Abstract
The application of radiogenomics in oncology has great prospects in precision medicine. Radiogenomics combines large volumes of radiomic features from medical digital images, genetic data from high-throughput sequencing, and clinical-epidemiological data into mathematical modelling. The amalgamation of radiomics and genomics provides an approach to better study the molecular mechanism of tumour pathogenesis, as well as new evidence-supporting strategies to identify the characteristics of cancer patients, make clinical decisions by predicting prognosis, and improve the development of individualized treatment guidance. In this review, we summarized recent research on radiogenomics applications in solid cancers and presented the challenges impeding the adoption of radiomics in clinical practice. More standard guidelines are required to normalize radiomics into reproducible and convincible analyses and develop it as a mature field.
Similar content being viewed by others
Background
Currently, tumour tissue biopsies are the protocol for cancer diagnosis [1]. However, the biopsy method has several significant limitations: it is high-risk, highly invasive, expensive, and difficult to perform in serial tests; it has technical limitations resulting from the tumour location and difficulty in extracting cellular subpopulations; and it is unavailable when tumour cells disseminate to other organs and tissues [2]. These limitations restrict the application of tissue biopsy in cancer screening, diagnosis, treatment, and follow-up.
Fortunately, radiomics, a medical imaging method, holds great promise to address the problem of spatially and temporally heterogeneous solid cancers in a noninvasive way. Radiomics was first presented by Lambin et al. in [3] and involves the high-throughput extraction of image features from large numbers of medical images. Radiomics hypothesizes that additional information not seen by the naked eye could be captured via advanced feature analysis from medical imaging. In detail, genomic and proteomic expression profiles can probably be visualized and expressed according to quantitative analysis of image-based features.
With the success of the Human Genome Project (HGP), the application of genomics in medicine shows strong feasibility and prospects, which has brought an anticipated transformation in the prevention, diagnosis, and treatment of diseases. Projects such as The Cancer Genome Atlas (TCGA) have witnessed tremendous strides towards comprehensively cataloguing human genes and mutations. Some specific ones were found to be capable of serving essentially as ‘drivers’ of oncogenesis, primarily via exome or genome sequencing of large numbers of tumour samples matched with normal tissues [4, 5]. According to the definition from the National Human Genome Research Institute, genomic medicine is an emerging medical discipline in which an individual’s genomic information is taken into account in their clinical care (for example, for diagnostic or therapeutic decision-making, as well as the health effects and policy ramifications of that clinical care) [6]. Radiogenomics is an approach that combines large volumes of radiomic features from medical digital images, genetic data from high-throughput sequencing, and clinical-epidemiological data into mathematical modelling [7]. The amalgamation of radiomics and genomics provides an approach to better study the molecular mechanism of tumour pathogenesis and develop new evidence-supporting strategies. It is beneficial to stratify patients, identify the characteristics of cancer patients, make clinical decisions, and guide treatment by predicting prognosis and assessing clinical outcomes, thus improving the development of precision medicine [8].
Process of radiomics
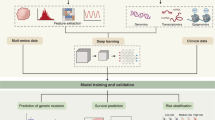
Generally, the radiomics workflow consists of the following main steps: imaging data collection, imaging preprocessing, identification and segmentation of the region or volume of interest, feature extraction, feature selection, model establishment, and model validation [9, 10].
Data collection
The original medical images are necessary for performing radiomics analysis. Computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET) have been extensively applied in the field of oncology to assist diagnosis and guide treatment. Images can provide detailed information about tumours’ anatomical and functional features. Medical imaging technology has made tremendous progress in the past, enabling the emergence of radiomics as a promising methodology for solving the complex problems of oncology. The development of medical imaging devices has moved from single-slice CT to multi-slice CT, which allows dynamic radiomics at multiple time points. In addition, dual-energy CT has been explored to improve the identification of tissue composition and density [3]. Diffusion-weighted MRI can reflect tumour density and cellularity, thereby having the capacity to monitor the response to cytotoxic treatment [11].
Imaging preprocessing
The preprocessing step is crucial to obtain medical images with preferred quality for subsequent analysis [12]. Imaging preprocessing steps generally include image normalization, denoising, bias field correction, imaging interpolation and resampling, motion correction, and imaging thresholding [13].
Image normalization is the process of changing the range of pixel intensity values, which is the foundation of medical image analysis. Conventional normalization approaches are generally used per dataset, such as generalized scale normalization, histogram normalization, and spatial normalization [14, 15]. Recent research proposed a joint normalization function across multiple datasets, which provided realism to the normalized images and improved image segmentation [16]. The crucial task of image denoising is keeping the most significant features of the images while simultaneously removing the nonimportant features. Classical denoising methods involve spatial domain filtering and variational denoising methods. The present transform domain methods, such as Fourier transform, cosine transform, wavelet domain methods [17], and sparse 3D filtering [18], were developed from the initial spatial domain methods [19]. Image resampling is divided into upsampling and downsampling. According to the Image Biomarker Standardization Initiative (IBSI), data from different modalities may fit different methods for image interpolation [20]. Motion correction is an approach to remove the motion artefacts caused by uncertain motion, which is also a vital preprocessing step to obtain reconstructed images with significantly improved quality [21].
Imaging segmentation
Three primary approaches are used to segment the region of interest (ROI) in two dimensions or the volume of interest (VOI) in three dimensions: manual, semiautomatic, and fully automatic. In comparing these approaches, semiautomatic segmentation is considered optimal [22]. ROIs/VOIs determine the region where radiomic features are extracted and calculated [23]. ROI/VOI segmentation through manual delineation is common in previous research and does not require the use of any sophisticated postprocessing methods or software. However, it is time-consuming and thus hard to use when analysing massive imaging data [24]. In addition, human delineation presents an inevitable bias resulting from different observers that cannot be ignored, leading to a lack of robustness due to intra- and interobserver variations [25]. Semiautomatic delineation uses computer algorithms to segment the ROIs/VOIs but usually needs to be corrected and calibrated manually [26]. Some primary open-source or commercial software could be applied to conduct semiautomatic segmentation, such as 3D Slicer [27], ITK-SNAP [28], LIFE [29], MITK [30], and ImageJ [31]. Fully automatic segmentation is based on the approach of deep learning using artificial neural networks. Deep learning has been universally capitalized in many fields associated with automated image recognition. Convolutional neural networks (CNNs), one of the deep neural networks designed for various computer vision tasks, have been successfully applied in imaging recognition and classification [32, 33]. CNN builds up the mapping directly from the input data and output labels, utilizing a hierarchical network to learn abstract features [34]. The advantages of CNN in radiomics are automatic feature extraction and accurate classification performance [35]. However, this method often requires a large sample size of medical images for deep learning. It is suitable when there is a substantial signal difference between the cancer region and the normal tissues [36].
Feature extraction
Quantitative radiomics features extracted from segmented ROIs/VOIs can be divided into several major categories, including shape, statistical, and texture features [37]. Shape features mainly include elongation, flatness, sphericity, surface area, surface volume, and voxel volume [38]. Statistical features are the first-order features obtained from a grey-level histogram, which presents the distribution of grey-level intensity values and reveals the tumour’s overall heterogeneity without considering spatial information. Texture features are second-order features that represent the spatial relationships between the voxels from matrices, including the grey-level run-length matrix (GLRLM), grey-level size zone matrix (GLSZM), grey-level co-occurrence matrix (GLCM), neighbourhood grey-tone difference matrix (NGTDM), size of homogeneous greyscale areas, length of greyscale ranges, and greyscale dependence. Feature extraction is commonly conducted using radiomic tools, such as PyRadiomics [39, 40], IBEX [41], MIRP [42], RaCaT [43], and RadiomiCRO [44]. These platforms are based on different languages of Python or MATLAB. However, the lack of standardization in the definition and calculation of radiomic features among these tools has hindered the application of radiomics in clinical practice. The IBSI addressed some of the questions and was thus recommended during feature extraction [45]. The IBSI is composed of a set of 169 standardized radiomics features, which could be verified and calibrated across different radiomics software. It provided standardized image biomarker nomenclature and definitions. Some new tools tend to conform to the IBSI, such as S-IBEX, an adaptation of IBEX to IBSI guidelines [46]. Recent research proposed a novel approach for evaluating the agreement among seven radiomics tools. They found that discrepancies still existed among standardized radiomic programs. Therefore, further efforts are needed to accelerate the use of radiomic models in clinical practice [47].
Analysis and modelling
The core of the radiomics approach is to establish a predictive model. The modelling process is based on the training data analysis. It is indispensable to apply the model on testing data to evaluate its extrapolation performance, proving the model's precision, fit, and feasibility. Generally, hundreds of radiomics features can be extracted from images. Therefore, feature selection needs to be performed to identify the most valuable traits to avoid overfitting, since the incorporation of redundant features into a model will degrade the performance of model extrapolation. Least absolute shrinkage and selection operator (LASSO) plays a prominent role in selecting informative features [48]. Principal component analysis (PCA) is also one of the preferable methods to reduce the dimensions of radiomics features but preserve their informative content [49]. Machine learning (ML) is a branch of artificial intelligence (AI) in which a machine is trained to learn from data without being explicitly programmed and can make decisions with minimal human intervention [50]. A variety of machine learning approaches are used for constructing radiomics models, such as support vector machine (SVM), random forest (RF), k-nearest neighbours (KNN), decision tree (DT), conditional inference trees (CIT), Bayes and back-propagation neural network (BPNet) [51]. One of the most technical steps is hyperparameter tuning and optimization for the learning algorithm to improve the performance of ML models [52]. The common approaches to tuning the hyperparameters [53] of the models included grid search, repeated stratified k-fold cross-validation, genetic algorithms (GAs), random search [54], sequential search, the Gaussian process approach (GP), tree-structured Parzen estimator (TPE), and Bayesian optimization [55, 56]. Recent research reported that model-based optimization (MBO) could be an excellent tuning strategy for RF. The R package tuneRanger was used to automatically tune RF with MBO [57]. Another study developed a metaheuristic algorithm using grey wolf optimization (GWO) and GA to tune the hyperparameters of ML algorithms and neural networks [58]. Notably, the metaheuristic methods displayed better and faster performance than other algorithms and are appropriate for datasets with unknown distributions. Traditional statistical methods, specifically univariate and multivariate analyses, are used to identify the statistically significant variables and radiomics signatures associated with the outcome [59]. Afterwards, logistic regression and Cox proportional hazards regression are often used to build a radiomics-clinical predictive signature that incorporates clinical risk factors and radiomics features, providing a basis for diagnosis, clinical treatment options, and prognostic prediction [60, 61].
In addition, deep learning provides a non-engineered radiomics modelling methods and has been applied in various medical image analysis tasks, including supervised learning, unsupervised learning, and semi-supervised learning [62]. Models are trained on labelled images in supervised learning, such as CNNs. In contrast, models in unsupervised learning analyse the underlying information in the images without any labels, such as autoencoders (AE) [63] and generative adversarial networks (GANs) [64]. In the semi-supervised learning, only a part of the training datasets is labelled, and the model will perform better through learning the unlabelled images [65]. Deep learning models based on artificial neural networks such as CNNs can be established to classify target lesions delineated in medical images, such as the discrimination of tumours as benign or malignant. The processes of data learning and classification are performed jointly in the deep learning approach [33]. Existing structures such as VGG [66], Resnet [67] or self-designed networks can be used for CNN. Transfer learning can be performed by tuning hyperparameter depending on the data size, providing a convenient approach to apply the existing deep learning algorithms.
The last step of the radiomics approach is assessing performance and external validation based on other datasets to prove the credibility of the established model. The performance can be evaluated using the receiver operating characteristic (ROC) curve, the area under the ROC curve (AUC), and the C-index to assess the discrimination of the model, and calibration curves can be used to appraise the calibration in the training and validation sets. The model's potential clinical value can be estimated by using decision curve analysis (DCA) [68]. In addition, resampling methods (for instance, cross-validation and bootstrapping) can also be applied to evaluate model performance [69].
Current applications of radiogenomics in cancer
The literature on radiogenomics has greatly expanded in the past decade. Radiogenomics has been applied in various types of cancer, including lung cancer [70,71,72], breast cancer [73,74,75], genitourinary cancer [76,77,78], digestive system cancer [79,80,81], and brain cancer [82,83,84]. The comprehensive analysis method of radiogenomics can be used to diagnose and predict patient outcomes such as recurrence, metastasis, and survival of patients with cancer; predict the tumour microenvironment; identify potential molecular biomarkers and targets for clinical diagnosis and treatment; and establish a molecular typing system of variable cancers. A selection of papers was compiled for illustrative purposes and the characteristics of the representative papers are displayed in Table 1.
Diagnostic value
Radiomics predictive models have diagnostic value capable of assisting clinicians in making decisions based on big data evidence. Chen et al. [85] established models to identify clear cell renal cell carcinomas (ccRCCs) and non-clear cell renal cell carcinomas (non-ccRCCs). The combined diagnostic model incorporated texture features extracted from CT images and other non-texture features. They found that the model significantly improved the predictive efficacy of ccRCC, with the AUC values for differentiating the two groups ranging from 0.864–0.900. The results showed a good capability for discriminating ccRCCs from non-ccRCCs. An integrative model combining imaging features and genomic information provides an insightful biological explanation from the molecular perspective for lesions on medical images directly visible to the naked eye. Smedley et al. [86] explored deep feedforward neural networks to identify gene expression profiles in an interpretable way, being capable of anticipating the quantitative radiomic features of CT and histological types (adenocarcinoma, squamous cell, and other) of non-small cell lung cancer (NSCLC). The study enrolled 262 and 89 patients from two public databases for training and testing, respectively. The results suggested that neural networks performed better than other classifiers, classifying histopathological types with AUCs of 0.86 (adenocarcinoma), 0.91 (squamous cell), and 0.71 (other) in the testing cohort. The AUCs for the classification performance of radiomics features ranged from 0.42 to 0.89. Gene sets of cardiac, immune system, and cell development processes were predictive (AUC > 0.70) of certain dissimilar radiomic features. In contrast, tumour necrosis factor (TNF), AKT signalling, and Rho gene sets could forecast the features of tumour textures. The research demonstrated that neural networks could be utilized to map the expression of genes to radiomic characteristics and further to histopathological types in NSCLC, which could explain how the models could recognize predictable genes related to various imaging features or histological types.
Predicting the patient outcome
Recurrence
Cancers can be devastating on account of the high recurrence rate. Recently, Shim et al. [87] proposed two different radiomics-based models through a neural network to predict the relapse pattern in glioblastoma, namely, local and distant recurrences. They used high-dimensional radiomics profiles based on dynamic susceptibility contrast-enhanced perfusion MRI. The AUCs were 0.969 (95% confidence interval: 0.903–1.000) for local recurrence and 0.864 (95% confidence interval: 0.726–0.976) for distant recurrence for patients in the validation cohort. Another recent study conducted by Kirienko et al. [88] evaluated the correlations of information from radiogenomics analyses with histotype and patient outcome in NSCLC using an ML algorithm. The results showed that two radiomics features, standardized uptake value and kurtosis, and the expression levels of TP63, EPHA10, FBN2, and IL1RAP were associated with the histology of NSCLC. They also identified robust PET radiomic features and gene expression profiles capable of predicting recurrence (with AUC = 0.87). Radiogenomics analysis may provide valuable evidence for predicting the histological type, invasiveness, and progression when making clinical decisions for patients with NSCLC.
Survival
Based on a radiomics cohort of medulloblastoma (MB) patients with MRI data, Yan et al. [89] constructed a radiomics signature combining the most prognostic radiomics features identified for survival prediction in a training cohort including 83 samples. Another radiogenomics cohort with matched MRI images and RNA sequencing (RNA-seq) data was used to identify nine vital biological pathways significantly related to radiomics characteristics. They also assessed the prognostic prediction value of the genes involved in the identified pathways based on an open-access dataset with RNA-seq and survival data on MB. The results indicated that the signature combining radiomics and clinicomolecular factors predicted overall survival (OS) (C-index of 0.762) and progression-free survival (PFS) (C-index of 0.697) more accurately than the separate radiomics model (C-indexes of 0.649 for OS and 0.593 for PFS) or the clinicomolecular model (C-indexes of 0.725 for OS and 0.691 for PFS). The research demonstrated that the radiomics signature was an independent predictive factor for survival in MB patients. The signature was related to dysregulated pathways and provided additive value over the clinicomolecular factors model. In a recent study of oesophageal squamous cell carcinoma (ESCC), Xie et al. [81] concluded that genomics association was advantageous for selecting meaningful radiomic features to establish CT-based radiomic signatures. This could contribute to predicting prognosis in terms of disease-free survival (DFS), particularly individualized long-term survival. The study involved 106 patients with ESCC who received neoadjuvant chemoradiation (nCRT) at two institutions. Radiomic features were selected according to their correlation with differentially expressed genes (DEGs) identified between relapsed and non-relapsed patients and the appended ML approach. A radiomic nomogram integrating the radiomic characteristics with prognostic clinical factors was established for DFS prediction, which significantly stratified patients into two groups, with high risk and low risk for DFS (p < 0.001) (AUCs for predicting 5-year DFS of training set: 0.912, internal test set: 0.852, and external test set: 0.769). The results showed that the radiomic prediction model utilizing genomics-assisted feature selection performed better in DFS prediction.
Contrast-enhanced CT is also a critical imaging examination method in clinical applications. Huang et al. [90] enrolled 205 patients with ccRCC with accessible contrast-enhanced CT images from The Cancer Imaging Archive (TCIA) database and matched transcriptomic data obtained from the TCGA database. They conducted research aiming to incorporate radiomic features extracted from medical images and corresponding genomics information to predict the OS of ccRCC patients. In the results, four prognosis-related imaging features (PRIFs) were selected from 107 extracted radiomics features by performing LASSO Cox regression and SVM-RFE. Four prognosis-related genes were identified through weighted gene coexpression network analysis (WGCNA). A mixed imaging-genomics prognostic factor (IGPF) signature was established by the RF algorithm, which displayed an enhanced prediction performance compared with the PRIF model alone (with average AUCs in the test dataset for 1-, 3-, and 5-year survival of 0.84 vs. 0.81, 0.81 vs. 0.74, and 0.75 vs. 0.68). The results suggested that the overall prognosis assessment may be credited to the involvement of imaging-genomics prognostic factors for ccRCC patients.
Metastasis
To assess the ability of a model constructed by integrating radiomics, genomics, and clinical features to predict the metastasis of colorectal cancer (CRC) patients, Liu et al. [80] retrospectively analysed 134 patients (primary cohort: 62, validation set: 28, independent test set: 44) clinicopathologically diagnosed with CRC and developed a multiscale preoperative model through multivariable logistic regression analysis for metastasis prediction, incorporating the above three types of features. Sixteen radiomics features and the expression levels of four identified genes were included in the multiscale nomogram model, which exhibited outstanding prediction performance, with AUCs of 0.981 in the primary dataset (95% CI 0.953–1.000), 0.822 in the validation dataset (95% CI 0.635–1.000), and 0.752 in the independent test dataset (95% CI 0.608–0.896), showing great potential to be applied clinically to assist in individualized preoperative metastasis evaluation in patients with CRC.
Predicting treatment responses
It is difficult to predict patient response to treatment on account of the heterogeneity of solid tumours, the absence of consistent and accurate biomarkers, and a vague understanding of resistance mechanisms. The approach of radiogenomics has shed light on addressing these challenges through the untapped and abundant resource of imaging data [91]. Aiming to predict the response to chemotherapy and metastasis probability, Kim et al. [92] recruited 73 paediatric osteosarcoma patients and constructed a prediction model using an ML algorithm. The model combined the gene expression data of KI67 and EZRIN with image texture features extracted from fluorodeoxyglucose positron emission tomography/computed tomography (18F-FDG PET/CT) images. The results showed that the best test accuracy and AUC for predicting chemotherapy response with KI67 and EZRIN were 0.85 and 0.89, respectively. In recent research for predicting platinum resistance of ovarian cancer (OC), Yi et al. [93] generated an ML model incorporating radiomics data based on pretreatment CT images, clinicopathological data, and genomic data of single-nucleotide polymorphisms (SNPs) of human sulfatase 1 (SULF1). This combined model showed better classification efficiency, high calibration, and promising clinical utility, with AUC values of 0.993 (95% CI 0.83–0.98) in the training dataset (n = 71) and 0.967 (95% CI 0.83–0.98) in the validation dataset (n = 31).
Predicting molecular characteristics
Through the combination of genomics data and CT radiomics features, it is likely feasible to predict the molecular characteristics of patients with cancer. In research on ccRCC, Zeng et al. [77] applied ML algorithms to predict gene mutations and different mRNA-based molecular subtypes according to radiomics features. The researchers collected the gene expression information of 207 ccRCC patients from TCGA and corresponding contrast-enhanced CT images from TCIA. The results proved excellent performance in detecting mutations of VHL (AUC: 0.971), PBRM1 (AUC: 0.972), BAP1 (AUC: 0.955), and SETD2 (AUC: 0.949), as well as the molecular subtypes, specifically m1 (AUC: 0.973), m2 (AUC: 0.968), m3 (AUC: 0.961), and m4 (AUC: 0.953), which were validated externally in another population containing 175 patients with ccRCC from West China Hospital. The study illustrated that integrative analysis of radiogenomics might be a practical approach to predict gene mutations or molecular characteristics in ccRCC patients, indicating a fast and noninvasive alternative to genetic testing.
Another multi-institutional retrospective study conducted by Park et al. [94] showed that preoperative MR imaging features may identify isocitrate dehydrogenase (IDH) wild-type lower-grade gliomas that have consistent molecular features as glioblastoma. The molecular features specifically included amplification of epidermal growth factor receptor (EGFR) or telomerase reverse transcriptase (TERT) promoter mutation. The research enrolled 64 patients in the training cohort and 57 patients in the validation cohort who were clinicopathologically diagnosed with IDH wild-type lower-grade gliomas. In the external test dataset, a model integrating both radiomic features and Visually AcceSAble Rembrandt Images revealed superior predictive performance (AUC of 0.854) than either that merely with clinical factors or Visually AcceSAble Rembrandt Images (AUCs of 0.514 and 0.648, respectively; P < 0.001, both). The predictive model showed the best performance when clinical features were added (AUCs of 0.514 vs. 0.863, P < 0.001).
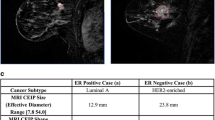
Radiomics analysis is also capable of helping with the establishment of a molecular typing system. By performing quantitative radiomics analysis, Li et al. [95] demonstrated that MR image-based tumour phenotypes could predict the molecular classification of invasive breast cancers. The results revealed that the tumour phenotypes extracted by the computer allowed the distinction between molecular prognostic indicators, with AUC values of 0.89, 0.69, 0.65, and 0.67 when distinguishing ER + vs. ER − , PR + vs. PR − , HER2 + vs. HER2 − , and triple-negative vs. others, respectively.
Challenges and future development
Despite the great potential shown by radiomics in clinical guidance, no developed radiomics signature is currently applied in the clinical field. There are still many challenges regarding the application of radiomics. A significant variation exists in every step during the whole analysis process derived from different sources, leading to the poor generalizability and reproducibility of the radiomics signatures previously published [96, 97]. For instance, the various settings in each stage of image acquisition, such as scanners, scanning techniques, and reconstruction parameters, might affect the extracted radiomics features. In recent research on renal clear cell carcinoma, Lu et al. [98] asserted that multiple uncontrolled confounders, including feature redundancy and image acquisition settings, especially tumour size and slice thickness, may bring about false or overvalued radiomics signatures. Considering the urgent need to standardize the radiomics process and accelerate its translation to clinical application, Lambin et al. [9] proposed the radiomics quality score (RQS) composed of sixteen criteria to evaluate both past and future radiomics studies, including well-documented image protocols that allow reproducibility or replicability, multiple segmentations, feature reduction or adjustment for multiple testing to decrease the overfitting risk, reporting discrimination statistics and calibration statistics, performing validation without retraining and adaptation of the cut-off value, etc. In summary, more standard guidelines are needed to normalize radiomics into reproducible and convincible analyses and to develop it as a mature field.
Conclusions
The application of radiogenomics in oncology shows great prospects in precision medicine. The methodology of radiogenomics integrates large volumes of quantitative yet significant imaging phenotypes extracted from medical digital images and tumour genetic profiles derived from high-throughput sequencing, together with clinical-epidemiological data, into mathematical modelling. The amalgamation of radiomics and genomics provides an approach to better study the molecular mechanism of tumour pathogenesis, as well as new evidence-supporting strategies to identify the characteristics of cancer patients, make clinical decisions by predicting prognosis, and improve the development of individualized treatment guidance. However, many challenges hinder the application of radiomics in real clinical situations, such as the poor generalizability and reproducibility of the previously published radiomics models. Therefore, more standard guidelines are needed in future research to normalize radiomics into reproducible and convincible analysis and develop it as a mature discipline.
Availability of data and materials
Not applicable.
Abbreviations
- HGP:
-
Human Genome Project
- CT:
-
Computed tomography
- MRI:
-
Magnetic resonance imaging
- PET:
-
Positron emission tomography
- ROI:
-
Region of interest
- VOI:
-
Volume of interest
- CNN:
-
Convolutional neural network
- LASSO:
-
Least absolute shrinkage and selection operator
- PCA:
-
Principal component analysis
- GLM:
-
Generalized linear model
- ML:
-
Machine learning
- RF:
-
Random forest
- LR:
-
Logistic regression
- SVM:
-
Support vector machine
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the curve
- DCA:
-
Decision curve analysis
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
- DFS:
-
Disease-free survival
- ccRCC:
-
Clear cell renal cell carcinoma
- NSCLC:
-
Non-small cell lung cancer
- MB:
-
Medulloblastoma
- ESCC:
-
Oesophageal squamous cell carcinoma
- CRC:
-
Colorectal cancer
- OC:
-
Ovarian cancer
- BC:
-
Breast cancer
References
Fernández-Lázaro D, Hernández JLG, García AC, Castillo ACD, Hueso MV, Cruz-Hernández JJ. Clinical perspective and translational oncology of liquid biopsy. Diagnostics (Basel). 2020;10(7):E443.
Mazouji O, Ouhajjou A, Incitti R, Mansour H. Updates on clinical use of liquid biopsy in colorectal cancer screening, diagnosis, follow-up, and treatment guidance. Front Cell Dev Biol. 2021;9: 660924.
Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RGPM, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441–6.
Shendure J, Findlay GM, Snyder MW. Genomic medicine-progress, pitfalls, and promise. Cell. 2019;177(1):45–57.
Singh A, Chitalia R, Kontos D. Radiogenomics in brain, breast, and lung cancer: opportunities and challenges. J Med Imaging (Bellingham). 2021;8(3): 031907.
Genomics and Medicine [Internet]. Genome.gov. [cited 2022 Aug 30]. https://www.genome.gov/health/Genomics-and-Medicine
Shui L, Ren H, Yang X, Li J, Chen Z, Yi C, et al. The era of radiogenomics in precision medicine: an emerging approach to support diagnosis, treatment decisions, and prognostication in oncology. Front Oncol. 2020;10: 570465.
Bodalal Z, Trebeschi S, Nguyen-Kim TDL, Schats W, Beets-Tan R. Radiogenomics: bridging imaging and genomics. Abdom Radiol (NY). 2019;44(6):1960–84.
Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–62.
Pinker K, Chin J, Melsaether AN, Morris EA, Moy L. Precision medicine and radiogenomics in breast cancer: new approaches toward diagnosis and treatment. Radiology. 2018;287(3):732–47.
Hsu JBK, Lee GA, Chang TH, Huang SW, Le NQK, Chen YC, et al. Radiomic immunophenotyping of GSEA-assessed immunophenotypes of glioblastoma and its implications for prognosis: a feasibility study. Cancers (Basel). 2020;12(10):E3039.
Smits M. MRI biomarkers in neuro-oncology. Nat Rev Neurol. 2021;17(8):486–500.
Shur JD, Doran SJ, Kumar S, ap Dafydd D, Downey K, O’Connor JPB, et al. Radiomics in oncology: a practical guide. Radiographics. 2021;41(6):1717–32.
Isaksson LJ, Raimondi S, Botta F, Pepa M, Gugliandolo SG, De Angelis SP, et al. Effects of MRI image normalization techniques in prostate cancer radiomics. Phys Med. 2020;71:7–13.
He H, Razlighi QR. Landmark-guided region-based spatial normalization for functional magnetic resonance imaging. Hum Brain Mapp. 2022;43(11):3524–44.
Delisle PL, Anctil-Robitaille B, Desrosiers C, Lombaert H. Realistic image normalization for multi-Domain segmentation. Med Image Anal. 2021;74: 102191.
Pizurica A, Philips W, Lemahieu I, Acheroy M. A versatile wavelet domain noise filtration technique for medical imaging. IEEE Trans Med Imaging. 2003;22(3):323–31.
Dabov K, Foi A, Katkovnik V, Egiazarian K. Image denoising by sparse 3-D transform-domain collaborative filtering. IEEE Trans Image Process. 2007;16(8):2080–95.
Fan L, Zhang F, Fan H, Zhang C. Brief review of image denoising techniques. Vis Comput Ind Biomed Art. 2019;2(1):7.
Zwanenburg A, Vallières M, Abdalah MA, Aerts HJWL, Andrearczyk V, Apte A, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295(2):328–38.
Jang S, Kim S, Kim M, Son K, Lee KY, Ra JB. Head motion correction based on filtered backprojection in helical CT scanning. IEEE Trans Med Imaging. 2020;39(5):1636–45.
El Ayachy R, Giraud N, Giraud P, Durdux C, Giraud P, Burgun A, et al. The role of radiomics in lung cancer: from screening to treatment and follow-up. Front Oncol. 2021;11: 603595.
van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging-"how-to" guide and critical reflection. Insights Imaging. 2020;11(1):91.
Kuo MD, Jamshidi N. Behind the numbers: decoding molecular phenotypes with radiogenomics–guiding principles and technical considerations. Radiology. 2014;270(2):320–5.
Baeßler B, Weiss K, Pinto Dos Santos D. Robustness and reproducibility of radiomics in magnetic resonance imaging: a phantom study. Invest Radiol. 2019;54(4):221–8.
Xie CY, Pang CL, Chan B, Wong EYY, Dou Q, Vardhanabhuti V. Machine learning and radiomics applications in esophageal cancers using non-invasive imaging methods-A critical review of literature. Cancers (Basel). 2021;13(10):2469.
Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, et al. 3D slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging. 2012;30(9):1323–41.
Yushkevich PA, Gao Y, Gerig G. ITK-SNAP: an interactive tool for semi-automatic segmentation of multi-modality biomedical images. Annu Int Conf IEEE Eng Med Biol Soc. 2016;2016:3342–5.
Nioche C, Orlhac F, Boughdad S, Reuzé S, Goya-Outi J, Robert C, et al. LIFEx: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res. 2018;78(16):4786–9.
Wolf I, Vetter M, Wegner I, Böttger T, Nolden M, Schöbinger M, et al. The medical imaging interaction toolkit. Med Image Anal. 2005;9(6):594–604.
Broeke J, Mateos Pérez JM, Pascau J. Image processing with ImageJ: extract and analyze data from complex images with ImageJ, the world’s leading image processing tool. Second edition. Birmingham Mumbai: Packt Publishing open source; 2015. 231 p. (Community experience distilled).
Binczyk F, Prazuch W, Bozek P, Polanska J. Radiomics and artificial intelligence in lung cancer screening. Transl Lung Cancer Res. 2021;10(2):1186–99.
Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging. 2018;9(4):611–29.
Zhang B, Qi S, Pan X, Li C, Yao Y, Qian W, et al. Deep CNN model using CT radiomics feature mapping recognizes EGFR gene mutation status of lung adenocarcinoma. Frontiers Oncol. 2021. https://doi.org/10.3389/fonc.2020.598721.
Zhou Y, Xu J, Liu Q, Li C, Liu Z, Wang M, et al. A radiomics approach with CNN for shear-wave elastography breast tumor classification. IEEE Trans Biomed Eng. 2018;65(9):1935–42.
Chaddad A, Kucharczyk MJ, Cheddad A, Clarke SE, Hassan L, Ding S, et al. Magnetic resonance imaging based radiomic models of prostate cancer: a narrative review. Cancers (Basel). 2021;13(3):552.
Arimura H, Soufi M, Kamezawa H, Ninomiya K, Yamada M. Radiomics with artificial intelligence for precision medicine in radiation therapy. J Radiat Res. 2019;60(1):150–7.
Bak SH, Park H, Lee HY, Kim Y, Kim HL, Jung SH, et al. Imaging genotyping of functional signaling pathways in lung squamous cell carcinoma using a radiomics approach. Sci Rep. 2018;8(1):3284.
van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104–7.
Mao B, Ma J, Duan S, Xia Y, Tao Y, Zhang L. Preoperative classification of primary and metastatic liver cancer via machine learning-based ultrasound radiomics. Eur Radiol. 2021;31(7):4576–86.
Zhang L, Fried DV, Fave XJ, Hunter LA, Yang J, Court LE. IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys. 2015;42(3):1341–53.
Zwanenburg A, Leger S, Agolli L, Pilz K, Troost EGC, Richter C, et al. Assessing robustness of radiomic features by image perturbation. Sci Rep. 2019;9(1):614.
Pfaehler E, Zwanenburg A, de Jong JR, Boellaard R. RaCaT: an open source and easy to use radiomics calculator tool. PLoS ONE. 2019;14(2): e0212223.
Avanzo M, Pirrone G, Vinante L, Caroli A, Stancanello J, Drigo A, et al. Electron density and biologically effective dose (BED) radiomics-based machine learning models to predict late radiation-induced subcutaneous fibrosis. Frontiers Oncol. 2020. https://doi.org/10.3389/fonc.2020.00490.
Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. Radiology. 2020;295(2):328–38.
Bettinelli A, Branchini M, De Monte F, Scaggion A, Paiusco M. Technical note: an IBEX adaption toward image biomarker standardization. Med Phys. 2020;47(3):1167–73.
Bettinelli A, Marturano F, Avanzo M, Loi E, Menghi E, Mezzenga E, et al. A novel benchmarking approach to assess the agreement among radiomic tools. Radiology. 2022;303(3):533–41.
Wu X, Li J, Mou Y, Yao Y, Cui J, Mao N, et al. Radiomics nomogram for identifying sub-1 cm benign and malignant thyroid lesions. Front Oncol. 2021;11: 580886.
Barabino E, Rossi G, Fedeli A, Cittadini G, Genova C. Radiomic-based diagnostics in oncology: challenges toward clinical practice. Oncoscience. 2021;8:72–3.
Mun SK, Wong KH, Lo SCB, Li Y, Bayarsaikhan S. Artificial intelligence for the future radiology diagnostic service. Front Mol Biosci. 2020;7: 614258.
Duan C, Liu F, Gao S, Zhao J, Niu L, Li N, et al. Comparison of radiomic models based on different machine learning methods for predicting intracerebral hemorrhage expansion. Clin Neuroradiol. 2022;32(1):215–23.
Valsecchi C, Consonni V, Todeschini R, Orlandi ME, Gosetti F, Ballabio D. Parsimonious optimization of multitask neural network hyperparameters. Molecules. 2021;26(23):7254.
Bergstra JS, Bardenet R, Bengio Y, Kégl B. Algorithms for hyper-parameter optimization:9.
Bergstra J, Bengio Y. Random Search for Hyper-Parameter Optimization:25.
Olisah CC, Smith L, Smith M. Diabetes mellitus prediction and diagnosis from a data preprocessing and machine learning perspective. Comput Methods Progr Biomed. 2022;220: 106773.
Martinez-Cantin R. Funneled Bayesian optimization for design, tuning and control of autonomous systems. IEEE Trans Cybern. 2019;49(4):1489–500.
Probst P, Wright MN, Boulesteix AL. Hyperparameters and tuning strategies for random forest. WIREs Data Min Knowl Discov. 2019;9(3): e1301.
Nematzadeh S, Kiani F, Torkamanian-Afshar M, Aydin N. Tuning hyperparameters of machine learning algorithms and deep neural networks using metaheuristics: a bioinformatics study on biomedical and biological cases. Comput Biol Chem. 2022;97: 107619.
Yang L, Gu D, Wei J, Yang C, Rao S, Wang W, et al. A radiomics nomogram for preoperative prediction of microvascular invasion in hepatocellular carcinoma. Liver Cancer. 2019;8(5):373–86.
Pei X, Wang P, Ren JL, Yin XP, Ma LY, Wang Y, et al. Comparison of different machine models based on contrast-enhanced computed tomography radiomic features to differentiate high from low grade clear cell renal cell carcinomas. Front Oncol. 2021;11: 659969.
Hong D, Zhang L, Xu K, Wan X, Guo Y. Prognostic value of Pre-treatment CT radiomics and clinical factors for the overall survival of advanced (IIIB-IV) lung adenocarcinoma patients. Front Oncol. 2021;11: 628982.
Avanzo M, Wei L, Stancanello J, Vallières M, Rao A, Morin O, et al. Machine and deep learning methods for radiomics. Med Phys. 2020;47(5):e185-202.
Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science. 2006;313(5786):504–7.
Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative Adversarial Networks [Internet]. arXiv; 2014 [cited 2022 Dec 16]. http://arxiv.org/abs/1406.2661
Chen X, Wang X, Zhang K, Fung KM, Thai TC, Moore K, et al. Recent advances and clinical applications of deep learning in medical image analysis. Med Image Anal. 2022;1(79): 102444.
Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition [Internet]. arXiv; 2015 [cited 2022 Dec 16]. http://arxiv.org/abs/1409.1556
He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) [Internet]. Las Vegas, NV, USA: IEEE; 2016 [cited 2022 Dec 16]. p. 770–8. http://ieeexplore.ieee.org/document/7780459/
Wu S, Zheng J, Li Y, Wu Z, Shi S, Huang M, et al. Development and validation of an MRI-based radiomics signature for the preoperative prediction of lymph node metastasis in bladder cancer. EBioMedicine. 2018;34:76–84.
Ligero M, Jordi-Ollero O, Bernatowicz K, Garcia-Ruiz A, Delgado-Muñoz E, Leiva D, et al. Minimizing acquisition-related radiomics variability by image resampling and batch effect correction to allow for large-scale data analysis. Eur Radiol. 2021;31(3):1460–70.
Xie D, Wang TT, Huang SJ, Deng JJ, Ren YJ, Yang Y, et al. Radiomics nomogram for prediction disease-free survival and adjuvant chemotherapy benefits in patients with resected stage I lung adenocarcinoma. Transl Lung Cancer Res. 2020;9(4):1112–23.
Ma DN, Gao XY, Dan YB, Zhang AN, Wang WJ, Yang G, et al. Evaluating solid lung adenocarcinoma anaplastic lymphoma kinase gene rearrangement using noninvasive radiomics biomarkers. Onco Targets Ther. 2020;13:6927–35.
Kim G, Kim J, Cha H, Park WY, Ahn JS, Ahn MJ, et al. Metabolic radiogenomics in lung cancer: associations between FDG PET image features and oncogenic signaling pathway alterations. Sci Rep. 2020;10(1):13231.
Arefan D, Hausler RM, Sumkin JH, Sun M, Wu S. Predicting cell invasion in breast tumor microenvironment from radiological imaging phenotypes. BMC Cancer. 2021;21(1):370.
Wu J, Mayer AT, Li R. Integrated imaging and molecular analysis to decipher tumor microenvironment in the era of immunotherapy. Semin Cancer Biol. 2022;84:310–28.
Fan M, Xia P, Clarke R, Wang Y, Li L. Radiogenomic signatures reveal multiscale intratumour heterogeneity associated with biological functions and survival in breast cancer. Nat Commun. 2020;11(1):4861.
Liu L, Yi X, Lu C, Pang Y, Zu X, Chen M, et al. Background, applications and challenges of radiogenomics in genitourinary tumor. Am J Cancer Res. 2021;11(5):1936–45.
Zeng H, Chen L, Wang M, Luo Y, Huang Y, Ma X. Integrative radiogenomics analysis for predicting molecular features and survival in clear cell renal cell carcinoma. Aging (Albany NY). 2021;13(7):9960–75.
Zhao Y, Liu G, Sun Q, Zhai G, Wu G, Li ZC. Validation of CT radiomics for prediction of distant metastasis after surgical resection in patients with clear cell renal cell carcinoma: exploring the underlying signaling pathways. Eur Radiol. 2021;31(7):5032–40.
Liu P, Tan XZ, Zhang T, Gu QB, Mao XH, Li YC, et al. Prediction of microvascular invasion in solitary hepatocellular carcinoma ≤ 5 cm based on computed tomography radiomics. World J Gastroenterol. 2021;27(17):2015–24.
Liu Q, Li J, Xu L, Wang J, Zeng Z, Fu J, et al. Individualized prediction of colorectal cancer metastasis using a radiogenomics approach. Front Oncol. 2021;11: 620945.
Xie CY, Hu YH, Ho JWK, Han LJ, Yang H, Wen J, et al. Using genomics feature selection method in radiomics pipeline improves prognostication performance in locally advanced esophageal squamous cell carcinoma-A pilot study. Cancers (Basel). 2021;13(9):2145.
Di Giannatale A, Di Paolo PL, Curione D, Lenkowicz J, Napolitano A, Secinaro A, et al. Radiogenomics prediction for MYCN amplification in neuroblastoma: a hypothesis generating study. Pediatr Blood Cancer. 2021;68(9): e29110.
Habib A, Jovanovich N, Hoppe M, Ak M, Mamindla P, R Colen R, et al. MRI-based radiomics and radiogenomics in the management of low-grade gliomas: evaluating the evidence for a paradigm shift. J Clin Med. 2021;10(7):1411.
Zheng S, Tao W. Identification of novel transcriptome signature as a potential prognostic biomarker for anti-angiogenic therapy in glioblastoma multiforme. Cancers (Basel). 2021;13(5):1013.
Chen M, Yin F, Yu Y, Zhang H, Wen G. CT-based multi-phase radiomic models for differentiating clear cell renal cell carcinoma. Cancer Imaging. 2021;21(1):42.
Smedley NF, Aberle DR, Hsu W. Using deep neural networks and interpretability methods to identify gene expression patterns that predict radiomic features and histology in non-small cell lung cancer. J Med Imaging (Bellingham). 2021;8(3): 031906.
Shim KY, Chung SW, Jeong JH, Hwang I, Park CK, Kim TM, et al. Radiomics-based neural network predicts recurrence patterns in glioblastoma using dynamic susceptibility contrast-enhanced MRI. Sci Rep. 2021;11(1):9974.
Kirienko M, Sollini M, Corbetta M, Voulaz E, Gozzi N, Interlenghi M, et al. Radiomics and gene expression profile to characterise the disease and predict outcome in patients with lung cancer. Eur J Nucl Med Mol Imaging. 2021;48(11):3643–55.
Yan J, Zhang S, Li KKW, Wang W, Li K, Duan W, et al. Incremental prognostic value and underlying biological pathways of radiomics patterns in medulloblastoma. EBioMedicine. 2020;61: 103093.
Huang Y, Zeng H, Chen L, Luo Y, Ma X, Zhao Y. Exploration of an integrative prognostic model of radiogenomics features with underlying gene expression patterns in clear cell renal cell carcinoma. Front Oncol. 2021;11: 640881.
Wang JH, Wahid KA, van Dijk LV, Farahani K, Thompson RF, Fuller CD. Radiomic biomarkers of tumor immune biology and immunotherapy response. Clin Transl Radiat Oncol. 2021;28:97–115.
Kim BC, Kim J, Kim K, Byun BH, Lim I, Kong CB, et al. Preliminary radiogenomic evidence for the prediction of metastasis and chemotherapy response in pediatric patients with osteosarcoma using 18F-FDF PET/CT, EZRIN and KI67. Cancers (Basel). 2021;13(11):2671.
Yi X, Liu Y, Zhou B, Xiang W, Deng A, Fu Y, et al. Incorporating SULF1 polymorphisms in a pretreatment CT-based radiomic model for predicting platinum resistance in ovarian cancer treatment. Biomed Pharmacother. 2021;133: 111013.
Park CJ, Han K, Kim H, Ahn SS, Choi D, Park YW, et al. MRI features may predict molecular features of glioblastoma in isocitrate dehydrogenase wild-type lower-grade gliomas. AJNR Am J Neuroradiol. 2021;42(3):448–56.
Li H, Zhu Y, Burnside ES, Huang E, Drukker K, Hoadley KA, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer. 2016;2:16012.
Zhao B. Understanding sources of variation to improve the reproducibility of radiomics. Front Oncol. 2021;11: 633176.
Miles K. Radiomics for personalised medicine: the long road ahead. Br J Cancer. 2020;122(7):929–30.
Lu L, Ahmed FS, Akin O, Luk L, Guo X, Yang H, et al. Uncontrolled confounders may lead to false or overvalued radiomics signature: a proof of concept using survival analysis in a multicenter cohort of kidney cancer. Front Oncol. 2021;11: 638185.
Acknowledgements
Not applicable.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Contributions
SML prepared the original manuscript; BSZ conceptualized and critiqued the draft. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, S., Zhou, B. A review of radiomics and genomics applications in cancers: the way towards precision medicine. Radiat Oncol 17, 217 (2022). https://doi.org/10.1186/s13014-022-02192-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13014-022-02192-2