Abstract
Background
Histone H3 gene clusters have been described as highly conserved chromosomal markers in invertebrates. Surprisingly, in bivalves remarkable interspecific differences were found among the eight mussels and between the two clams in which histone H3 gene clusters have already been located. Although the family Veneridae comprises 10 % of the species of marine bivalves, their chromosomes are poorly studied. The clams belonging to this family present 2n = 38 chromosomes and similar karyotypes showing chromosome pairs gradually decreasing in length. In order to assess the evolutionary behavior of histone and rRNA multigene families in bivalves, we mapped histone H3 and ribosomal RNA probes to chromosomes of ten species of venerid clams.
Results
In contrast with the reported conservation of histone H3 gene clusters and their intercalary location in invertebrates, these loci varied in number and were mostly subterminal in venerid clams. On the other hand, while a single 45S rDNA cluster, highly variable in location, was found in these organisms, 5S rDNA clusters showed interspecific differences in both number and location. The distribution patterns of these sequences were species-specific and mapped to different chromosomal positions in all clams but Ruditapes decussatus, in which one of the minor rDNA clusters and the major rDNA cluster co-located.
Conclusion
The diversity in the distribution patterns of histone H3 gene, 5S rDNA and 28S rDNA clusters found in venerid clams, together with their different evolutionary behaviors in other invertebrate taxa, strongly suggest that the control of the spreading of these multigene families in a group of organisms relies upon a combination of evolutionary forces that operate differently depending not only on the specific multigene family but also on the particular taxa. Our data also showed that H3 histone gene and rDNA clusters are useful landmarks to integrate nex-generation sequencing (NGS) and evolutionary genomic data in non-model species.
Similar content being viewed by others
Background
The analysis of the chromosome changes encompassing the evolution of a group of organisms relies on the accurate identification of their chromosomes. When chromosome-specific painting probes are not available, as frequently happens in invertebrates, and karyotypes are composed by chromosomes gradually decreasing in length, chromosomal identification turns into a very difficult task. In those cases, the hybridization of highly conserved repetitive sequences, among which ribosomal RNA (rRNA) and histone genes are paramount, usually constitutes a first step in finding chromosome-specific probes.
Eukaryotic genomes present multiple copies of genes encoding histones, the basic proteins responsible of packaging DNA into chromatin. The histone multigene family includes five main types of genes, those encoding the histones of the nucleosome core particle (H2A, H2B, H3 and H4) and those for the linker histones (H1) [1]. rRNA genes are also organized in multigene families, one expressing for the 18S, 5.8S and 28S rRNAs (45S rDNA) and the other for the 5S rRNA [2]. Histone and rRNA genes in invertebrates are usually organized in tandem arrays clustered in one or more chromosomal positions, although other organizations have also been described [3, 4]. The evolutionary dynamics of both histone gene and rDNA clusters has been analyzed in only a few groups of these organisms, i.e. grasshoppers [5, 6], beetles [7, 8], aphids [9] and moths [10]. Whereas histone gene clusters were extremely conserved in number and location in all these groups, 45S and 5S rDNAs showed high degrees of variation.
In bivalves, the genomic organization of the histone genes has been studied using molecular methodologies in species belonging to the families Mytilidae [11–14], Pectinidae [15] and Veneridae [16], showing, in all of them, a tandemly arranged organization. Usually, the clusters comprise only core histone genes but repeated clusters including linker histone and/or other genes have also been reported. In these organisms histone H3 genes have been mapped to chromosomes in only eight mussels [13, 14, 17–20], four scallops [21], one oyster [22] and two clams [23]. In comparison with 45S and 5S rDNA clusters, surprisingly remarkable differences in number and location of the histone H3 gene clusters were found, more outstandingly among mussels and clams.
The venerid clams of the family Veneridae (Rafinesque 1815) represent almost 10 % of the species of marine bivalves [24]. Phylogenetic relationships among the species of this family were the subject of many recent investigations using DNA sequences whose results, in some cases, challenged the traditional, morphologically based classification [25–29]. In contrast, the chromosomal characterization of the venerid clams lags far beyond the knowledge achieved for other bivalve families. Classical venerid cytogenetics was limited to determine chromosome numbers and karyotypes in a few species [30–32]. More recently, a restriction endonuclease banding pattern was described in Ruditapes decussatus [33] and some repetitive DNA sequences were mapped by fluorescent in situ hybridization (FISH). The location of telomeric sequences and/or major and minor rDNA was reported for Mercenaria mercenaria [34, 35], Dosinia exoleta [36, 37], Ruditapes decussatus and Ruditapes philippinarum [37, 38], Polititapes aureus and Polititapes rhomboides [23] and Venerupis corrugata and Venus verrucosa [37]. On the other hand, histone gene clusters, as indicated above, were only mapped to chromosomes of Polititapes aureus and Polititapes rhomboides [23].
In order to get a better understanding of the evolutionary behavior of these multigene families in these organisms, we have hybridized H3 gene, 5S rDNA and 28S rDNA probes to mitotic and meiotic chromosomes of ten species of clams of the family Veneridae, Ruditapes philippinarum, Ruditapes decussatus, Venerupis corrugata, Clausinella fasciata, Chamelea gallina, Venus verrucosa, Venus casina, Dosinia exoleta, Dosinia lupinus and Petricola litophaga.
Results
FISH experiments identified a total of 14 loci for the histone H3 gene in the 10 species analyzed (Figs. 1 and 2). We detected a single core histone gene cluster in six of the species and two clusters in the remaining four. Regarding their chromosomal location, 11 of the histone clusters were close to the telomeres, two were intercalary, and the remaining one close to the centromere. The subterminal location of the clusters was confirmed by FISH on synaptonemal complex spreads (Fig. 3). A summary of the data obtained in this work, together with the other currently available FISH mapping data for the family Veneridae, is presented in Table 1. The species were arranged following the molecular phylogenetic tree suggested by [29] (Chen et al.) and assigned both to their proposed clade groups (A1, A2, A3, A4, B1) and the subfamilies of the traditional classification (Tapetinae, Chioninae, Venerinae, Dosininae and Petricolinae). Histone H3 gene clusters mapped to a single locus in two of the three analyzed species included in clade A1 (Tapetinae: Ruditapes philippinarum and Ruditapes decussatus), to two of the four species in clade A2 (Venerinae: Venus verrucosa and Venus casina), and in the two species in clade A4 (Dosininae: Dosinia exoleta and Dosinia lupinus). In the remaining four species, one in clade A1 (Tapetinae: Venerupis corrugata), two in clade A2 (Chioninae: Clausinella fasciata and Chamelea gallina), and one in clade B1 (Petricolinae: Petricola litophaga), core histone gene clusters mapped to two loci situated in different chromosome pairs.
Chromosomal location of H3 histone genes in venerid clams. H3 histone gene (H3) probes mapped by FISH to mitotic chomosomes of Ruditapes philippinarum (RPH), Ruditapes decussatus (RDE), Venerupis corrugata (VCO), Venus verrucosa (VVE) and Dosinia exoleta (DEX). To ascertain the chromosomal position of core histone gene clusters in relation to rDNA clusters, the same metaphases were rehybridized using 5S rDNA (5S) and major rDNA (28S) probes. Excluding 5S and major rDNA in RDE (arrows), all signals are on different chromosome pairs. Scale bars, 5 μm
Chromosomal location of H3 histone genes (H3), 5S rDNA (5S) and major rDNA (28S) in venerid clams. Single FISH using H3 histone gene probes mapped to chromosomes of Clausinella fasciata (CFA), Chamelea gallina (CGA), Venus casina (VCA), Dosinia lupinus (DLU), and Petricola litophaga (PLI), followed by double-FISH using 5S rDNA (5S) and major rDNA (28S) probes on the same metaphase plates. All signals for the different probes appear at different chromosome pairs with the exception of H3 histone gene and major rDNA in Clausinella fasciata (CFA, first row). Scale bars, 5 μm
Subterminal H3 histone gene clusters in venerid clams. Examples of FISH to surface spread synaptonemal complexes of Ruditapes philippinarum (RPH) and Venerupis corrugata (VCO) clearly denote the subterminal location of the H3 histone gene clusters (H3, green). 5S rDNA clusters (5S, red) are also subterminal in VCO but intercalary in RPH. Scale bars, 5 μm
In order to investigate the location of the core histone gene clusters in relation to rDNAs, we performed double and sequential FISH experiments using core histone gene, major rDNA and 5S rDNA probes in the five species of clams in which the location of rDNA sequences was already known (Fig. 1) and in the other five in which not previous data were available (Fig. 2). Whereas all species presented a single major rDNA cluster per haploid genome, the number of 5S rDNA clusters was one in five of the species (Venerupis corrugata, Clausinella fasciata, Venus verrucosa, Venus casina and Dosinia lupinus), two in four (Ruditapes philippinarum, Ruditapes decussatus, Chamelea gallina and Dosinia exoleta) and three in the remaining one, Petricola litophaga (Table 1). Taking into account that two of the three 5S rDNA clusters detected in Petricola litophaga were close together, sometimes giving double FISH signals and other times a single signal, we performed FISH on prophase I meiotic bivalents to clarify the true nature of the signals. As shown in Fig. 4, three clearly different signals were detected, two of them on the same bivalent.
Clausinella fasciata was the only species in which a single chromosome harbors both histone gene and rDNA cluster; in this species, a subterminal histone gene cluster and a subcentromeric major rDNA cluster appeared in the long arm of chromosome 10 (Fig. 2). Confirming previously published results, in Ruditapes decussatus the signals for one of the 5S rDNA clusters and the major rDNA cluster overlap on chromosome 3 (Fig. 1).
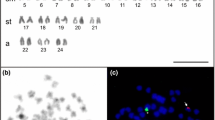
Figure 5 presents an ideogrammatic representation of the karyotypes of the 13 species of Veneridae for which histone gene and/or rDNA mapping results are currently available.
Ideograms showing the chromosomal location of H3 histone genes, 5S rDNA and major rDNA in thirteen species of Veneridae. The green areas represent the H3 histone gene clusters, the red areas the 5S rDNA clusters and the yellow areas the major rDNA clusters. The magenta area in Ruditapes decussatus indicates overlapping of major and 5S rDNA signals
FISH experiments using a vertebrate telomeric (C3TA2)3 peptide nucleic acid (PNA) probe gave terminal signals at the ends of the sister chromatids of every mitotic chromosome in all clam species. No intercalary signals were detected.
Discussion
Multigene families are useful cytogenetic markers not only for studying chromosomal evolution but also for the correct interpretation of the data obtained via NGS. Aligning and assembling NGS data is a hard task in many non-model organisms mainly due to the obstacles posed by the abundance of repetitive DNA sequences. The physical location of repetitive gene families will help in this task.
In this work we have demonstrated the presence of remarkable interspecific differences in the physical location of H3 histone gene clusters in venerid clams. These clusters have been described as highly conserved chromosomal markers in other invertebrate groups. Our data, together with previously published results [23], showed variation for the number of core histone gene sites in venerid clams, with six species carrying a single cluster, five presenting two clusters and one showing four clusters (Table 1). The observed variation in the number of core histone gene clusters did not present any obvious relationship with the currently taxonomic classification of the family Veneridae or its most represented clades. In clade A1 (Tapetinae) there were species showing one (Ruditapes philippinarum and Ruditapes decussatus), two (Venerupis corrugata and Polititapes rhomboides) and four (Polititapes aureus) histone gene clusters. A similar divergence also applied to clade A2, including species of the subfamilies Venerinae and Chioninae, in which two species (Venus verrucosa and Venus casina) showed a single cluster and the other two species (Clausinella fasciata and Chamelea gallina) presented two clusters.
These differences in the number of core histone gene clusters in related species are coincident with results reported for other bivalve families. While three species of scallops and one mussel showed a single core histone gene cluster [17, 21], one scallop and six mussels presented two [17, 18, 20, 21] and one mussel had four [19]. This behavior differs with those described for other invertebrate groups in which histone gene clusters have been reported to be a highly conservative cytogenetic marker [5–10].
In contrast with the variability in number, 75 % of the H3 histone gene clusters detected in venerid clams (15 of a total of 20) were located at subterminal chromosome positions. This is unusual for bivalves and also for other invertebrates; in fact, barely a 30 % of the core histone gene clusters detected in mussels of the family Mytilidae (5 of 17) [17–20] and only a 40 % of those reported in the scallops of the family Pectinidae (2 of 5) [21] were subterminal. For other invertebrates, the subterminal position of the histone gene signal has only been described in three grasshoppers [5].
Taking into account the above mentioned data, the presumably ancestral situation in venerids is a single subterminal core histone gene cluster. Although the mechanisms that allowed increasing the number of clusters remain to be determined, the presence of most of these sequences in close proximity to the telomeres might facilitate their spreading to non-homologous chromosomes. Subtelomeric chromosomal regions are characterized in many eukaryotes by accumulating repeat sequences and harbor many breakpoints [39, 40]. These features, together with the telomere clustering in meiotic cells, probably favors their implication in sequence exchanges between non-homologous chromosomes and contribute to their highly dynamic behavior [41].
Regarding rDNA, the clams of the family Veneridae showed variation in both number and chromosomal location of the 5S rDNA clusters but only in location of the 45S rDNA clusters. Whereas all species had a single 45S rDNA cluster, the number of 5S rDNA clusters varied; seven species showed a single cluster, six species two and Petricola litophaga three. The conservation in the number of major rDNA clusters in Veneridae was not paired by their chromosomal location; subterminal, intercalary and subcentromeric locations were found. The 5S rDNA clusters were either subterminal or intercalary. As happened with H3 histone gene clusters, the variant 45S and 5S rDNA arrangements did not concord with the taxonomic distribution of the species of the family Veneridae; species belonging to the same clade or subfamily showed 45S and 5S rDNA clusters differing in number and chromosome location (Table 1).
These results partially differ with those found in other bivalve families. While mytilid mussels showed one to four major rDNA and two to five 5S rDNA clusters [17–20, 42], both Pectinidae and Ostreidae species presented one or two major and 5S rDNA clusters [31, 43–47].
The evolutionary dynamics of the major rDNA clusters in venerid clams is similar to the one reported for tortricid moths [10] but the opposite to the described in other invertebrate groups in which both the number and the position of these sequences has been reported as highly variable [5–8]. In contrast, the behavior of the 5S rDNA is common to some other invertebrate taxa in which it has been described as a highly variable chromosomal marker [5–8] whose movement has been attributed to transposition and/or unequal crossover [48].
In conclusion, the diversity in the distribution patterns of histone H3 gene, 5S rDNA and 28S rDNA clusters found in venerid clams, together with their different evolutionary behaviors in other invertebrate taxa, strongly suggest that the control of the spreading of these multigene families in a group of organisms relies upon a combination of evolutionary forces that operate differently depending not only on the specific multigene family but also on the particular taxa. On the other hand, our data clearly showed that the number and position of the H3 histone gene and rDNA clusters are species-specific in venerid clams and that the complexity of their evolutionary patterns make them useful landmarks that can contribute to integrate NGS and evolutionary genomic data in non model species.
Methods
Venus clam specimens
Samples of the Japanese carpet shell Ruditapes philippinarum (Adams and Reeve 1850), the grooved carpet shell Ruditapes decussatus (Linnaeus 1758), the pullet carpet shell Venerupis corrugata (pullastra) (Gmelin 1791), the banded venus Clausinella fasciata (da Costa 1778), the warty venus Venus verrucosa (Linnaeus 1758), the pale venus Venus casina (Linnaeus 1758), the rayed artemis Dosinia exoleta (Linnaeus 1758), the smooth artemis Dosinia lupinus (Linnaeus 1758), and the boring petricola Petricola litophaga (Retzius1788) were collected from natural and cultured populations in Ría de Pontevedra and Ría de Vigo (NW Spain). Samples of the striped venus Chamelea gallina (Linnaeus 1758) were collected from natural populations in the Gulf of Valencia (E Spain). The nomenclature used for these taxa follows the World Register of Marine Species database [49].
Chromosome preparation
Mitotic metaphase and meiotic prophase I spreads were prepared as previously described [50]. In brief, after exposing the clams to colchicine (0.005 %, 12 h), gills and gonads were removed. The tissues were treated with diluted sea water (50 %, 25 %, 1 h) and fixed in ethanol/acetic acid (3:1, 1 h). The cell suspension obtained after dissociating the tissue (60 % acetic acid) was dropped onto heated slides.
Synaptonemal complexes were spread as indicated by Hurtado and Pasantes [36]. Suspensions of male gonadic cells were spread on slides using 0.1 M sucrose and 0.5 % Triton X-100, fixed with paraformaldehyde (4 %), rinsed in distilled water and air-dried.
DNA extraction, PCR amplification and probe labeling
DNA was extracted using the method published by Winnepenninckx et al. [51]. The tissue was homogenized in hexadecyltrimethylammoniumbromide (CTAB) buffer and digested with pronase (1.5 mg/mL, 60 °C). The DNA was extracted with chloroform/isoamyl alcohol (24/1).
FISH probes were amplified by polymerase chain reaction (PCR). Reactions used 50 ng DNA, 1x PCR buffer, 0.5 mM each dNTP, 2.5 mM MgCl2, 1 μM each primer and 1 U BIOTAQ DNA polymerase (Bioline) in a volume of 20 μl. A fragment of the 28S rRNA gene of the major rDNA repeat was amplified using universal primers [52]. Primers designed from the sequence of the 5S rRNA of M. edulis [53] were used to amplify the whole repeat of the 5S rDNA. The amplification of the histone H3 genes used primers described by Giribet and Distel [54].
After an initial denaturation at 95 °C, 30 cycles (95 °C, 20 s; 48 °C, 20 s; 72 °C, 30 s) of amplification and a final extension step of 7 min at 72 °C were applied in a GeneAmp PCR system 9700 (Applied Biosystems). Electrophoresis on 2 % agarose gels demonstrated that single PCR products were obtained. 28S rDNA probes were labeled by nick translation (Roche Applied Science) with biotin-16-dUTP (Roche Applied Science) and/or digoxigenin-11-dUTP (10x DIG Labeling Mix, Roche Applied Science). Histone H3 gene and 5S rDNA probes were labeled by PCR either with biotin-16-dUTP (20 μM) or digoxigenin-11-dUTP (5 μM). The labeled PCR products were precipitated before FISH.
Fluorescent in situ hybridization (FISH)
Single and double FISH experiments using biotin and digoxigenin labeled histone H3 gene and 28S and 5S rDNA probes were performed following previously published methods [19, 37]. Slides were pre-treated with RNase and pepsin before denaturating them for 2 min at 70 °C (mitotic chromosomes) or 80 °C (meiotic chromosomes). Hybridizations were performed overnight at 37 °C. Signal detection was carried out with fluorescein avidin and biotinylated anti-avidin for the biotinylated probes and with mouse anti-digoxigenin and anti-mouse TRITC for the probes labeled with digoxigenin. Slides were counterstained for 8 min with 4′-6-diamidin-2-fenilindol (DAPI: 0.14 μg mL−1 in 2xSSC) and mounted in antifade (Vectashield, Vector). In addition we also performed FISH with a vertebrate telomeric (C3TA2)3 peptide nucleic acid (PNA) probe (Applied Biosystems) following the protocol indicated by the supplier.
A Nikon Eclipse-800 microscope equipped with an epifluorescence system was used to record a minimum of 20 metaphase plates per probe or combination of probes in 10 specimens (5 male, 5 female) per species. Separated images for each fluorochrome were obtained with a DS-Qi1Mc CCD camera (Nikon) controlled by the NIS-Elements software (Nikon). Merging of the images was performed with Adobe Photoshop.
For each species, karyotypes were constructed from 10 complete metaphase plates showing FISH signals. Relative lengths and centromeric indices were determined. Chromosomes nomenclature follows Levan et al. [55].
Abbreviations
- CFA:
-
Clausinella fasciata
- CGA:
-
Chamelea gallina
- DAPI:
-
4′,6-diamidino-2-phenylindole
- DEX:
-
Dosinia exoleta
- DLU:
-
Dosinia lupinus
- FISH:
-
Fluorescence in situ hybridization
- NGS:
-
Next-generation sequencing
- PCR:
-
Polymerase chain reaction
- PLI:
-
Petricola litophaga
- PNA probe:
-
Peptide nucleic acid probe
- RDE:
-
Ruditapes decussatus
- rDNA:
-
Ribosomal DNA
- RPH:
-
Ruditapes philippinarum
- rRNA:
-
Ribosomal RNA
- TRITC:
-
Tetramethyl rhodamine isothiocyanate
- VCA:
-
Venus casina
- VCO:
-
Venerupis corrugata
- VVE:
-
Venus verrucosa
References
Kedes LH. Histone genes and histone messengers. Annu Rev Biochem. 1979;48:837–70.
Long EO, Dawid IB. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–64.
Eirín-López JM, González-Romero R, Dryhurst D, Méndez J, Ausió J. Long-term evolution of histone families: old notions and new insights into their mechanisms of diversification across eukaryotes. In: Pontarotti P, editor. Evolutionary Biology. Berlin: Springer; 2009. p. 139–62.
Drouin G, Moniz-de-Sá M. The concerted evolution of 5S ribosomal genes linked to the repeat units of other multigene families. Mol Biol Evol. 1995;12:481–93.
Cabrero J, López-León MD, Teruel M, Camacho JPM. Chromosome mapping of H3 and H4 histone gene clusters in 35 species of acridid grasshoppers. Chromosome Res. 2009;17:397–404.
Cabral-de-Mello DC, Cabrero J, López-León MD, Camacho JPM. Evolutionary dynamics of 5S rDNA location in acridid grasshoppers and its relationship with H3 histone gene and 45S rDNA location. Genetica. 2011;139:921–31.
Cabral-de-Mello DC, Moura RC, Martins C. Cytogenetic mapping of rRNAs and histone H3 genes in 14 species of Dichotomius (Coleoptera, Scarabaeidae, Scarabaeinae) beetles. Cytogenet Genome Res. 2011;134:127–35.
Cabral-de-Mello DC, Oliveira SG, de Moura RC, Martins C. Chromosomal organization of the 18S and 5S rRNAs and histone H3 genes in Scarabaeinae coleopterans: insights into the evolutionary dynamics of multigene families and heterochromatin. BMC Genet. 2011;12:88.
Mandrioli M, Manicardi GC. Chromosomal mapping reveals a dynamic organization of the histone genes in aphids (Hemiptera: Aphididae). Entomología. 2013;1:e2. doi:10.4081/entomologia.2013.e22013.
Šíchová J, Nguyen P, Dalíková M. Marec F (2013) Chromosomal evolution in tortricid moths: Conserved karyotypes with diverged features. PLoS One. 2013;8(5):e64520. doi:10.1371/journal.pone.00645203.
Drabent B, Kim JS, Albig W, Prats E, Cornudella L, Doenecke D. Mytilus edulis histone gene clusters containing only H1 genes. J Mol Evol. 1999;49:645–55.
Albig W, Warthorst U, Drabent B, Prats E, Cornudella L, Doenecke D. Mytilus edulis core histone genes are organized in two clusters devoid of linker histone genes. J Mol Evol. 2003;56:597–606.
Eirín-López JM, González-Tizón AM, Martínez A, Méndez J. Molecular and evolutionary analysis of mussel histone genes (Mytilus spp): possible evidence of an “orphon origin” for H1 histone genes. J Mol Evol. 2002;55:272–83.
Eirín-López JM, Ruiz MF, González-Tizón AM, Martínez A, Sánchez L, Méndez J. Molecular evolutionary characterization of the mussel Mytilus histone multigene family: first record of a tandemly repeated unit of a five histone genes containing an H1 subtype whit “orphon” features. J Mol Evol. 2004;58:131–44.
Li C, Song L, Zhao J, Zou H, Su J, Zhang H. Genomic organization, nucleotide sequence analysis of the core histone genes cluster in Chlamys farreri and molecular evolution assessment of the H2A and H2B. DNA Seq. 2006;17:440–51.
González-Romero R, Ausió J, Méndez J, Eirín-López JM. Early evolution of histone genes: Prevalence of an ‘orphon’ H1 lineage in Protostomes and birth-and-death process in the H2A family. J Mol Evol. 2008;66:505–18.
Pérez-García C, Cambeiro JM, Morán P, Pasantes JJ. Chromosomal mapping of rDNAs, core histone genes and telomeric sequences in Perumytilus purpuratus (Bivalvia: Mytilidae). J Exp Mar Biol Ecol. 2010;395:199–205.
Pérez-García C, Guerra-Varela J, Morán P, Pasantes JJ. Chromosomal mapping of rRNA genes, core histone genes and telomeric sequences in Brachidontes puniceus and Brachidontes rodriguezi (Bivalvia: Mytilidae). BMC Genet. 2010;11:109.
Pérez-García C, Morán P, Pasantes JJ. Cytogenetic characterization of the invasive mussel species Xenostrobus securis Lmk. (Bivalvia: Mytilidae). Genome. 2011;54:771–8.
Pérez-García C, Morán P, Pasantes JJ. Karyotypic diversification in Mytilus mussels (Bivalvia: Mytilidae) inferred from chromosomal mapping of rRNA and histone gene clusters. BMC Genet. 2014;15:84.
Zhang L, Bao Z, Wang S, Huang X, Hu J. Chromosome rearrangements in Pectinidae (Bivalvia; Pteriomorphia) implied based on chromosomal localization of histone H3 gene in four scallops. Genetica. 2007;130:193–8.
Bouilly K, Chaves R, Fernandes M, Guedes-Pinto H. Histone H3 gene in the Pacific oyster, Crassostrea gigas Thunberg, 1793: molecular and cytogenetic characterisations. Comp Cytogen. 2010;4:111–21.
Carrilho J, Pérez-García C, Leitão A, Malheiro I, Pasantes JJ. Cytogenetic characterization and mapping of rDNAs, core histone genes and telomeric sequences in Venerupis aurea and Tapes rhomboides (Bivalvia: Veneridae). Genetica. 2011;139:823–30.
Huber M. Compendium of bivalves. A full-color guide to 3300 of the world’s marine bivalves. A status on Bivalvia after 250 years of research. Hackenheim: ConchBooks; 2010.
Canapa A, Marota I, Rollo F, Olmo E. Phylogenetic analysis of Veneridae (Bivalvia): comparison of molecular and paleontological data. J Mol Evol. 1996;43:517–22.
Canapa A, Schiaparelli S, Marota I, Barucca M. Molecular data from the 16S rRNA gene for the phylogeny of Veneridae. Mar Biol. 2003;142:1125–30.
Kappner I, Bieler R. Phylogeny of venus clams (Bivalvia: Venerinae) as inferred from nuclear and mitochondrial gene sequences. Mol Phylogenet Evol. 2006;40:317–31.
Mikkelsen PM, Bieler R, Kappner I, Rawlings TA. Phylogeny of Veneroidea (Mollusca: Bivalvia) based on morphology and molecules. Zool J Linnean Soc. 2006;148:439–521.
Chen J, Li Q, Kong L, Zheng X. Molecular phylogeny of venus clams (Mollusca, Bivalvia, Veneridae) with emphasis on the on the systematic position of taxa along the coast of mainland China. Zoologica Scripta. 2011;40:260–71.
Thiriot-Quiévreux C. Advances in cytogenetics of aquatic organisms. In: Beaumont AR, editor. Genetics and evolution of aquatic organisms. London: Chapman and Hall; 1994. p. 369–88.
Thiriot-Quiévreux C. Review of the literature on bivalve cytogenetics in the last ten years. Cah Biol. 2002;43:17–26.
Ebied ABM, Aly FM. Cytogenetic studies on metaphase chromosomes of six bivalve species of families Mytilidae and Veneridae (Nucinelloidea, Mollusca). Cytologia. 2004;69:261–73.
Leitão A, Chaves R, Matias D, Joaquim S, Ruano F, Guedes-Pinto H. Restriction enzyme digestion chromosome banding on two commercially important veneroid bivalve species: Ruditapes decussatus and Cerastoderma edule. J Shellfish Res. 2006;25:857–64.
Wang Y, Guo X. Chromosomal mapping of the vertebrate telomeric sequence (TTAGGG)n in four bivalve molluscs by fluorescence in situ hybridization. J Shellfish Res. 2001;20:1187–90.
Wang Y, Guo X. Chromosomal mapping of major ribosomal rRNA genes in the hard clam (Mercenaria mercenaria) using fluorescent hybridization. Mar Biol. 2007;150:1183–9.
Hurtado N, Pasantes JJ. Surface spreading of synaptonemal complexes in the clam Dosinia exoleta (Mollusca, Bivalvia). Chromosome Res. 2005;13:575–80.
Pérez-García C, Hurtado N, Morán P, Pasantes JJ. Evolutionary dynamics of rDNA clusters in chromosomes of five clam species belonging to the family Veneridae (Mollusca, Bivalvia). BioMed Res Int. 2014;2014:754012.
Hurtado N, Pérez-García C, Morán P, Pasantes JJ. Genetic and cytological evidence of hybridization between native Ruditapes decussatus and introduced Ruditapes philippinarum (Mollusca, Bivalvia, Veneridae) in NW Spain. Aquaculture. 2011;311:123–8.
Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–81.
Torres GA, Gong Z, Iovene M, Hirsh CD, Buell CR, Bryan GJ, et al. Organization and evolution of subtelomeric satellite repeats in the potato genome. G3 (Bethesda). 2011;1:85–92.
Linardopoulou EV, Williams EM, Fan YX, Friedman C, Young JM, Trask BJ. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100.
Insua A, Freire R, Ríos R, Méndez J. The 5S rDNA of mussels Mytilus galloprovincialis and M. edulis: sequence, variation and chromosomal location. Chromosome Res. 2001;9:495–505.
Guo X, Wang Y, Xu Z. Genomic analyses using fluorescence in situ hybridization. In: Liu Z, editor. Aquaculture genome technologies. Oxford: Blackwell Publishing; 2007. p. 289–311.
Leitão A, Chaves R. Banding for chromosomal identification in bivalves. A 20-year history. In Russo R, editor. Aquaculture 1. Dynamic Biochemistry, Process Biotechnology and Molecular Biology 2 (Special Issue 1). Global Science Books; 2008. p. 44–49. http://www.globalsciencebooks.info/Journals/images/GSB_Journal_Information_Stats.pdf
Insua A, López-Piñón MJ, Méndez J. Characterization of Aequipecten opercularis (Bivalvia: Pectinidae) chromosomes by different staining techniques and fluorescent in situ hybridization. Genes Genet Syst. 1998;73:193–200.
Wang Y, Guo X. Chromosomal rearrangement in Pectinidae revealed by rRNA loci and implications for bivalve evolution. Biol Bull. 2004;207:247–56.
López-Piñón MJ, Insua A, Méndez J. Chromosome analysis and mapping of ribosomal genes by one- and two-color fluorescent in situ hybridization in Hinnites distortus (Bivalvia: Pectinidae). J Hered. 2005;96:52–8.
Eickbush TH, Eickbush DG. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics. 2007;175:477–85.
WoRMS Editorial Board. World Register of Marine Species. http://www.marinespecies.org/. Accessed 25 Mar 2015.
Martínez-Expósito MJ, Pasantes JJ, Méndez J. Proliferation kinetics of mussel (Mytilus galloprovincialis) gill cells. Mar Biol. 1994;120:41–5.
Winnepenninckx B, Backeljau T, Wachter R. Extraction of high molecular weight DNA from molluscs. Trends Genet. 1993;9:407.
Vilgalys R. http://www.biology.duke.edu/fungi/mycolab/primers.htm. Accessed 25 Mar 2015.
Fang BL, De Baere R, Vandenberghe A, De Wachter R. Sequences of three molluscan 5S ribosomal RNAs confirm the validity of a dynamic secondary structure model. Nucleic Acids Res. 1982;10:4679–85.
Giribet G, Distel D. Bivalve phylogeny and molecular data. In: Lydeard C, Lindberg DR, editors. Systematics and Phylogeography of Molluscks. Washington: Smithsonian Books; 2003. p. 45–90.
Levan A, Fredga K, Sandberg AA. Nomenclature for centromeric position on chromosomes. Hereditas. 1964;52:201–20.
Acknowledgements
We wish to thank N. Santamaría for her technical assistance and M. Lastra, A. Simón and S. Pereira for kindly providing the clams. This work was partly funded by grants from Xunta de Galicia and Fondos FEDER: “Unha maneira de facer Europa” (08MMA023310PR; Grupos de Referencia Competitiva, 2010/80; Grupos con Potencial Crecimiento, GPC2013-011). D. García-Souto was partially supported by a FPU fellowship from Ministerio de Educación (Spain).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
DGS did most part of the cytogenetic procedures and collaborated on the molecular work, the bibliographic review and the writing of this paper. CPG helped with the cytogenetic procedures and the writing of the manuscript. PM participated in developing the molecular techniques and helped in the writing. JJP coordinated the study, helped in developing the laboratory techniques and cytogenetic analyses and coordinated the writing of the manuscript. All authors read and approved the final manuscript.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
García-Souto, D., Pérez-García, C., Morán, P. et al. Divergent evolutionary behavior of H3 histone gene and rDNA clusters in venerid clams. Mol Cytogenet 8, 40 (2015). https://doi.org/10.1186/s13039-015-0150-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-015-0150-7