Abstract
Radiomics is a quantitative approach to medical imaging, which aims at enhancing the existing data available to clinicians by means of advanced mathematical analysis. Through mathematical extraction of the spatial distribution of signal intensities and pixel interrelationships, radiomics quantifies textural information by using analysis methods from the field of artificial intelligence. Various studies from different fields in imaging have been published so far, highlighting the potential of radiomics to enhance clinical decision-making. However, the field faces several important challenges, which are mainly caused by the various technical factors influencing the extracted radiomic features.
The aim of the present review is twofold: first, we present the typical workflow of a radiomics analysis and deliver a practical “how-to” guide for a typical radiomics analysis. Second, we discuss the current limitations of radiomics, suggest potential improvements, and summarize relevant literature on the subject.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Key points
-
Radiomics represents a method for the quantitative description of medical images.
-
A step-by-step “how-to” guide is presented for radiomics analyses.
-
Throughout the radiomics workflow, numerous factors influence radiomic features.
-
Guidelines and quality checklists should be used to improve radiomics studies’ quality.
-
Digital phantoms and open-source data help to improve the reproducibility of radiomics.
Background
Like many other areas of human activity in the last decades, medicine has seen a constant increase in the digitalization of the information generated during clinical routine. As more medical data became available in digital format, new and always more sophisticated software was developed to analyze them. At the same time, the research on artificial intelligence (AI) has long reached a point where its methods and software tools have become not only powerful, but also accessible enough to leave the computer science departments and find applications in an increasing variety of domains. As a consequence, the recent years have witnessed a continuous increase of AI applications in the medical sector, aiming at facilitating repetitive tasks clinicians encounter in their daily clinical workflows and to support clinical decision-making.
The different techniques used in AI—i.e., mainly machine learning and deep learning algorithms—are especially useful when it comes to the emerging field of “big data”. Big data is defined as “a term that describes large volumes of high velocity, complex and variable data that require advanced techniques and technologies to enable the capture, storage, distribution, management, and analysis of the information.” Footnote 1 Due to the high amount of multi-dimensional information, techniques from the field of AI are needed to extract the desired information from these data.
In medicine, various ways to generate big data exist, including the widely known fields of genomics, proteomics, or metabolomics. Similar to these “omics” clusters, imaging has been used increasingly to generate a dedicated omics cluster itself called “radiomics”. Radiomics is a quantitative approach to medical imaging, which aims at enhancing the existing data available to clinicians by means of advanced, and sometimes non-intuitive mathematical analysis. The concept of radiomics, which has most broadly (but not exclusively) been applied in the field of oncology, is based on the assumption that biomedical images contain information of disease-specific processes [1] that are imperceptible by the human eye [2] and thus not accessible through traditional visual inspection of the generated images. Through mathematical extraction of the spatial distribution of signal intensities and pixel interrelationships, radiomics quantifies textural information [3, 4] by using analysis methods from the field of AI. In addition, visual appreciable differences in image intensity, shape, or texture can be quantified by means of radiomics, thus overcoming the subjective nature of image interpretation. Thus, radiomics does not imply any automation of the diagnostic processes, rather it provides existing ones with additional data.
Radiomics analysis can be performed on medical images from different modalities, allowing for an integrated cross-modality approach using the potential additive value of imaging information extracted, e.g., from magnetic resonance imaging (MRI), computed tomography (CT), and positron-emission-tomography (PET), instead of evaluating each modality by its own. However, the current state-of-the-art of the research still shows lack of stability and generalization, and the specific study conditions and the authors’ choices have still a great influence on the results.
In this work, we present the typical workflow of a radiomics analysis, discussing the current limitations of this approach, suggesting potential improvements, and commenting relevant literature on the subject.
Radiomics–how to?
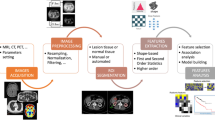
The following section will give a practical advice on “how to do radiomics” by illustrating each of the required steps in the radiomics pipeline (illustrated in Fig. 1) and highlighting important points.
The radiomics workflow. Schematic illustration of the patient journey including image acquisition, analysis utilizing radiomics, and derived patient-specific therapy and prognosis. After image acquisition and segmentation, radiomic features are extracted. High-level statistical modeling involving machine learning is applied for disease classification, patient clustering, and individual risk stratification
Step 1: image segmentation
For any radiomics approach, delineation of the region of interest (ROI) in two-dimensional (2D) or of the volume of interest (VOI) in three-dimensional (3D) approaches is the crucial first step in the pipeline. ROIs/VOIs define the region in which radiomic features are calculated.
Image segmentation might be done manually, semi-automatically (using standard image segmentation algorithms such as region-growing or thresholding), or fully automatically (nowadays using deep learning algorithms). A variety of different software solutions—either open-source or commercial—are available, such as 3D Slicer Footnote 2 [5], MITK Footnote 3, ITK-SNAP Footnote 4, MeVisLab Footnote 5, LifEx Footnote 6, or ImageJ Footnote 7 [6], to name only some frequently used open-source tools. For reviews on various different tools for image segmentation, please refer to [7, 8].
Manual and semi-automated image segmentation (usually with manual correction) are the most often encountered methods but have several drawbacks. Firstly, manual segmentation is time-consuming – depending on how many images and datasets have to be segmented. Second, manual and semi-automated segmentation introduce a considerable observer-bias, and studies have shown that many radiomic features are not robust against intra- and inter-observer variations concerning ROI/VOI delineation [9]. Consequently, studies using manual or semi-automated image segmentation with manual correction should perform assessments of intra- and inter-observer reproducibility of the derived radiomic features and exclude non-reproducible features from further analyses.
Deep learning-based image segmentation (often using some sort of U-Net [10]) is rapidly emerging and many different algorithms have already been trained for image segmentation tasks of various organs (currently, most of them being useful for the segmentation of entire organs, but not for segmentation of dedicated tumor regions), several of them being published as open-source. Since recently, there are also several possibilities for integration of such algorithms in platforms like 3D Slicer or MITK. Automated image segmentation certainly is the best option, since it avoids intra- and inter-observer variability of radiomic features. However, generalizability of trained algorithms currently is a major limitation, and applying those algorithms on a different dataset often results in complete failure. Thus, further research has to be devoted to the development of robust and generalizable algorithms for automated image segmentation.
Step 2: image processing
Image processing is located between the image segmentation and feature extraction step. It represents the attempt to homogenize images from which radiomic features will be extracted with respect to pixel spacing, grey-level intensities, bins of the grey-level histogram, and so forth. Preliminary results have shown that the test-retest robustness of radiomic features extracted largely depends on the image processing settings used [11,12,13,14,15]. In order to allow for reproducible research, it is therefore important to report each detail of the image processing step.
Several of the above-mentioned software platforms (namely, 3D Slicer and LifEx) have integrations for radiomics analyses. 3D Slicer has incorporated an installable plugin for the open-source pyRadiomics package [16] (which can otherwise be used within a solo Python framework), whereas LifEx is a stand-alone platform with integrated segmentation and texture analysis tools and a graphical user interface. The image processing step in the pyRadiomics package (which currently is one of the most commonly used packages for radiomics analyses) can be defined by writing a so-called parameter file (in a YAML or JSON structured text file). This parameter file can be loaded into 3D Slicer or be incorporated into a Python framework. Example parameter files for different modalities can be found in the pyRadiomics GitHub repositoryFootnote 8.
Interpolation to isotropic voxel spacing is necessary for most texture feature sets to become rotationally invariant and to increase reproducibility between different datasets [17]. Currently, there is no clear recommendation whether upsampling or downsampling should be the preferred method. In addition, data from different modalities might need different approaches for image interpolation. CT, for example, usually delivers isotropic datasets, whereas MRI often delivers non-isotropic data with need for different approaches to interpolation. After applying interpolation algorithms to the image, the delineated ROI/VOI should also be interpolated. For a detailed description of image interpolation and different interpolation algorithms, please refer to [17].
Range re-segmentation and intensity outlier filtering (normalization) are performed to remove pixels/voxels from the segmented region that fall outside of a specified range of grey-levels [17]. Whereas range re-segmentation usually is required for CT and PET data (e.g., for excluding pixels/voxels of air or bone within a tumor ROI/VOI), range re-segmentation is not possible for data with arbitrary intensity units such as MRI. For MRI data, intensity outlier filtering is applied. The most commonly used method is to calculate the mean μ and standard deviation σ of grey-levels within the ROI/VOI and to exclude grey-levels outside the range μ ± 3σ [17,18,19].
The last image processing step is discretization of image intensities inside the ROI/VOI (Fig. 2). Discretization consists in grouping the original values according to specific range intervals (bins); the procedure is conceptually equivalent to the creation of a histogram. This step is required to make feature calculation tractable [20].
Three parameters characterize discretization: the range of the discretized quantity, the number of bins, and their width (size). The range equals the product of the bin number times the bin width; therefore, only two of the parameters can be freely set. Different combinations can lead to different results; the choice of the three parameters is usually influenced by the context, e.g., to simplify the comparison with other works using a particular binning:
-
The range is usually preserved from the original data, but exceptions are not uncommon, e.g. when the discretized data is to be compared with some reference dataset or when ROIs with much smaller range than the original have to be analyzed. It is worth mentioning that when the range is not preserved and if the number of bins is particularly small, the choice of the range boundaries can have a strong impact on the results;
-
Fixing the bin number (as is the case of discretizing grey-level intensities) normalizes images and is especially beneficial in data with arbitrary intensity units (e.g., MRI) and where contrasts are considered important [17]. Thus, it is the recommended discretization method for MRI data, although this recommendation is not without controversies (for further discussion, please refer to the relative pyRadiomics documentationFootnote 9). The use of a fixed bin number discretization is thought to make radiomic features more reproducible across different samples, since the absolute values of many features depend on the number of grey levels within the ROI/VOI;
-
Fixing the bin size results in having direct control on the absolute range represented on each bin, therefore allowing the bin sequence to have an immediate relationship with the original intensity scale (such as Hounsfield units or standardized uptake values). This approach makes it possible to compare discretized data with different ranges, since the bins belonging to the overlapping range will represent the same data interval. For that reason, previous work recommends the use of a fixed bin size for PET images [14]. It is recommended to use identical minimum values for all samples, defined by the lower bound of the re-segmentation range
A still open question is the optimal bin number/bin width which should be used in this discretization step. This question becomes particularly important when considering that the discretization is equivalent to averaging the values within each bin, and the effect is similar to applying a smoothing filter on the data distribution. When the bins are too wide (too few), features can be averaged out and lost; when the bins are too small (too many), features can become indistinguishable from noise. A balance is reached when discretization can filter out the noise while preserving the interesting features; unfortunately, this implies that the optimal choice of binning is highly dependent from the both data acquisition parameters (noise) and content (features). As an example, previous preliminary work has shown that different MRI sequences might need different bin numbers for obtaining robust and reproducible radiomics features [11]. Moreover, small number of bins can generate undesired dependencies on the particular choice of range and bin boundaries, thus undermining the robustness of the analysis. The present recommendation is to always start by inspecting the histogram of the data from which radiomic features are to be extracted and to decide upon a reasonable set of parameters for the discretization step based on the experience.
Step 3: feature extraction
After image segmentation and processing, extraction of radiomic features can finally be performed. Feature extraction refers to the calculation of features as a final processing step, where feature descriptors are used to quantify characteristics of the grey levels within the ROI/VOI [17]. Since many different ways and formulas exist to calculate those features, adherence to the Image Biomarker Standardization Initiative (IBSI) guidelines [17] is recommended. These guidelines offer a consensus for standardized feature calculations from all radiomic feature matrices. Different types (i.e., matrices) of radiomic features exist, the most often encountered ones being intensity (histogram)-based features, shape features, texture features, transform-based features, and radial features. In addition, different types of filters (e.g., wavelet or Gaussian filters) are often applied during the feature extraction step. In practice, feature extraction means simply pressing the “run” button and waiting for the computation to be finished.
Step 4: feature selection/dimension reduction
Depending on the software package used for feature extraction and the number of filters applied during the process, the number of extracted features to deal with during the following step of statistical analysis and machine learning ranges between a few and, in theory, unlimited. The higher the number of features/variables in a model and/or the lower the number of cases in the groups, e.g., for a classification task, the higher the risk of model overfitting.
As a consequence, reducing the number of features to build statistical and machine learning models during a step called feature selection or dimension reduction is of crucial importance for generating valid and generalizable results. Several “rules of thumb” may exist for defining the optimal number of features for a given sample size, but no true evidence for these rules exists in the literature. For some guidance regarding study design or sample size calculation, please consider reference [21]. The dimension reduction is a multi-step process, leading to exclusion of non-reproducible, redundant, and non-relevant features from the dataset.
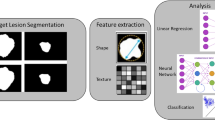
Multiple ways for dimension reduction and feature selection exist among researchers. The following steps reflect our personal experience and have been performed in several clinical studies so far [2, 22,23,24,25,26,27] (Fig. 3).
The first step should involve exclusion of non-reproducible features, if manual or semi-automated ROI/VOI delineation was used during the image segmentation step. A feature which suffers from higher intra- or interobserver variability is not likely to be informative, e.g., for assessing therapeutic response. Similarly, the test-retest robustness of the extracted features should be assessed (e.g., using a phantom). Non-robust features should also be excluded if the study aim is the evaluation of longitudinal data, although it is important that the relevant change of features over time is incorporated into the selection procedure [28]. Simply assessing reproducibility/robustness by calculation of intra-class-correlation coefficients (ICCs) might not be sufficient since ICCs are known to depend on the natural variance of the underlying data. Recommendations for assessing reproducibility, repeatability, and robustness can be found in [29].
The second step in the feature selection process is the selection of the most relevant variables for the respective task. Various approaches often relying on machine learning techniques can be used for this initial feature selection step, such as knock-off filters, recursive feature elimination methods, or random forest algorithms.
Since these algorithms often do not account for collinearities and correlations in the data, building correlation clusters represents the logical next—third—step in the dimension reduction workflow. In some cases, this step might be combined with the previous (second) step since few machine learning techniques are able to account for correlations within the data. The majority, however, is not. Correlation clusters (for an example, see Fig. 3) visualize clusters of highly correlated features in the data and allow selection of only one representative feature per correlation cluster. This selection process again might be based on machine learning algorithms and/or on conventional statistical methods and data visualization. As a general principle, the variable with the highest biological-clinical variability in the dataset should be selected since it might be most representative of the variations within the specific patient cohort. The data visualization step is also of high importance once the dimensionality of the data has been reduced.
Finally, the remaining, non-correlated and highly relevant features can be used to train the model for the respective classification task. Although the present review does not aim to cover the model training and selection process, the importance of splitting the dataset into a training and at least an independent testing dataset (for optimal conditions even an additional validation dataset) cannot be stressed enough [30]. This is especially relevant given the limitations currently encountered in the field of radiomics as discussed in the following section.
Current limitations in radiomics
Although radiomics has shown its potential for diagnostic, prognostic, and predictive purposes in numerous studies, the field is facing several challenges. The existing gap between knowledge and clinical needs results in studies lacking clinical utility. In case a clinically relevant question is considered, the reproducibility of radiomic studies is often poor, due to lack of standardization, insufficient reporting, or limited open source code and data. Also, the lack of proper validation and the subsequent risk of false-positive results hampers the translation to clinical practice [31]. Moreover, the interpretability of the features, especially those derived from texture matrices and/or after filtering, mistakes in the interpretation of the results (e.g., causation vs. correlation), or the lack of comparison with well-established prognostic and predictive factors, results in reservation towards its use in clinical decision support systems. Furthermore, radiomics studies are often based on retrospectively collected data and thus have low level of evidence and mainly serve as proof-of-concept, whereas prospective studies are required to confirm the value of radiomics.
Due to the retrospective nature of radiomic studies, imaging protocols, including acquisition, and reconstruction settings, are often not controlled or standardized. For each image modality, multiple studies have assessed the impact of these settings on radiomic features or attempted to minimize their influence by eliminating features that are sensitive to these variabilities. Although these studies are relevant to create awareness of the influencing factors, it should be noted that the information is often not directly helpful to future studies. The reproducibility of radiomic features is not necessarily generalizable to different disease sites, modalities, or scanners, e.g., robust features in one disease site are not necessarily robust in another disease site [32]. Moreover, in case robust radiomic features are assessed using cut-off values of correlation coefficients, one should be aware that these cut-offs are often arbitrarily chosen and the number of “robust” features depend on the number of subjects involved. Furthermore, for the generalizability of robustness studies, it is important that radiomic feature calculations are compliant with the IBSI guidelines [17].
Apart from the variations in scanners and settings, radiomic feature values are also influenced by patient variabilities, e.g., geometry, which impact the levels of noise and presence of artifacts in an image. Therefore, the aim of a recent study was to quantify these so-called “non-reducible technical variations” and stabilize the radiomic features accordingly [33].
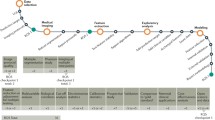
The next sections summarize the studies that assessed radiomic feature robustness for different acquisition and reconstruction settings of CT, PET, and MRI, as well as for ROI delineation and image pre-processing steps. Figure 4 provides an overview of factors that have been investigated in literature for their influence on radiomic feature values. In Tables 1, 2, and 3, the studies are collected in one overview for all three modalities considered in this review: CT, MRI, and PET, respectively. A recent review provides an overview of existing phantoms that have been used for radiomics for all three modalities [120].
CT and PET CT
Multiple studies (16 were identified in this review) have investigated the stability over test-retest scenarios for CT radiomics (Table 1), where the publicly available RIDER Lung CT collection was often evaluated [121]. For PET, only a few test-retest studies were performed, which were either on a phantom or lung cancer data (Table 2). Recently, an extensive review on factors influencing PET radiomics was published [122].
The voxel size was the mostly investigated influencing reconstruction factor for CT, whereas this was the full-width half maximum (FWHM) of the Gaussian filter for PET. Four and 12 studies were identified that studied the influence of image discretization on CT and PET radiomic features, respectively. Figure 4 provides an overview of factors that have been investigated in literature for their influence on radiomic feature values.
MRI
The impact of test-retest, acquisition and reconstruction settings, segmentation, and image pre-processing has been explored less extensively to date than for PET and CT. Only four studies were found that investigated the influence of reconstruction settings, one of these studies included patient images. The influence of segmentation on MRI radiomic features has been more extensively studied for a variety of tumor sites. Table 3 summarizes the present literature for influencing factors on radiomic features in MRI. Figure 4 provides an overview of factors that have been investigated in literature for their influence on radiomic feature values.
Reduce radiomics’ dependency
Recent literature regarding the robustness for different acquisition and reconstruction settings, ROI delineation, and image pre-processing steps shows that the most commonly used approach to deal with this is to eliminate radiomic features that are not robust against these factors. The drawback of this method is that potentially relevant information could be removed, whereas stability not necessarily means informativity. A few solutions have been proposed in order to reduce the influence of the aforementioned factors on radiomics studies. One proposed solution is to eliminate the dependency of features on a certain factor by modeling the relationship and applying corrections accordingly. This had been explored recently for different CT exposure settings [123]. Another method to eliminate the dependency is to convert images using deep learning, in order to simulate reconstruction with different settings, which was shown to improve CT radiomics’ reproducibility for images reconstructed with different kernels [62]. This approach has the potential to solve other radiomics dependencies to improve robustness in the future. Different than image-wise dependency corrections, post-reconstruction batch harmonization has been proposed in order to harmonize radiomic feature sets originating from different institutes, which is a method called ComBat [124,125,126]. Furthermore, a recent study investigated the performance of data augmentation instead of feature elimination to incorporate the knowledge on influencing factors on radiomic features [127].
Open-source data
Publicly available datasets like the RIDER dataset Footnote 10 help to gain knowledge about the impact of varying factors in radiomics [121]. Also, the availability of a public phantom dataset, intended for radiomics reproducibility tests on CT, could help to further assess the influence of acquisition settings in order to eliminate non-robust radiomic features [128]. However, studies are needed to show if robustness data acquired on a phantom can be translated to the human. Similar initiatives for PET and MRI would help to understanding of the impact of changes in settings on radiomics. In other words, open-source data plays an important role in the future improvement of radiomics.
Solution: quality control and standardization
In order to increase the chance of clinically relevant and valuable radiomics studies, we would recommend verifying whether the following questions could be answered with “yes,” prior to commencement of the study:
-
Is there an actual clinical need which could potentially be answered with (the help of) radiomics?
-
Is there enough expertise in the research team, preferably from at least two different disciplines, to ensure high quality of the study and potential of clinical implementation?
-
Is there access to enough data to support the conclusions with sufficient power, including external validation datasets?
-
Is it possible to retrieve all other non-imaging data that is known to be relevant for the research question (e.g., from biological information, demographics)?
-
Is information on the acquisition and reconstruction of the images available?
-
Are the imaging protocols standardized and if not, is there a solution to harmonize images or to ensure minimal influence of varying settings on the modeling?
Besides these general questions, which should been asked before the start of a study, there are some recent contributions in the field that aim to facilitate the execution of radiomics studies with higher quality: (1) IBSI: harmonization of radiomics implementations and guidelines on reporting of radiomic studies [17, 129], (2) Radiomics Quality Score (RQS): checklist to ensure quality of radiomics studies [130], and (3) Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) statement—guidelines for reporting of prediction models for prognosis or diagnosis [30]. For the radiomic feature calculation, we recommend to use an implementation that is IBSI compliant, which could be verified using the publicly available digital phantom [129, 130]. Also, regarding choices for image discretization and resampling, we recommend following the IBSI guidelines. Besides that, it is important to be consistent and transparent, and detailed reporting on the pre-processing steps applied to improve reproducibility and repeatability of radiomic studies need to be ensured.
A recent study evaluated the quality of 77 oncology-related radiomics studies using RQS and TRIPOD, and concluded that “the overall scientific quality and reporting of radiomics studies is insufficient,” showing the importance of guidelines and criteria for future studies [131].
Outlook: workflow integration
While currently many research efforts aim towards standardization of radiomics, translation into clinical practice also requires adequate implementation of radiomics analyses into the clinical workflow once the standardization issue has been adequately addressed and clinical utility has been proven in prospective clinical trials.
A useful radiomics tool should seamlessly integrate into the clinical radiological workflow and be incorporated into or interfaced with existing RIS/PACS systems. Such systems should provide segmentation tools or ideally deep learning-based automated segmentation methods as well as standardized feature extraction algorithms and modality-adjusted image processing adhering to the standards described above. In case of fully automated segmentation, the possibility to inspect and manually correct the segmentation results should be incorporated.
In a future workflow, known important radiomics features could then be displayed alongside other quantitative imaging biomarkers and the images themselves. The radiologist could then use all these information to support his clinical judgement or—where possible—estimate, e.g., prognostic factors.
It is, however, important to note, that radiomics should only be viewed as an additional tool and not as a standalone diagnostic algorithm. Certainly, many challenges lie ahead until radiomics can be integrated in our daily routine: from the above-mentioned issues surrounding image standardization to legal issues that will certainly arise regarding regulatory issues. Nonetheless, it could prove a valuable if not critical step towards a more integrated approach to healthcare.
Conclusions
Throughout the radiomics workflow, multiple factors have been identified that influence the feature values, including random variations in scanner and patients, image acquisition and reconstruction settings, ROI segmentation, and image preprocessing. Several studies have proposed to either eliminate unstable features, correct for influencing factors, or harmonize datasets in order to improve the robustness of radiomics. Recently published guidelines and checklists aim to improve the quality of future radiomics studies, but transparency has been recognized as the most important factor for reproducibility. Assessment of clinical relevance and impact prior to study commencement, increased level of evidence using studies with large enough datasets and external validation, and its combination with established methods will help moving the field towards clinical implementation.
Availability of data and materials
Not applicable.
Notes
TechAmerica Foundation’s Federal Big Data Commission, 2012 https://bigdatawg.nist.gov/_uploadfiles/M0068_v1_3903747095.pdf
https://slicer.org
https://mitk.org
https://itksnap.org
https://mevislab.de
https://lifexsoft.org
https://imagej.nih.gov
https://github.com/Radiomics/pyradiomics/tree/master/examples/exampleSettings
https://pyradiomics.readthedocs.io/en/latest/faq.html#radiomics-fixed-bin-width
https://wiki.cancerimagingarchive.net/display/Public/RIDER+Lung+CT
Abbreviations
- AI:
-
Artificial intelligence
- CT :
-
Computed tomography
- DL:
-
Deep learning
- IBSI:
-
Imaging Biomarker Standardization Initiative
- ICC:
-
Intra-class-correlation coefficient
- ML:
-
Machine learning
- MRI:
-
Magnetic resonance imaging
- PET:
-
Positron emission tomography
- ROI:
-
Region of interest
- RQS:
-
Radiomics Quality Score
- TRIPOD:
-
Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis
- VOI:
-
Volume of interest
References
Neisius U, El-Rewaidy H, Nakamori S, Rodriguez J, Manning WJ, Nezafat R (2019) Radiomic analysis of myocardial native T1 imaging discriminates between hypertensive heart disease and hypertrophic cardiomyopathy. JACC Cardiovasc Imaging 12:1946–1954 https://doi.org/10.1016/j.jcmg.2018.11.024
Mannil M, von Spiczak J, Manka R, Alkadhi H (2018) Texture analysis and machine learning for detecting myocardial infarction in noncontrast low-dose computed tomography: unveiling the invisible. Invest Radiol 53:338–343 https://doi.org/10.1097/RLI.0000000000000448
Castellano G, Bonilha L, Li LM, Cendes F (2004) Texture analysis of medical images. Clin Radiol 59:1061–1069 https://doi.org/10.1016/j.crad.2004.07.008
Tourassi GD (1999) Journey toward computer-aided diagnosis: role of image texture analysis. Radiology 213:317–320 https://doi.org/10.1148/radiology.213.2.r99nv49317
Fedorov A, Beichel R, Kalpathy-Cramer J et al (2012) 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 30:1323–1341 https://doi.org/10.1016/j.mri.2012.05.001
Abràmoff MD, Magalhães PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics Int 7:36–42
Kresanova Z, Kostolny J. Comparison of Software for Medical Segmentation, p 15
Lay-Khoon Lee, Siau-Chuin Liew (2015) A survey of medical image processing tools. https://doi.org/10.13140/RG.2.1.3364.4241
Baeßler B, Weiss K, Pinto dos Santos D (2019) Robustness and reproducibility of radiomics in magnetic resonance imaging: a phantom study. Invest Radiol 54:221–228 https://doi.org/10.1097/RLI.0000000000000530
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation. arXiv:1505.04597
Wichtmann B, Attenberger U, Harder FM et al (2018) Influence of image processing on the robustness of radiomic features derived from magnetic resonance imaging—a phantom study. In: ISMRM 2018, p 5
Altazi BA, Zhang GG, Fernandez DC et al (2017) Reproducibility of F18-FDG PET radiomic features for different cervical tumor segmentation methods, gray-level discretization, and reconstruction algorithms. J Appl Clin Med Phys 18:32–48 https://doi.org/10.1002/acm2.12170
Bailly C, Bodet-Milin C, Couespel S et al (2016) Revisiting the robustness of PET-based textural features in the context of multi-centric trials. PLoS One 11:e0159984 https://doi.org/10.1371/journal.pone.0159984
Leijenaar RTH, Nalbantov G, Carvalho S et al (2015) The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep 5:11075 https://doi.org/10.1038/srep11075
Shafiq-ul-Hassan M, Zhang GG, Latifi K et al (2017) Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys 44:1050–1062 https://doi.org/10.1002/mp.12123
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107 https://doi.org/10.1158/0008-5472.CAN-17-0339
Zwanenburg A, Leger S, Vallières M, Löck S (2016) Image biomarker standardisation initiative. arXiv:1612.07003
Collewet G, Strzelecki M, Mariette F (2004) Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn Reson Imaging 22:81–91 https://doi.org/10.1016/j.mri.2003.09.001
Vallières M, Freeman CR, Skamene SR, Naqa IE (2015) A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol 60:5471–5496 https://doi.org/10.1088/0031-9155/60/14/5471
Yip SSF, Aerts HJWL (2016) Applications and limitations of radiomics. Phys Med Biol 61:R150–R166 https://doi.org/10.1088/0031-9155/61/13/R150
Riley RD, Snell KI, Ensor J et al (2019) Minimum sample size for developing a multivariable prediction model: PART II - binary and time-to-event outcomes. Stat Med 38:1276–1296 https://doi.org/10.1002/sim.7992
Baessler B, Mannil M, Oebel S, Maintz D, Alkadhi H, Manka R (2018) Subacute and chronic left ventricular myocardial scar: accuracy of texture analysis on nonenhanced Cine MR images. Radiology 286:103–112 https://doi.org/10.1148/radiol.2017170213
Baessler B, Luecke C, Lurz J et al (2018) Cardiac MRI texture analysis of T1 and T2 maps in patients with infarctlike acute myocarditis. Radiology 289:357–365 https://doi.org/10.1148/radiol.2018180411
Baessler B, Luecke C, Lurz J et al (2019) Cardiac MRI and texture analysis of myocardial T1 and T2 maps in myocarditis with acute versus chronic symptoms of heart failure. Radiology 292:608–617 https://doi.org/10.1148/radiol.2019190101
Baeßler B, Mannil M, Maintz D, Alkadhi H, Manka R (2018) Texture analysis and machine learning of non-contrast T1-weighted MR images in patients with hypertrophic cardiomyopathy-preliminary results. Eur J Radiol 102:61–67 https://doi.org/10.1016/j.ejrad.2018.03.013
Baessler B, Nestler T, Pinto dos Santos D et al (2020) Radiomics allows for detection of benign and malignant histopathology in patients with metastatic testicular germ cell tumors prior to post-chemotherapy retroperitoneal lymph node dissection. Eur Radiol 30:2334–2345 https://doi.org/10.1007/s00330-019-06495-z
Di Noto T, von Spiczak J, Mannil M et al (2019) Radiomics for distinguishing myocardial infarction from myocarditis at late gadolinium enhancement at MRI: comparison with subjective visual analysis. Radiol Cardiothorac Imaging 1:e180026 https://doi.org/10.1148/ryct.2019180026
van Timmeren JE, Leijenaar RTH, van Elmpt W, Reymen B, Lambin P (2017) Feature selection methodology for longitudinal cone-beam CT radiomics. Acta Oncol 56:1537–1543 https://doi.org/10.1080/0284186X.2017.1350285
Sullivan DC, Obuchowski NA, Kessler LG et al (2015) Metrology standards for quantitative imaging biomarkers. Radiology 277:813–825 https://doi.org/10.1148/radiol.2015142202
Collins GS, Reitsma JB, Altman DG, Moons KGM (2015) Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ 350:g7594–g7594 https://doi.org/10.1136/bmj.g7594
Chalkidou A, O’Doherty MJ, Marsden PK (2015) False discovery rates in PET and CT studies with texture features: a systematic review. PLoS One 10:e0124165 https://doi.org/10.1371/journal.pone.0124165
van Timmeren J, Leijenaar RTH, van Elmpt W et al (2016) Test–retest data for radiomics feature stability analysis: generalizable or study-specific? Tomography 2:361–365 https://doi.org/10.18383/j.tom.2016.00208
Mühlberg A, Katzmann A, Heinemann V et al (2020) The technome - a predictive internal calibration approach for quantitative imaging biomarker research. Sci Rep 10:1103 https://doi.org/10.1038/s41598-019-57325-7
Du Q, Baine M, Bavitz K et al (2019) Radiomic feature stability across 4D respiratory phases and its impact on lung tumor prognosis prediction. PLoS One 14:e0216480 https://doi.org/10.1371/journal.pone.0216480
Mahon RN, Hugo GD, Weiss E (2019) Repeatability of texture features derived from magnetic resonance and computed tomography imaging and use in predictive models for non-small cell lung cancer outcome. Phys Med Biol 64:145007 https://doi.org/10.1088/1361-6560/ab18d3
Tanaka S, Kadoya N, Kajikawa T et al (2019) Investigation of thoracic four-dimensional CT-based dimension reduction technique for extracting the robust radiomic features. Phys Med 58:141–148 https://doi.org/10.1016/j.ejmp.2019.02.009
Tunali I, Hall LO, Napel S et al (2019) Stability and reproducibility of computed tomography radiomic features extracted from peritumoral regions of lung cancer lesions. Med Phys 46:5075–5085 https://doi.org/10.1002/mp.13808
Zwanenburg A, Leger S, Agolli L et al (2019) Assessing robustness of radiomic features by image perturbation. Sci Rep 9:614 https://doi.org/10.1038/s41598-018-36938-4
Berenguer R, Pastor-Juan MDR, Canales-Vázquez J et al (2018) Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology 288:407–415 https://doi.org/10.1148/radiol.2018172361
Desseroit M-C, Tixier F, Weber WA et al (2017) Reliability of PET/CT shape and heterogeneity features in functional and morphologic components of non–small cell lung cancer tumors: a repeatability analysis in a prospective multicenter cohort. J Nucl Med 58:406–411 https://doi.org/10.2967/jnumed.116.180919
Larue RTHM, van Timmeren JE, de Jong EEC et al (2017) Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: a comprehensive phantom study. Acta Oncol 56:1544–1553 https://doi.org/10.1080/0284186X.2017.1351624
Larue RTHM, Van De Voorde L, van Timmeren JE et al (2017) 4DCT imaging to assess radiomics feature stability: An investigation for thoracic cancers. Radiother Oncol 125:147–153 https://doi.org/10.1016/j.radonc.2017.07.023
Hu P, Wang J, Zhong H et al (2016) Reproducibility with repeat CT in radiomics study for rectal cancer. Oncotarget 7 https://doi.org/10.18632/oncotarget.12199
Aerts HJWL, Velazquez ER, Leijenaar RTH et al (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006 https://doi.org/10.1038/ncomms5006
Balagurunathan Y, Gu Y, Wang H et al (2014) Reproducibility and prognosis of quantitative features extracted from CT images. Transl Oncol 7:72–87 https://doi.org/10.1593/tlo.13844
Balagurunathan Y, Kumar V, Gu Y et al (2014) Test–retest reproducibility analysis of lung CT image features. J Digit Imaging 27:805–823 https://doi.org/10.1007/s10278-014-9716-x
Fried DV, Tucker SL, Zhou S et al (2014) Prognostic value and reproducibility of pretreatment ct texture features in stage III non-small cell lung cancer. Int J Radiat Oncol 90:834–842 https://doi.org/10.1016/j.ijrobp.2014.07.020
Hunter LA, Krafft S, Stingo F et al (2013) High quality machine-robust image features: Identification in nonsmall cell lung cancer computed tomography images: Robust quantitative image features. Med Phys 40:121916 https://doi.org/10.1118/1.4829514
Hepp T, Othman A, Liebgott A, Kim JH, Pfannenberg C, Gatidis S (2020) Effects of simulated dose variation on contrast-enhanced CT-based radiomic analysis for Non-Small Cell Lung Cancer. Eur J Radiol 124:108804 https://doi.org/10.1016/j.ejrad.2019.108804
Piazzese C, Foley K, Whybra P, Hurt C, Crosby T, Spezi E (2019) Discovery of stable and prognostic CT-based radiomic features independent of contrast administration and dimensionality in oesophageal cancer. PLoS One 14:e0225550 https://doi.org/10.1371/journal.pone.0225550
Robins M, Solomon J, Hoye J, Abadi E, Marin D, Samei E (2019) Systematic analysis of bias and variability of texture measurements in computed tomography. J Med Imaging 6:033503 https://doi.org/10.1117/1.JMI.6.3.033503
Ger RB, Zhou S, Chi P-CM et al (2018) Comprehensive investigation on controlling for CT imaging variabilities in radiomics studies. Sci Rep 8:13047 https://doi.org/10.1038/s41598-018-31509-z
Mackin D, Ger R, Dodge C et al (2018) Effect of tube current on computed tomography radiomic features. Sci Rep 8:2354 https://doi.org/10.1038/s41598-018-20713-6
Shafiq-ul-Hassan M, Latifi K, Zhang G, Ullah G, Gillies R, Moros E (2018) Voxel size and gray level normalization of CT radiomic features in lung cancer. Sci Rep 8:10545 https://doi.org/10.1038/s41598-018-28895-9
Buch K, Li B, Qureshi MM, Kuno H, Anderson SW, Sakai O (2017) Quantitative assessment of variation in CT parameters on texture features: pilot study using a nonanatomic phantom. AJNR Am J Neuroradiol 38:981–985 https://doi.org/10.3174/ajnr.A5139
Mackin D, Fave X, Zhang L et al (2017) Harmonizing the pixel size in retrospective computed tomography radiomics studies. PLoS One 12:e0178524 https://doi.org/10.1371/journal.pone.0178524
Shafiq-ul-Hassan M, Zhang GG, Hunt DC et al (2017) Accounting for reconstruction kernel-induced variability in CT radiomic features using noise power spectra. J Med Imaging 5:1 https://doi.org/10.1117/1.JMI.5.1.011013
Lo P, Young S, Kim HJ, Brown MS, McNitt-Gray MF (2016) Variability in CT lung-nodule quantification: Effects of dose reduction and reconstruction methods on density and texture based features: Variability in CT lung-nodule quantification. Med Phys 43:4854–4865 https://doi.org/10.1118/1.4954845
Solomon J, Mileto A, Nelson RC, Choudhury KR, Samei E (2016) Quantitative features of liver lesions, lung nodules, and renal stones at multi–detector row CT examinations: dependency on radiation dose and reconstruction algorithm. Radiology 279:185–194 https://doi.org/10.1148/radiol.2015150892
Fave X, Cook M, Frederick A et al (2015) Preliminary investigation into sources of uncertainty in quantitative imaging features. Comput Med Imaging Graph 44:54–61 https://doi.org/10.1016/j.compmedimag.2015.04.006
Oliver JA, Budzevich M, Zhang GG, Dilling TJ, Latifi K, Moros EG (2015) Variability of image features computed from conventional and respiratory-gated PET/CT images of lung cancer. Transl Oncol 8:524–534 https://doi.org/10.1016/j.tranon.2015.11.013
Choe J, Lee SM, Do K-H et al (2019) Deep learning–based image conversion of CT reconstruction kernels improves radiomics reproducibility for pulmonary nodules or masses. Radiology 292:365–373 https://doi.org/10.1148/radiol.2019181960
Ligero M, Torres G, Sanchez C, Diaz-Chito K, Perez R, Gil D (2019) Selection of radiomics features based on their reproducibility. In: 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). IEEE, Berlin, pp 403–408
Varghese BA, Hwang D, Cen SY et al (2019) Reliability of CT-based texture features: Phantom study. J Appl Clin Med Phys 20:155–163 https://doi.org/10.1002/acm2.12666
Bogowicz M, Riesterer O, Bundschuh RA et al (2016) Stability of radiomic features in CT perfusion maps. Phys Med Biol 61:8736–8749 https://doi.org/10.1088/1361-6560/61/24/8736
Kim H, Park CM, Lee M et al (2016) Impact of reconstruction algorithms on CT radiomic features of pulmonary tumors: analysis of intra- and inter-reader variability and inter-reconstruction algorithm variability. PLoS One 11:e0164924 https://doi.org/10.1371/journal.pone.0164924
Lu L, Ehmke RC, Schwartz LH, Zhao B (2016) Assessing agreement between radiomic features computed for multiple CT imaging settings. PLoS One 11:e0166550 https://doi.org/10.1371/journal.pone.0166550
Zhao B, Tan Y, Tsai W-Y et al (2016) Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep 6:23428 https://doi.org/10.1038/srep23428
Kim HG, Chung YE, Lee YH et al (2015) Quantitative analysis of the effect of iterative reconstruction using a phantom: determining the appropriate blending percentage. Yonsei Med J 56:253 https://doi.org/10.3349/ymj.2015.56.1.253
Zhao B, Tan Y, Tsai WY, Schwartz LH, Lu L (2014) Exploring Variability in CT characterization of tumors: a preliminary phantom study. Transl Oncol 7:88–93 https://doi.org/10.1593/tlo.13865
Qiu Q, Duan J, Duan Z et al (2019) Reproducibility and non-redundancy of radiomic features extracted from arterial phase CT scans in hepatocellular carcinoma patients: impact of tumor segmentation variability. Quant Imaging Med Surg 9:453–464 https://doi.org/10.21037/qims.2019.03.02
Pavic M, Bogowicz M, Würms X et al (2018) Influence of inter-observer delineation variability on radiomics stability in different tumor sites. Acta Oncol 57:1070–1074 https://doi.org/10.1080/0284186X.2018.1445283
Kalpathy-Cramer J, Mamomov A, Zhao B et al (2016) Radiomics of lung nodules: a multi-institutional study of robustness and agreement of quantitative imaging features. Tomography 2:430–437 https://doi.org/10.18383/j.tom.2016.00235
Parmar C, Rios Velazquez E, Leijenaar R et al (2014) Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS ONE 9:e102107 https://doi.org/10.1371/journal.pone.0102107
Lee S-H, Cho H, Lee HY, Park H (2019) Clinical impact of variability on CT radiomics and suggestions for suitable feature selection: a focus on lung cancer. Cancer Imaging 19:54 https://doi.org/10.1186/s40644-019-0239-z
Bagher‐Ebadian H, Siddiqui F, Liu C, Movsas B, Chetty IJ (2017) On the impact of smoothing and noise on robustness of CT and CBCT radiomics features for patients with head and neck cancers. Med Phys 44:1755–1770 https://doi.org/10.1002/mp.12188
Konert T, Everitt S, La Fontaine MD et al (2020) Robust, independent and relevant prognostic 18F-fluorodeoxyglucose positron emission tomography radiomics features in non-small cell lung cancer: Are there any? PLoS One 15:e0228793 https://doi.org/10.1371/journal.pone.0228793
Vuong D, Tanadini-Lang S, Huellner MW et al (2019) Interchangeability of radiomic features between [18F]- FDG PET / CT and [18F]- FDG PET / MR. Med Phys 46:1677–1685 https://doi.org/10.1002/mp.13422
Gallivanone F, Interlenghi M, D’Ambrosio D, Trifirò G, Castiglioni I (2018) Parameters influencing PET imaging features: a phantom study with irregular and heterogeneous synthetic lesions. Contrast Media Mol Imaging 2018:1–12 https://doi.org/10.1155/2018/5324517
Leijenaar RTH, Carvalho S, Velazquez ER et al (2013) Stability of FDG-PET Radiomics features: An integrated analysis of test-retest and inter-observer variability. Acta Oncol 52:1391–1397 https://doi.org/10.3109/0284186X.2013.812798
Pfaehler E, Beukinga RJ, de Jong JR et al (2019) Repeatability of 18 F-FDG PET radiomic features: A phantom study to explore sensitivity to image reconstruction settings, noise, and delineation method. Med Phys 46:665–678 https://doi.org/10.1002/mp.13322
Branchini M, Zorz A, Zucchetta P et al (2019) Impact of acquisition count statistics reduction and SUV discretization on PET radiomic features in pediatric 18F-FDG-PET/MRI examinations. Phys Med 59:117–126 https://doi.org/10.1016/j.ejmp.2019.03.005
Carles M, Torres-Espallardo I, Alberich-Bayarri A et al (2017) Evaluation of PET texture features with heterogeneous phantoms: complementarity and effect of motion and segmentation method. Phys Med Biol. 62(2):652–668 https://doi.org/10.1088/1361-6560/62/2/652
Lovat E, Siddique M, Goh V, Ferner RE, Cook GJ, Warbey VS (2017) The effect of post-injection 18F-FDG PET scanning time on texture analysis of peripheral nerve sheath tumours in neurofibromatosis-1. EJNMMI Res 7:35 https://doi.org/10.1186/s13550-017-0282-3
Reuzé S, Orlhac F, Chargari C et al (2017) Prediction of cervical cancer recurrence using textural features extracted from 18F-FDG PET images acquired with different scanners. Oncotarget 8 https://doi.org/10.18632/oncotarget.17856
Shiri I, Rahmim A, Ghaffarian P, Geramifar P, Abdollahi H, Bitarafan-Rajabi A (2017) The impact of image reconstruction settings on 18F-FDG PET radiomic features: multi-scanner phantom and patient studies. Eur Radiol 27:4498–4509 https://doi.org/10.1007/s00330-017-4859-z
Forgacs A, Pall Jonsson H, Dahlbom M et al (2016) A study on the basic criteria for selecting heterogeneity parameters of F18-FDG PET images. PLoS One 11:e0164113 https://doi.org/10.1371/journal.pone.0164113
Grootjans W, Tixier F, van der Vos CS et al (2016) The impact of optimal respiratory gating and image noise on evaluation of intratumor heterogeneity on 18F-FDG PET imaging of lung cancer. J Nucl Med 57:1692–1698 https://doi.org/10.2967/jnumed.116.173112
Nyflot MJ, Yang F, Byrd D, Bowen SR, Sandison GA, Kinahan PE (2015) Quantitative radiomics: impact of stochastic effects on textural feature analysis implies the need for standards. J Med Imaging 2:041002 https://doi.org/10.1117/1.JMI.2.4.041002
Cheng NM, Fang YH, Tsan DL, Hsu CH, Yen TC (2016) Respiration-averaged CT for attenuation correction of PET images – impact on pet texture features in non-small cell lung cancer patients. PLoS One 11:e0150509 https://doi.org/10.1371/journal.pone.0150509
Lasnon C, Majdoub M, Lavigne B et al (2016) 18F-FDG PET/CT heterogeneity quantification through textural features in the era of harmonisation programs: a focus on lung cancer. Eur J Nucl Med Mol Imaging 43:2324–2335 https://doi.org/10.1007/s00259-016-3441-2
van Velden FHP, Kramer GM, Frings V et al (2016) Repeatability of radiomic features in non-small-cell lung cancer [18F]FDG-PET/CT studies: impact of reconstruction and delineation. Mol Imaging Biol 18:788–795 https://doi.org/10.1007/s11307-016-0940-2
Doumou G, Siddique M, Tsoumpas C, Goh V, Cook GJ (2015) The precision of textural analysis in 18F-FDG-PET scans of oesophageal cancer. Eur Radiol 25:2805–2812 https://doi.org/10.1007/s00330-015-3681-8
Yan J, Chu-Shern JL, Loi HY et al (2015) Impact of image reconstruction settings on texture features in 18F-FDG PET. J Nucl Med 56:1667–1673 https://doi.org/10.2967/jnumed.115.156927
Yang F, Simpson G, Young L, Ford J, Dogan N, Wang L (2020) Impact of contouring variability on oncological PET radiomics features in the lung. Sci Rep 10:369 https://doi.org/10.1038/s41598-019-57171-7
Hatt M, Laurent B, Fayad H, Jaouen V, Visvikis D, Le Rest CC (2018) Tumour functional sphericity from PET images: prognostic value in NSCLC and impact of delineation method. Eur J Nucl Med Mol Imaging 45:630–641 https://doi.org/10.1007/s00259-017-3865-3
Lu L, Lv W, Jiang J et al (2016) Robustness of Radiomic Features in [11C]Choline and [18F]FDG PET/CT Imaging of Nasopharyngeal Carcinoma: Impact of Segmentation and Discretization. Mol Imaging Biol 18:935–945 https://doi.org/10.1007/s11307-016-0973-6
Hatt M, Tixier F, Le Rest CC, Pradier O, Visvikis D (2013) Robustness of intratumour 18F-FDG PET uptake heterogeneity quantification for therapy response prediction in oesophageal carcinoma. Eur J Nucl Med Mol Imaging 40:1662–1671 https://doi.org/10.1007/s00259-013-2486-8
Whybra P, Parkinson C, Foley K, Staffurth J, Spezi E (2019) Assessing radiomic feature robustness to interpolation in 18F-FDG PET imaging. Sci Rep 9:9649 https://doi.org/10.1038/s41598-019-46030-0
Presotto L, Bettinardi V, De Bernardi E et al (2018) PET textural features stability and pattern discrimination power for radiomics analysis: An “ad-hoc” phantoms study. Phys Med 50:66–74 https://doi.org/10.1016/j.ejmp.2018.05.024
Yip SS, Parmar C, Kim J, Huynh E, Mak RH, Aerts HJ (2017) Impact of experimental design on PET radiomics in predicting somatic mutation status. Eur J Radiol 97:8–15 https://doi.org/10.1016/j.ejrad.2017.10.009
Bianchini L, Botta F, Origgi D et al (2020) PETER PHAN: An MRI phantom for the optimisation of radiomic studies of the female pelvis. Phys Med 71:71–81 https://doi.org/10.1016/j.ejmp.2020.02.003
Fiset S, Welch ML, Weiss J et al (2019) Repeatability and reproducibility of MRI-based radiomic features in cervical cancer. Radiother Oncol 135:107–114 https://doi.org/10.1016/j.radonc.2019.03.001
Peerlings J, Woodruff HC, Winfield JM et al (2019) Stability of radiomics features in apparent diffusion coefficient maps from a multi-centre test-retest trial. Sci Rep 9:4800 https://doi.org/10.1038/s41598-019-41344-5
Schwier M, van Griethuysen J, Vangel MG et al (2019) Repeatability of Multiparametric Prostate MRI Radiomics Features. Sci Rep 9:1–16 https://doi.org/10.1038/s41598-019-45766-z
Bologna M, Corino V, Mainardi L (2019) Technical Note: Virtual phantom analyses for preprocessing evaluation and detection of a robust feature set for MRI-radiomics of the brain. Med Phys 46:5116–5123 https://doi.org/10.1002/mp.13834
Cattell R, Chen S, Huang C (2019) Robustness of radiomic features in magnetic resonance imaging: review and a phantom study. Vis Comput Ind Biomed Art 2:19 https://doi.org/10.1186/s42492-019-0025-6
Um H, Tixier F, Bermudez D, Deasy JO, Young RJ, Veeraraghavan H (2019) Impact of image preprocessing on the scanner dependence of multi-parametric MRI radiomic features and covariate shift in multi-institutional glioblastoma datasets. Phys Med Biol 64(16):165011 Published 2019 Aug 21. https://doi.org/10.1088/1361-6560/ab2f44
Yang F, Dogan N, Stoyanova R, Ford JC (2018) Evaluation of radiomic texture feature error due to MRI acquisition and reconstruction: A simulation study utilizing ground truth. Phys Med 50:26–36 https://doi.org/10.1016/j.ejmp.2018.05.017
Traverso A, Kazmierski M, Zhovannik I et al (2020) Machine learning helps identifying volume-confounding effects in radiomics. Phys Med 71:24–30 https://doi.org/10.1016/j.ejmp.2020.02.010
Duron L, Balvay D, Vande Perre S et al (2019) Gray-level discretization impacts reproducible MRI radiomics texture features. PLoS One 14:e0213459 https://doi.org/10.1371/journal.pone.0213459
Tixier F, Um H, Young RJ, Veeraraghavan H (2019) Reliability of tumor segmentation in glioblastoma: Impact on the robustness of MRI-radiomic features. Med Phys:mp.13624 https://doi.org/10.1002/mp.13624
Zhang X, Zhong L, Zhang B et al (2019) The effects of volume of interest delineation on MRI-based radiomics analysis: evaluation with two disease groups. Cancer Imaging 19:89 https://doi.org/10.1186/s40644-019-0276-7
Saha A, Harowicz MR, Mazurowski MA (2018) Breast cancer MRI radiomics: An overview of algorithmic features and impact of inter-reader variability in annotating tumors. Med Phys 45:3076–3085 https://doi.org/10.1002/mp.12925
Veeraraghavan H, Dashevsky BZ, Onishi N et al (2018) Appearance constrained semi-automatic segmentation from DCE-MRI is reproducible and feasible for breast cancer radiomics: a feasibility study. Sci Rep 8:4838 https://doi.org/10.1038/s41598-018-22980-9
Isaksson LJ, Raimondi S, Botta F et al (2020) Effects of MRI image normalization techniques in prostate cancer radiomics. Phys Med 71:7–13 https://doi.org/10.1016/j.ejmp.2020.02.007
Scalco E, Belfatto A, Mastropietro A et al (2020) T2w-MRI signal normalization affects radiomics features reproducibility. Med Phys:14038 https://doi.org/10.1002/mp.14038
Moradmand H, Aghamiri SMR, Ghaderi R (2020) Impact of image preprocessing methods on reproducibility of radiomic features in multimodal magnetic resonance imaging in glioblastoma. J Appl Clin Med Phys 21:179–190 https://doi.org/10.1002/acm2.12795
Um H, Tixier F, Bermudez D, Deasy JO, Young RJ, Veeraraghavan H (2019) Impact of image preprocessing on the scanner dependence of multi-parametric MRI radiomic features and covariate shift in multi-institutional glioblastoma datasets. Phys Med Biol 64:165011 https://doi.org/10.1088/1361-6560/ab2f44
Valladares A, Beyer T, Rausch I (2020) Physical imaging phantoms for simulation of tumor heterogeneity in PET, CT, and MRI: An overview of existing designs. Med Phys:mp.14045 https://doi.org/10.1002/mp.14045
Zhao B, James LP, Moskowitz CS et al (2009) Evaluating variability in tumor measurements from same-day repeat CT scans of patients with non–small cell lung cancer. Radiology 252:263–272 https://doi.org/10.1148/radiol.2522081593
Zwanenburg A (2019) Radiomics in nuclear medicine: robustness, reproducibility, standardization, and how to avoid data analysis traps and replication crisis. Eur J Nucl Med Mol Imaging 46:2638–2655 https://doi.org/10.1007/s00259-019-04391-8
Zhovannik I, Bussink J, Traverso A et al (2019) Learning from scanners: bias reduction and feature correction in radiomics. Clin Transl Radiat Oncol 19:33–38 https://doi.org/10.1016/j.ctro.2019.07.003
Orlhac F, Boughdad S, Philippe C et al (2018) A postreconstruction harmonization method for multicenter radiomic studies in PET. J Nucl Med 59:1321–1328 https://doi.org/10.2967/jnumed.117.199935
Orlhac F, Frouin F, Nioche C, Ayache N, Buvat I (2019) Validation of A Method to Compensate Multicenter Effects Affecting CT Radiomics. Radiology 291:53–59 https://doi.org/10.1148/radiol.2019182023
Mahon RN, Ghita M, Hugo GD, Weiss E (2020) ComBat harmonization for radiomic features in independent phantom and lung cancer patient computed tomography datasets. Phys Med Biol 65:015010 https://doi.org/10.1088/1361-6560/ab6177
Götz M, Maier-Hein KH (2020) Optimal statistical incorporation of independent feature stability information into radiomics studies. Sci Rep 10:737 https://doi.org/10.1038/s41598-020-57739-8
Kalendralis P, Traverso A, Shi Z et al (2019) Multicenter CT phantoms public dataset for radiomics reproducibility tests. Med Phys 46:1512–1518 https://doi.org/10.1002/mp.13385
Zwanenburg A, Vallières M, Abdalah MA et al (2020) The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology:191145 https://doi.org/10.1148/radiol.2020191145
Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–762 https://doi.org/10.1038/nrclinonc.2017.141
Park JE, Kim D, Kim HS et al (2020) Quality of science and reporting of radiomics in oncologic studies: room for improvement according to radiomics quality score and TRIPOD statement. Eur Radiol 30:523–536 https://doi.org/10.1007/s00330-019-06360-z
Funding
None.
Author information
Authors and Affiliations
Contributions
All authors helped in writing and revising the manuscript, drafting of figures and tables. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
van Timmeren, J., Cester, D., Tanadini-Lang, S. et al. Radiomics in medical imaging—“how-to” guide and critical reflection. Insights Imaging 11, 91 (2020). https://doi.org/10.1186/s13244-020-00887-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13244-020-00887-2