Abstract
Background
Scarabaeinae beetles show a high level of macro-chromosomal variability, although the karyotypic organization of heterochromatin and multigene families (rDNAs and histone genes) is poorly understood in this group. To better understand the chromosomal organization and evolution in this group, we analyzed the karyotypes, heterochromatin distribution and chromosomal locations of the rRNAs and histone H3 genes in beetles belonging to eight tribes from the Scarabaeinae subfamily (Coleoptera, Scarabaeidae).
Results
The number of 18S rRNA gene (a member of the 45S rDNA unit) sites varied from one to 16 and were located on the autosomes, sex chromosomes or both, although two clusters were most common. Comparison of the 45S rDNA cluster number and the diploid numbers revealed a low correlation value. However, a comparison between the number of 45S rDNA sites per genome and the quantity of heterochromatin revealed (i) species presenting heterochromatin restricted to the centromeric/pericentromeric region that contained few rDNA sites and (ii) species with a high quantity of heterochromatin and a higher number of rDNA sites. In contrast to the high variability for heterochromatin and 45S rDNA cluster, the presence of two clusters (one bivalent cluster) co-located on autosomal chromosomes with the 5S rRNA and histone H3 genes was highly conserved.
Conclusions
Our results indicate that the variability of the 45S rDNA chromosomal clusters is not associated with macro-chromosomal rearrangements but are instead related to the spread of heterochromatin. The data obtained also indicate that both heterochromatin and the 45S rDNA loci could be constrained by similar evolutionary forces regulating spreading in the distinct Scarabaeinae subfamily lineages. For the 5S rRNA and the histone H3 genes, a similar chromosomal organization could be attributed to their association/co-localization in the Scarabaeinae karyotypes. These data provide evidence that different evolutionary forces act at the heterochromatin and the 45S rDNA loci compared to the 5S rRNA and histone H3 genes during the evolution of the Scarabainae karyotypes.
Similar content being viewed by others
Background
Repetitive DNA elements constitute a large portion of eukaryote genomes and include satellites, minisatellites, microsatellites, transposable elements and some multigene families with high copy numbers [1, 2]. Among them, ribosomal RNAs (rRNA) and histone genes are grouped into distinct multigene families that are organized in tandem with hundreds to thousands of copies of each [3, 4]. The major ribosomal DNA cluster (tandem arrayed 45S rDNA repeating units) encodes for the 28S, 18S and 5.8S rRNAs, while the minor rDNA cluster (tandemly arrayed 5S rDNA repeating units) is responsible for the transcription of 5S rRNA [3]. The histone genes may be clustered into distinct chromosomal regions, and among invertebrates, these genes are typically clustered as quartets (H2A, H2B, H3, and H4) or quintets (H2A, H2B, H3, and H4 plus H1), although scattered solitary genes have also been reported [4–6].
Ribosomal RNA and multigene histone families are very useful cytogenetic markers for studying chromosomal diversification and genome organization. In insects, these sequences have been cytogenetically mapped more frequently in species belonging to the orders Coleoptera, Lepidoptera and Orthoptera, although studies are still incipient and have concentrated on mapping the major rDNA locus [7–11]. The mapping of other multigene families, such as the 5S rDNA locus and the histone genes, was primarily performed in specific groups, e.g., Acrididae [9, 12] and Proscopiidae grasshoppers [10], chironomid midges [13] and fruit flies [14, 15].
Scarabaeidae beetles comprise more than 25,000 species that are distributed worldwide and represent the largest group of the Scarabaeiodea superfamily [16, 17]. Notably, the macro karyotypic structure of the Scarabaidae family is highly diverse in the representatives of subfamily Scarabaeinae (dung beetles), with variations in the diploid number that range from 2n = 8 to 2n = 24, distinct sex chromosome systems and chromosomal morphologies [18]. Scarabaeidae have been poorly studied with regards to chromosomal mapping of DNA sequences, and the analyses were primarily performed to describe the major rDNA clusters [19–27]. Only species from the genus Dichotomius have been analyzed with regard to the histone genes and the 5S rDNA locus [28].
Due to the high chromosomal diversity of Scarabaeinae beetles, the chromosomal organization and evolution of the group, the diploid number, the distribution of heterochromatin, the location of the 18S and 5S rRNA loci, and the histone H3 gene arrays were analyzed in several species that belong to different lineages, in order to understand the patterns of karyotypic evolution in the group. Our results showed distinct evolutionary patterns for the multigene families studied. The 5S rRNA and histone H3 gene clusters were primarily associated and were highly conserved in number, while the major rDNA locus and heterochromatin regions showed an intense turnover in their number and location in the Scarabaeinae karyotypes. These results are also discussed in the light of the possible mechanisms that are involved in diversification of repeated DNA elements.
Results
Karyotypes and heterochromatin distribution
Chromosomal complements and heterochromatin distribution patterns of 13 Scarabaeinae species were determined, including new descriptions and re-descriptions of previously published data (Table 1). The variation in the diploid number ranged from 2n = 8 to 2n = 20. The species were classified in three distinct groups of karyotypes based on their heterochromatin distribution pattern: (i) heterochromatin restricted to the centromeric/pericentromeric regions (non-spread pattern), (ii) heterochromatin in the centromeric/pericentromeric regions with additional terminal or subterminal heterochromatic blocks (moderately spread pattern), and (iii) high amount of heterochromatin, primarily represented by diphasic chromosomes (with heterochromatin blocks occupying one entire chromosomal arm) and large paracentromeric blocks of heterochromatin (highly spread pattern) (Table 1).
Mapping of multigene families
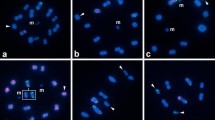
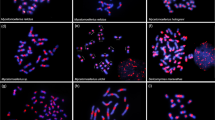
Chromosomal mapping of the 18S and 5S rDNA loci primarily revealed conspicuous clusters on distinct chromosomes (Figures 1, 2). The most common pattern for the 18S rDNA clusters was the presence of two sites (one bivalent). However, the number of clusters for this repeated gene ranged from one to 16, with an average of 4.2 sites per diploid genome, that were located on the autosomes, sex chromosomes or both. The autosomes were the most common location (~89.0% of the sites) (Figures 1, 2, 3 and Table 1). For the 5S rRNA gene almost all species presented only two sites (one bivalent) that were located on one autosomal pair (Figures 1, 2 and Table 1). Distinct 5S rDNA location patterns were observed for several species. In Eurysternus caribaeus (Figure 2a), the 5S rDNA was restricted to the × chromosome; in Diabroctis mimas (Figure 1c), six clusters were observed (five on the autosomes and one on the × chromosome). In Coprophanaeus ensifer (Figure 1a), the clusters were located on the sex chromosomes. In general, the two rRNA genes presented distinct location, although in three species the sites of the 18S and 5S rDNA loci occurred in the same chromosomal region (Figures 1, 2). In D. mimas, the sites of the 18S and 5S rDNA loci co-occurred on one autosomal bivalent pair and on the × chromosome (Figure 1c). For Digitonthophagus gazella and E. caribaeus, these genes were collocated on one autosomal bivalent or on the × chromosome, respectively (Figure 2a, b).
Fluorescent in situ hybridization in metaphase I using 5S rDNA (red) and 18S rDNA (green) in nine representative species of Scarabaeinae that belong to three distinct tribes ([a-d] Phanaeini, [e-h] Canthonini, [i] Ateuchini). (a) Coprophanaeus ensifer, (b) C. cyanescens, (c) Diabroctis mimas, (d) Phanaeus splendidulus, (e) Canthon staigi, (f) Deltochilum calcaratum, (g) D. verruciferum, (h) D. elevatum and (i) Atheuchus sp. The arrows indicate the sex chromosome bivalents. Note the co-localization of the two gene clusters in (c) at two chromosomes. Bar = 5 μm.
Cytogenetic mapping of the 5S (red) and 18S (green) rDNA clusters in four species of Scarabaeinae belonging to the (a) Oniticellini, (b) Onthophagini, and (c, d) Coprini tribes. (a) Eurysternus caribaeus, (b) Digitonthophagus gazella, (c) Ontherus apendiculatus and (d) O. sulcator. The arrows indicate the sex bivalents. Note the co-localization of the two gene clusters in (a) and (b). Bar = 5 μm.
FISH for the 5S rRNA and histone H3 genes revealed that these two markers show overlapping signals in the karyotypes of six species randomly selected and in all other previously studied species, indicating that the two genes co-locate (Figure 4, Table 1). In most species, these two sequences were located only on one autosomal bivalent (Figure 4, Table 1). However, in C. ensifer, the 5S rDNA and histone H3 gene elements were located on the × and Y chromosomes (Figure 4b), while in E. caribaeus, the H3 gene elements were exclusively found on the × chromosome (Figure 4e). In D. mimas, these two loci were located on five autosomal chromosomes and the × chromosome (results not shown).
FISH for the 5S rRNA and histone H3 genes in five Scarabaeinae representatives. (a) Coprophanaeus cyanescens, (b) C. ensifer, (c) Phanaeus splendidulus, (d) Deltochilum verruciferum and (e) Eurysternus caribaeus. The arrows indicate the sex bivalents. Note the co-localization of the two clusters in all cells, and the presence of only one cluster for the two genes on the × chromosome in (e). Bar = 5 μm.
The analysis of interphasic nuclei and early meiotic cells (with less condensed chromosomes/chromatin) hybridized with the 18S and 5S rDNA and histone H3 probes revealed that the 5S rRNA and histone H3 genes overlapped (additional file 1), as observed in the condensed metaphasic chromosomes (Figure 4). However, the 18S rDNA loci were located in a distinct region of the cell from the 5S and histone sites, and only the species that showed co-location of the 5S/18S rDNAs (E. caribaeus and D. gazella) (Figure 2a, b), showed overlapping signals (additional file1e, f).
An interesting characteristic for the three multigene families studied was the presence of only one site per chromosome (Figures 1, 2, 4). In addition to the variability observed for the 18S rDNA loci in the species studied here for the first time, we identified polymorphisms regarding the number of sites in species that had previously described, such as in Deltochilum calcaratum, C. ensifer, C. cyanescens and Diabroctis mimas (Table 1).
Heteromorphism related to the cytogenetic mapping of the three genes in several species was observed with regard to the size and presence/absence of the clusters in the homologous chromosomes, as in, for example, Ontherus sulcator (Figure 2d), D. mimas (Figure 1c) and Phanaeus splendidulus (Figure 1d). For E. caribaeus it was observed variability of 18S rDNA cluster size between × and Y chromosomes, and 5S rRNA/H3 histone genes were restrict to × chromosome. Although we were able to define the number of clusters for the genes mapped, the precise positions along the chromosomes were sometimes difficult to determine due the small size and the high condensation level of the chromosomes. It was also difficult to determine the specific chromosome pair that contained the sequences studied in some species due the karyotypic symmetry.
The data obtained regarding the number of major rDNA sites for 31 species (including both the present work and previous published data) were compared to the diploid numbers (2n) and heterochromatin distribution patterns to evaluate the relationship between these karyotype features. The variations in the diploid number and the number of rDNA sites showed no apparent relationship as indicated by a low correlation value (r = 0.21, P = 0.199) between the data (Figure 5a). However, a higher number of major rDNA sites were observed in species with increased amounts of heterochromatin (Figure 5b).
Comparison of the distribution of the number of 18S rDNA loci and the diploid number in 31 species of Scarabaeinae (a) or with the distribution of heterochromatin in 21 species (b). Each symbol indicated below the name of species represents one of the distinct tribes: (T with stroke) Oniticellini, (phi) Ateuchini, (black circle) Coprini, (empty triangle) Onthophagini, (plus sign) Gymnopleurini, (asterisk) Canthonini, (empty circle) Onitini and (black square) Phaneini. Species with intraspecific variation were considered twice (see Table 1). The color of the bars in (a) corresponds to each defined class regarding the diploid number, 2n = 8 (black), 2n = 14 (light green), 2n = 16 (orange), 2n = 18 (red), 2n = 20 (blue), and in (b) the distribution of heterochromatin, pericentromeric blocks (purple), pericentromeric and terminal blocks (brown), pericentromeric blocks and diphasic chromosomes (dark green). In (b), the colored squares correspond to each heterochromatin distribution pattern.
Discussion
High diversity of the chromosomal distribution of the 18S rDNA clusters and heterochromatin
Considering the major rRNA genes, two main patterns of distribution were detected (i) two rDNA sites (one chromosomal bivalent) harboring these genes, as observed in Dichotomius, Canthon staigi, Deltochilum elevatum and Ontherus apendiculatus; (ii) increased numbers of 18S rDNA clusters (ranging from 3 to 16 sites), as observed in Bubas bison, C. ensifer, C. cyanescens, D. mimas, Ontherus sulcator and three Deltochilum species. This suggests that the major rRNA genes are under a distinct evolutionary mechanism regarding cluster spreading.
The two main patterns for the major rDNA distribution were primarily observed for three tribes, Canthonini, Coprini and Phanaeini, which include several species analyzed. Among Coprini species, the clusters of the major rDNA clusters have not suffered intense chromosomal reorganization, as they are primarily associated with only one bivalent, as observed in Dichotomius species and Ontherus apendiiculatus. Phanaeini is characterized by an intense movement of the major rDNA clusters that resulted in the generation of different numbers of sites on several chromosomes, as observed in C. ensifer, which presents the highest number of rDNA clusters (16 sites) within the subfamily, and in Coleoptera [27]. Intraspecific polymorphism with regard to the number of rDNA clusters was observed in Coprophaneus ensifer, C. cyanescens, and D. mimas. In the Canthonini tribe, variable patterns of rDNA clusters were observed, with species presenting with either no spreading of the major rDNA clusters, such as Canthon staigi and Deltochilum elevatum, or with scattered rDNA clusters, as observed for three of the Deltochilum representatives. These results indicate that the major rDNA clusters' ability to move is independent of taxonomic units and may be related to the heterochromatin dispersion (see discussion below).
The ancient condition in Scarabaeinae appears to be the occurrence of one autosomal bivalent as a nucleolar organizer. This theory is corroborated by the presence of the pattern in a large number of species within the group and the sister groups of the subfamily. That distribution pattern is also the most common pattern for Coleoptera as a whole, at least for representatives of Polyphaga [29]. In addition to this common pattern that consists of only one chromosomal pair of major rDNA clusters, an intense repositioning of the major rDNA clusters in Scarabaeinae was involved in the increasing number of rDNA sites and the movement to different autosomes and sex chromosomes. The presence of major rDNA clusters associated with the sex chromosomes in different species could be related to either (i) the occurrence of large chromosomal rearrangements, such as fusions, as observed in Deltochilum calcaratum, D. morbillosum and E. caribaeus, species that have a derived neo-XY sex system, or (ii) the occurrence of transpositions, as observed in Coprophanaeus, D. mimas and Deltochilum verruciferum, which are species with the ancient Scarabaeinae diploid number (2n = 20). Although the occurrence of chromosomal fusions were proposed in some species with a reduced diploid number, the presence of rDNA clusters on the sex chromosomes could also be a result of transpositions if fusion involving only autosomes is considered, as in Dichotomius [28].
Although the variation in the number of major rDNA clusters can be attributed to chromosomal rearrangements in some species, there is no correlation between the variation in the rDNA sites and the diploid number. There are examples of species that have a reduction in the diploid number without a modification to the number of rDNA sites, while species with conservation of the ancestral diploid number and extensive repositioning and expansion of major rDNA clusters number have also been identified. There is evidence of the "movement" and "multiplication" of the major rDNA clusters without fusions or other chromosomal rearrangements [30]. In Scarabaeinae, these modifications could be attributed to an ectopic recombination and transposition and to inversions and translocations within the genome. Similar mechanisms are responsible for intra- and interspecific variations in other insects, such as Acrididae grasshoppers [8] and in Lepidoptera [11]. These results indicate distinct evolutionary trends that are related to the macro-chromosomal structure (diploid number, chromosome morphology and sex chromosomes) and the organization of the major rDNA genes in some insect genomes.
The analysis of heterochromatin and major rDNA dispersion revealed an interesting relationship pattern. Species with heterochromatin restricted to the centromeric/pericentromeric regions were primarily characterized as having a stable number of major rDNA that were restricted to one chromosomal bivalent. Only Ateuchus sp. had four clusters, while the presence of three clusters in D. semisquamosus was a polymorphic condition. However, extensive variability in the number of major rDNA sites was observed in the majority of representatives (except for Isocopris inhiata) in which heterochromatin was dispersed and occurred in large quantities within the karyotypes, e.g., large paracentromeric heterochromatic blocks and diphasic chromosomes. In species that showed a moderate dispersion of heterochromatin, the major rDNA clusters spread in two species and was restricted to one autosomal bivalent in another, Ontherus appendiculatus. Interestingly in species whose the relationship in position for heterochromatic blocks and major rDNA was possible to determine it was observed a general pattern for non co-localization in some representatives without dispersion for these two chromosomal markers, such as in Dichotomius [28]. In species with spreading of these elements in general they were co-located, such as in Deltochilum and Coprophaneus [26, 27]. Our results indicate that the same evolutionary forces might be acting on these two components of the Scarabaeinae genome, resulting in the spreading of the major rDNA clusters along with heterochromatin. This hypothesized pattern of evolution might be favored by ectopic paring during chromocenter formation during the initial meiotic stage. Ectopic pairing is a common behavior in this insect group that appears to play an important role in nucleolar organization and chromosomal segregation [31, 32].
The restriction or spreading of the number of rDNA clusters might be associated with the presence or absence of an appropriate molecular mechanism associated with heterochromatin and involved in the ectopic recombination possibly caused by repeated DNAs. The ability of rDNA clusters to move and vary in number was first observed by Schubert (1984) [33] in Allium. Since then, some additional evidence has accumulated concerning the ability of rDNA to move within the genome. Recent studies have proposed that transposable elements are a potential source for the movement of rDNA [34, 35] and other genes [36, 37] to different regions of the genome.
The conservation of the 5S rRNA and histone H3 genes in Scarabaeinae karyotypes
In contrast to the variability in the number of major rDNA clusters, a high conservation in the number of 5S rRNA and histone H3 gene clusters was observed. For invertebrates, the mapping of these sequences was previously restricted to few species of mollusks, insects, crustaceans, annelids and echinoderms [9, 10, 12, 28, 38–42]. In insects, these types of studies have been mainly focused on grasshoppers [9, 10, 12, 42], and only 14 species of beetles had been previously studied, all of which belong to the genus Dichotomius [28]. The co-localized clusters (one bivalent) for these two genes in some Scarabaeinae species could indicate that this is the ancient organization for these sequences, and they have not extensively changed in number since the origin of Scarabaeinae [43], despite the diversification of the species. An intense conservation of the number of histone gene clusters, with only one or two chromosomes containing clusters, has also been described in grasshoppers [9, 10], mollusks [40, 41] and fish species [44, 45], although variability has also been reported in these groups. These results might indicate that a strong purifying selection acts on the histone clusters, preventing the spread of these genes through the genome, as was proposed for the grasshoppers [9].
The 5S rDNA gene is highly conserved in Scarabaeinae representatives, and all species examined showed an overlap between the 5S rDNA and the histone H3 genes' signal at the same chromosomal region. This overlap was corroborated by the observation of overlapped signals in cells that were in the initial stages of meiosis and that had interphasic nuclei containing less condensed chromosomes. This indicates that these two multigene families could have a linked organization in the Scarabaeinae genomes. The associated dispersion of the 5S rRNA/histone H3 genes in D. mimas and the restriction of these two sequences to the × chromosome of E. caribaeus, likely due to unequal cross-over events between the × and Y chromosome in this species [24], reinforces the hypothesis that the 5S rDNA/histone H3 gene clusters are associated in the genome. Additional molecular studies are necessary to fully confirm this hypothesis. An associated or co-localized organization has also been described in mollusks [46], crustaceans [47–49], Dichotomius coleopterans [28] and Proscopiidae grasshoppers [10]. Our results reinforce the idea that the association of the 5S rDNA and histone H3 clusters is not sporadic in coleopterans and that it appears to be common. Besides the association of 5S rDNA and histone H3 genes, co-localization or linked organization of major rDNA and histone genes were also reported in insect as described recently for example in Diuraphis noxia (Hemiptera) [50], Anthonomus grandis and A. texanus (Coleoptera) [51].
Unlike the results observed among the representatives of Scarabaeinae, the 5S rDNA cluster is highly dynamic among chromosomes and the genomic dynamism in some animal groups, such as in fish and Acrididae grasshoppers [12, 52, 53]. This stability in Scarabaeidae beetles could be the result of its association with the histone genes, which may result in the same purifying selection that appears to act against the spread of histone clusters.
In contrast to the co-localization of the 5S rDNA/histone H3 clusters, the 18S rDNA is not co-localized in the genomes of the Scarabaeinae species studied. Only Diabroctis mimas and Digitonthophagus gazella showed a co-localization of these sequences. These results could be explained by a transposition of the 18S rDNA cluster due to its intense movement in the genome of some species. This physical separation could be result in a functional advantage for these ribosomal sequences. The disassociation of the two multigene families that encode for rRNAs is a common pattern for eukaryotic and vertebrate chromosomes, including those in fishes [54–56]. However, some invertebrate species have co-localized rRNA clusters, including representatives of the annelids, mollusks and crustaceans; however, a non co-localized organization has also been described [38, 48, 57, 58].
The association/co-localization of multigene families in animal genomes has been reported for some sequences, including rRNAs, the histone genes and small nuclear RNAs (snRNAs). These associations/co-localizations are poorly understood, and their biological effect is still unclear. According to studies by Dover (1986) [59] and Liu and Fredga (1999) [60], the linkage is important to maintain multiple, conserved arrays. Kaplan et al. (1993) [61] hypothesized that the association of the repetitive multigene families might play a functional role in the organization of the nucleus. In the case of the 18S rDNA, 5S rDNA and histone H3 histone, the separation of the 18S and 5S rDNA arrays might convey a functional advantage, since the 18S rRNA gene is transcribed by RNA polymerase I and the 5S rRNA gene is transcribed by RNA polymerase III. However, the association of the histone H3 and 5S rRNA genes cannot be explained by a transcriptional advantage because these two sequences are transcribed by different polymerases.
Conclusions
The high variability in the karyotype organization previously observed in Scarabaeinae representatives resulted from distinct chromosome rearrangements during their evolution [18] is also observed for heterochromatin and the 18S rDNA clusters. Our results indicate that these two genomic elements (18S rDNA and heterochromatin) likely are subjected to similar evolutionary forces that regulate their spreading in the distinct Scarabaeinae subfamily lineages. The conservation of the location and number of the 5S rRNA and histone H3 gene clusters indicates that these multigene families are likely under the control of different evolutionary forces than the 18S rDNA clusters and heterochromatin. This separation reinforces the idea that evolutionary spreading mechanism might operate differently at multigene families and other repeat elements.
Methods
Samples from adult males of 13 species of Scarabaeinae beetles were collected in distinct cities in the Minas Gerais, Paraná, Pernambuco, and São Paulo states in Brazil. The testes were fixed in Carnoy (3:1 ethanol:acetic acid) and were stored in the freezer at -20°C. To check the male karyotypes, the slides were stained with 2% lacto-acetic orcein. The chromosome preparations for the C-banding and fluorescence in situ hybridization (FISH) experiments were made by squashing using a drop of 45% acetic acid and subsequently removing the coverslip after immersion in liquid nitrogen. The C-banding experiments were performed according the protocol described by Sumner (1972) [62].
DNA probes for 5S rDNA and H3 histone sequences were obtained from cloned fragments obtained from the genome of the beetle Dichotomius semisquamosus [63]. The 18S rRNA and histone H3 gene probes were labeled by nick translation using biotin-11-dATP (Invitrogen, San Diego, CA, USA), and the 5S rRNA gene was labeled with digoxigenin-11-dUTP (Roche, Mannheim, Germany). The FISH procedures were performed according to the method adapted by Cabral-de-Mello et al. (2010b) [63]. Preparations were counterstained with 4,6-diamidino-2-phenylindole (DAPI) and mounted in Vectashield (Vector, Burlingame, CA, USA). Images were captured using an Olympus DP71 digital camera coupled to a BX61 Olympus microscope and were optimized for brightness and contrast using Adobe Photoshop CS2.
Statistical analysis was performed using the Pearson rank test to analyze the degree of correlation between the number of 45S rDNA sites and the diploid number. A comparative analysis between the distribution of heterochromatin and the number of 45S rDNA sites was also performed.
Abbreviations
- 2n:
-
diploid number
- DAPI:
-
4, 6-diamidino-2-phenylindole
- FISH:
-
fluorescence in situ hybridization
- rDNA:
-
ribosomal DNA
- rRNA:
-
ribosomal RNA
- snRNA:
-
small nuclear RNA
References
Biémont C, Vieira C: Junk DNA as an evolutionary force. Nature. 2006, 443: 521-524. 10.1038/443521a.
Charlesworth B, Sniegowski P, Stephan W: The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994, 371: 215-220. 10.1038/371215a0.
Long EO, Dawid ID: Repeated genes in eukaryotes. Annu Rev Biochem. 1980, 49: 727-764. 10.1146/annurev.bi.49.070180.003455.
Maxon R, Cohn R, Kedes L: Expression and organization of histone genes. Ann Rev Gen. 1983, 17: 239-277. 10.1146/annurev.ge.17.120183.001323.
Lifton RP, Goldberg ML, Karp RW, Hogness DS: The organization of the histone genes in Drosophila melanogaster: Functional and evolutionary implications. Cold Spring Harbor Symp Quant Biol. 1977, 42: 1047-1051.
Engel JD, Dodgson JB: Histone genes are clustered but not tandemly repeated in the chicken genome. Proc Natl Acad Sci USA. 1981, 78: 2856-2860. 10.1073/pnas.78.5.2856.
De La Rúa P, Serrano J, Hewitt GM, Galián J: Physical mapping of rDNA genes in the ground beetle Carabus and related genera (Coleoptera: Carabidae). J Zool Syst Evol Res. 1996, 34: 95-101.
Cabrero J, Camacho JPM: Location and expression of ribosomal RNA genes in grasshoppers: Abundance of silent and cryptic loci. Chromosome Res. 2008, 16: 595-607. 10.1007/s10577-008-1214-x.
Cabrero J, López-León MD, Teruel M, Camacho JPM: Chromossome mapping of H3 and H4 histone gene clusters in 35 species of acridid grasshoppers. Chromosome Res. 2009, 17: 397-404. 10.1007/s10577-009-9030-5.
Cabral-de-Mello DC, Martins C, Souza MJ, Moura RC: Cytogenetic mapping of 5S and 18S rRNAs and H3 histone genes in 4 ancient Proscopiidae grasshopper species: contribution to understanding the evolutionary dynamics of multigene families. Cytogenet Genome Res. 2011, 132: 89-93. 10.1159/000317476.
Nguyen P, Sahara K, Yoshido A, Marec F: Evolutionary dynamics of rDNA clusters on chromosomes of moths and butterflies (Lepidoptera). Genetica. 2010, 138: 343-354. 10.1007/s10709-009-9424-5.
Cabral-de-Mello DC, Cabrero J, López-León MD, Camacho JPM: Evolutionary dynamics of 5S rDNA location in acridid grasshoppers and its relationship with H3 histone gene and 45S rDNA location. Genetica. 2011, 139: 921-931. 10.1007/s10709-011-9596-7.
Hankeln T, Keyl HG, Ross R, Schmidt ER: Evolution of histone gene loci in chironomid midges. Genome. 1993, 36: 852-862. 10.1139/g93-113.
Schienman JE, Lozovskaya ER, Strausbaugh LD: Drosophila virilis has atypical kinds and arrangements of histone repeats. Chromosoma. 1998, 107: 529-539. 10.1007/s004120050339.
Ranz JM, González J, Casals F, Ruiz A: Low occurrence of gene transposition events during the evolution of the genus Drosophila. Evolution. 2003, 57: 1325-1335.
Costa C: Estado de conocimiento de los Coleoptera Neotropicales. Hacia un Proyecto CYTED para el inventario y estimación de la diversidad Entomológica en Iberoamérica: PrIBES-2000, m3m. Edited by: Martin Piera F, Morrone JJ, Melic A. 2000, Monografias Tercer Milênio v.1. Zaragoza: SEA, 99-114.
Vaz-de-Mello FZ: Estado atual de conhecimento dos Scarabaeidae s. str. (Coleoptera: Scarabaeoidea) do Brasil. Hacia un Proyecto CYTED para el inventario y estimación de la diversidad Entomológica en Iberoamérica: PrIBES-2000, m3m. Edited by: Martin Piera F, Morrone JJ, Melic A. 2000, Monografias Tercer Milênio v.1. Zaragoza: SEA, 183-195.
Cabral-de-Mello DC, Oliveira SG, Ramos IC, Moura RC: Karyotype differentiation patterns in species of the subfamily Scarabaeinae (Scarabaeidae: Coleoptera). Micron. 2008, 39: 1243-1250. 10.1016/j.micron.2008.04.002.
Moura RC, Souza M, Mel NF, Lira-Neto AC: Karyotypic characterization of representatives from Melolonthinae (Coleoptera, Scarabaeidae): Karyotypic analysis, banding and fluorescent in situ hybridization (FISH). Hereditas. 2003, 138: 200-206. 10.1034/j.1601-5223.2003.01611.x.
Bione E, Moura RC, Carvalho R, Souza MJ: Karyotype, C-and fluorescence banding pattern, NOR location and FISH study of five Scarabaeidae (Coleoptera) species. Gen Mol Biol. 2005, 26: 376-381.
Bione E, Camparoto ML, Simões ZLP: A study of the constitutive heterochromatin and nucleolus organizer regions of Isocopris inhiata and Diabroctis mimas (Coleoptera: Scarabaeidae, Scarabaeinae) using C-banding, AgNO3 staining and FISH techniques. Gen Mol Biol. 2005, 28: 111-116. 10.1590/S1415-47572005000100019.
Colomba MS, Vitturi R, Zunino M: Karyotype analyzes, banding, and fluorescent in situ hybridization in the Scarab beetle Gymnopleurus sturmi McLeady (Coleoptera, Scarabaeoidea, Scarabaeidae). J Heredity. 2000, 91: 260-264. 10.1093/jhered/91.3.260.
Colomba MS, Vitturi R, Libertini A, Gregorini A, Zunino M: Heterochromatin of the scarab beetles, Bubas bison (Coleoptera: Scarabaeidae) II. Evidence for AT-rich compartmentalization and a high amount of rDNA copies. Micron. 2006, 37: 47-51. 10.1016/j.micron.2005.06.004.
Arcanjo AP, Cabral-de-Mello DC, Silva AEB, Moura RC: Cytogenetic characterization of Eurysternus caribaeus (Coleoptera: Scarabaeidae): evidence of sex-autosome fusion and diploid number reduction prior to species dispersion. J Gen. 2009, 88: 177-182. 10.1007/s12041-009-0025-y.
Silva GM, Bione EG, Cabral-de-Mello DC, Moura RC, Simões ZLP, Souza MJ: Comparative cytogenetics study in three species of the genus Dichotomius (Coleoptera: Scarabaeidae). Gen Mol Biol. 2009, 32: 276-280. 10.1590/S1415-47572009005000040.
Cabral-de-Mello DC, Moura RC, Carvalho R, Souza MJ: Cytogenetic analysis of two related Deltochilum (Coleoptera, Scarabaeidae) species: diploid number reduction, extensive heterochromatin addition and differentiation. Micron. 2010, 41: 112-117. 10.1016/j.micron.2009.10.005.
Oliveira SG, Moura RC, Silva AEB, Souza MJ: Cytogenetic analysis of two Coprophanaeus species (Scarabaeidae) revealing wide constitutive heterochromatin variability and the largest number of 45S rDNA sites among Coleoptera. Micron. 2010, 41: 960-965. 10.1016/j.micron.2010.06.015.
Cabral-de-Mello DC, Moura RC, Martins C: Cytogenetic mapping of rRNAs and histone H3 genes in 14 species of Dichotomius (Coleoptera, Scarabaeidae, Scarabaeinae) beetles. Cytogenet Genome Res. 2011, 134: 127-135. 10.1159/000326803.
Schneider MC, Rosa SP, Almeida MC, Costa C, Cella DM: Chromosomal similarities and differences among four Neotropical Elateridae (Conoderini and Pyrophorini) and other related species, with comments on the NOR patterns in Coleoptera. J Zool Syst Evol Res. 2007, 45: 308-316. 10.1111/j.1439-0469.2006.00398.x.
Dubkovsky J, Dvorak J: Ribosomal RNA multigene loci: Nomads of the Triticeae genomes. Genetics. 1995, 140: 1367-1377.
Drets ME, Corbella E, Panzera F, Folle GA: C-banding and non-homologous association II. The "parachute" Xyp sex bivalent and the behavior of heterochromatic segments in Epilachna paenulata. Chromosoma. 1983, 88: 249-255. 10.1007/BF00292901.
Smith SG, Virkki N: Coleoptera. Animal Cytogenetics. Edited by: John B. 1978, Borntraeger, Berlin, Stuttgard, 366-
Schubert I: Mobile nucleolus organizing regions (NORs) in Allium (Liliaceae S-Lat)- inferences from the specifity of silver staining. Plant Syst Evol. 1984, 144: 291-305. 10.1007/BF00984139.
Raskina O, Barber JC, Nevo E, Belyayev A: Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogenet Genome Res. 2008, 120: 351-357. 10.1159/000121084.
Zhang X, Eickbush MT, Eickbush TH: Role of recombination in the long-term retention of transposable elements in rRNA gene loci. Genetics. 2008, 180: 1617-1626. 10.1534/genetics.108.093716.
Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR: Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004, 431: 569-573. 10.1038/nature02953.
Lai Z, Nakazato T, Salmaso M, Burke JM, Tang S, Knapp SJ, Rieseberg LH: Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics. 2005, 171: 291-303. 10.1534/genetics.105.042242.
Vitturi R, Colomba M, Mandrioli M, Pirrone AM: rDNA (18S-28S and 5S) co-localization and linkage between ribosomal genes and (TTAGGG) n telomeric sequence in the earthworm Octodrilus complanatus (Annelida: Oligochaeta: Lumbricidae) revealed by single- and double-colour FISH. J Hered. 2002, 93: 279-282. 10.1093/jhered/93.4.279.
Gornung E, Kartavenko T, Kurchashova S, Kireev I, Fais D: Physical mapping of the 5S rRNA in the common sea urchin, Paracentrotus lividus (Echinodermata: Echinoidea), by in situ hybridization. Cytogenet Genome Res. 2005, 111: 186C-10.1159/000086394.
Zhang L, Bao Z, Wang S, Huang X, Hu J: Chromosome rearrangements in Pectinidae (Bivalvia: Pteriomorphia) implied based on chromosomal localization of histone H3 gene in four scallops. Genetica. 2007, 130: 193-198. 10.1007/s10709-006-9006-8.
Pérez-García C, Guerra-Varela J, Morán P, Pasantes JJ: Chromosomal mapping of rRNA genes, core histone genes and telomeric sequences in Brachidontes puniceus and Brachidontes rodriguezi (Bivalvia, Mytilidae). BMC Genetics. 2010, 11: 109-
Teruel M, Cabrero J, Perfectti F, Camacho JPM: B chromosome ancestry revealed by histone genes in the migratory locust. Chromosoma. 2010, 119: 217-225. 10.1007/s00412-009-0251-3.
Krell F-T: Fossil record and evolution of Scarabaeoidea (Coleoptera: Polyphaga). Coleopterists Sciety Monograph. 2006, 5: 120-143.
Pendás AM, Morán P, García-Vázquez E: Organization and chromosomal location of the major histone cluster in brown trout, Atlantic salmon and rainbow trout. Chromosoma. 1994, 103: 147-152. 10.1007/BF00352324.
Hashimoto DT, Ferguson-Smith MA, Rens W, Foresti F, Porto-Foresti F: Chromosome Mapping of H1 Histone and 5S rRNA Gene Clusters in Three Species of Astyanax (Teleostei, Characiformes). Cytogenet Genome Res. 2011, 134: 64-71. 10.1159/000323512.
Eirín-López JM, Ruiz MF, González-Tizón AM, Martínez A, Sánchez L, Méndez J: Molecular evolutionary characterization of the mussel Mytilus histone multigene family: First record of a tandemly repeated unit of five histone genes containing an H1 subtype with "orphon" features. J Mol Evol. 2004, 58: 131-144. 10.1007/s00239-003-2531-5.
Andrews MT, Vaughn JC, Perry BA, Bagshaw JC: Interspersion of histone and 5S RNA genes in Artemia. Gene. 1987, 51: 61-67. 10.1016/0378-1119(87)90474-4.
Drouin G, Moniz de Sá M: The concerted evolution of 5S ribosomal genes linked to the repeated units of other multigene families. Mol Biol Evol. 1995, 12: 481-493.
Barzotti R, Pelliccia F, Bucciarelli E, Rocchi A: Organization, nucleotide sequence, and chromosomal mapping of a tandemly repeated unit containing the four core histone genes and a 5S rRNA gene in an isopod crustacean species. Genome. 2000, 43: 341-345. 10.1139/g99-142.
Novotná J, Havelka J, Starý P, Koutecký P, Vítková M: Karyotype analysis of the Russian wheat aphid, Diuraphis noxia (Kurdjumov) (Hemiptera: Aphididae) reveals a large × chromosome with rRNA and histone gene families. Genetica. 2011, 139: 281-289. 10.1007/s10709-011-9546-4.
Roehrdanz R, Heilmann L, Senechal P, Sears S, Evenson P: Histone and ribosomal RNA repetitive gene clusters of the boll weevil are linked in a tandem array. Insect Mol Biol. 2010, 19: 463-471.
Martins C, Ferreira IA, Oliveira C, Foresti F, Galetti PM: A tandemly repetitive centromeric DNA sequence of the fish Hoplias malabaricus (Characiformes: Erythrinidae) is derived from 5S rDNA. Genetica. 2006, 127: 133-141. 10.1007/s10709-005-2674-y.
da Silva M, Matoso DA, Vicari MR, Almeida MC, Margarido VP, Artoni RF: Physical Mapping of 5S rDNA in Two Species of Knifefishes: Gymnotus pantanal and Gymnotus paraguensis (Gymnotiformes). Cytogenet Genome Res. 2011,
Sola L, De Innocentiis S, Gornung E, Papalia S, Rossi AR, Marino G, De Marco P, Cataudella S: Cytogenetic analysis of Epinephelus marginatus (Pisces: Serranidae), with the chromosome localization of the 18S and 5S rRNA genes and of the (TTAGGG) n telomeric sequence. Mar Biol. 2000, 137: 47-51. 10.1007/s002270000334.
Martins C, Galetti PM: Two rDNA arrays in Neotropical fish species: is it a general rule for fishes?. Genetica. 2001, 111: 439-446. 10.1023/A:1013799516717.
Pisano E, Ghigliotti L: Ribosomal genes in notothenioid fishes: Focus on the chromosomal organization. Mar Genomics. 2009, 2: 75-80. 10.1016/j.margen.2009.03.006.
Vitturi R, Sineo L, Volpe N, Lannino A, Colomba M: Repetitive DNAs in the slug Milax nigricans: association of ribosomal (18S-28S and 5S rDNA) and (TTAGGG) n telomeric sequences in the slug M. nigricans (Mollusca: Gastropoda: Pulmonata). Micron. 2004, 35: 255-260. 10.1016/j.micron.2003.11.011.
Wang Y, Guo X: Chromosomal rearrangement in Pectinidae revealed by rRNA loci and implications for bivalve evolution. Biol Bull. 2004, 207: 247-256. 10.2307/1543213.
Dover GA: Linkage disequilibrium and molecular drive in the rDNA gene family. Genetics. 1986, 122: 249-252.
Liu WS, Fredga K: Telomeric (TTAGGG) n sequences are associated with nucleolus organizer regions (NORs) in the wood lemming. Chromosome Res. 1999, 7: 235-240. 10.1023/A:1009255517764.
Kaplan FS, Murray J, Sylvester JE, Gonzalez IL, O'Connor JP, Doering JL, Muenke M, Emanuel BS, Zasloff MA: The topographic organization of repetitive DNA in the human nucleolus. Genomics. 1993, 15: 123-132. 10.1006/geno.1993.1020.
Sumner AT: A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res. 1972, 75: 304-306. 10.1016/0014-4827(72)90558-7.
Cabral-de-Mello DC, Moura RC, Martins C: Chromosomal mapping of repetitive DNAs in the beetle Dichotomius geminatus provides the first evidence for an association of 5S rRNA and histone H3 genes in insects, and repetitive DNA similarity between the B chromosome and A complement. Heredity. 2010, 104: 393-400. 10.1038/hdy.2009.126.
Acknowledgements
The authors are grateful to Fernando Silva and Cristiane Costa for taxonomic identification of the species studied. The study was supported by the Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP), the Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and the Fundação de Amparo a Pesquisa do Estado de Pernambuco (FACEPE).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
DCCM, CM and RCM designated and coordinated the study and drafted and revised the manuscript. DCCM and SGO carried out the experimental analysis. All authors read and approved the final manuscript.
Electronic supplementary material
12863_2011_949_MOESM1_ESM.JPEG
Additional file 1: Fluorescence in situ hybridization using as probes 18S and 5S rDNAs and histone H3 in initial meiotic and interphasic nucleus of Scarabaeinae beetles species. Initial meiotic cells (a-e, h, i) and interphasic nuclei (f, g) hybridized with probes for the 18S (green) and 5S rDNA (red) genes (a-f) and the histone H3 (green) and 5S rRNA (red) genes (g-i). (a) Deltochilum elevatum, (b) Deltochilum calcaratum, (c) Dichotomius crinicollis, (d) Coprophanaeus cyanescens, (e, f) Diabroctis mimas, (g) Dichotomius bos, (h) Dichotomius laevicollis and (i) Deltochilum verruciferum. Note the separation of the 18S and 5S rDNA signals in (a-e). In (f), two small signals for 18S and 5S rDNA overlap, and note the overlapped configuration of 5S rRNA and histone H3 genes in (g-i). Also note the formation of the chromocenter by heterochromatic sequences (a, b, d, e and i). A scale bar is not shown. (JPEG 1 MB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Cabral-de-Mello, D.C., Oliveira, S.G., de Moura, R.C. et al. Chromosomal organization of the 18S and 5S rRNAs and histone H3 genes in Scarabaeinae coleopterans: insights into the evolutionary dynamics of multigene families and heterochromatin. BMC Genet 12, 88 (2011). https://doi.org/10.1186/1471-2156-12-88
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2156-12-88