161(4):1199–1206
Vanaja DK et al (2003) Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res 63(14):3877–3882
Haqq C et al (2005) The gene expression signatures of melanoma progression. Proc Natl Acad Sci USA 102(17):6092–6097
Su H et al (2003) Gene expression analysis of esophageal squamous cell carcinoma reveals consistent molecular profiles related to a family history of upper gastrointestinal cancer. Cancer Res 63(14):3872–3876
Su X et al (2010) TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 467(7318):986–990
Muller PAJ et al (2009) Mutant p53 drives invasion by promoting integrin recycling. Cell 139(7):1327–1341
Adorno M et al (2009) A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell 137(1):87–98
Fukushima H et al (2009) Loss of DeltaNp63alpha promotes invasion of urothelial carcinomas via N-cadherin/Src homology and collagen/extracellular signal-regulated kinase pathway. Cancer Res 69(24):9263–9270
Higashikawa K et al (2009) DeltaNp63alpha-dependent expression of Id-3 distinctively suppresses the invasiveness of human squamous cell carcinoma. Int J Cancer 124(12):2837–2844
Kommagani R et al (2009) Regulation of VDR by {Delta}Np63{alpha} is associated with inhibition of cell invasion. J Cell Sci
Ihrie RA et al (2005) Perp is a p63-regulated gene essential for epithelial integrity. Cell 120(6):843–856
Leonard MK et al (2011) ΔNp63α regulates keratinocyte proliferation by controlling PTEN expression and localization. Cell Death Differ
Girardini JE et al (2011) A Pin1/Mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell 20(1):79–91
Acknowledgments
This work was supported by the National Key Basic Research Program (973 Program) of China (2012CB910700) and National Science Foundation of China (#31171362) to ZX. X., and United States Department of Defense Congressionally Directed Medical Research Programs grant W81XWH-10-1-0161 to J.B.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bergholz, J., Xiao, ZX. Role of p63 in Development, Tumorigenesis and Cancer Progression. Cancer Microenvironment 5, 311–322 (2012). https://doi.org/10.1007/s12307-012-0116-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12307-012-0116-9