Abstract
Background
Hyperspectral imaging (HSI) is a promising non-contact approach to tissue diagnostics, generating large amounts of raw data for whose processing computer vision (i.e. deep learning) is particularly suitable. Aim of this proof of principle study was the classification of hyperspectral (HS)-reflectance values into the human-oral tissue types fat, muscle and mucosa using deep learning methods. Furthermore, the tissue-specific hyperspectral signatures collected will serve as a representative reference for the future assessment of oral pathological changes in the sense of a HS-library.
Methods
A total of about 316 samples of healthy human-oral fat, muscle and oral mucosa was collected from 174 different patients and imaged using a HS-camera, covering the wavelength range from 500 nm to 1000 nm. HS-raw data were further labelled and processed for tissue classification using a light-weight 6-layer deep neural network (DNN).
Results
The reflectance values differed significantly (p < .001) for fat, muscle and oral mucosa at almost all wavelengths, with the signature of muscle differing the most. The deep neural network distinguished tissue types with an accuracy of > 80% each.
Conclusion
Oral fat, muscle and mucosa can be classified sufficiently and automatically by their specific HS-signature using a deep learning approach. Early detection of premalignant-mucosal-lesions using hyperspectral imaging and deep learning is so far represented rarely in in medical and computer vision research domain but has a high potential and is part of subsequent studies.
Similar content being viewed by others
Background
The detection of pathological tissue changes at the macroscopic and microscopic level is one of the foundations of any diagnosis of disease. However, the starting point for any microscopic examination and assessment is the collection of tissue samples. Depending on the aetiology and the affected body region, the precision of diagnosis often differs depending on the invasiveness of sample collection. For example, the gold standard for diagnosis of suspicious oral lesions still consists of incision biopsy with subsequent histopathological examination.
However, this yields disadvantages such as invasiveness and cost intensity. Aiming for a less or non-invasive diagnosis, different procedures (e.g. 1brush biopsy [1,2,3], 2in-vivo staining with toluidine blue or 5-aminolevulinic acid or the use of 3tissue autofluorescence [4, 5]) have been developed during the last years, showing sensitivities of 191 and 391% (77 to 97%) as well as specificities of 191 and 358% (22 to 87%) [6] and providing limited applicability to multifocal lesions with an increased false negative rate [7]. The abovementioned methods have not shown a demonstrable improvement in early cancer detection which is reflected in a consistently high incidence rate of advanced oral squamous cell carcinoma (OSCC). A prerequisite for microscopic assessment of malignancy is the recognition of the physiological status and discrimination against different types of tissue from and to each other. HSI is a promising non-invasive and non-ionizing technique that supports rapid acquisition and analysis of diagnostic information in several fields of clinical medicine [8,9,10,11,12,13]. For life sciences, the various applications of HSI (monitoring of wound healing [14], perfusion monitoring of microvascular flaps [15], assessment on in-sano resection of oesophageal and oropharyngeal carcinomas [16, 17]) have already been successfully demonstrated. HSI includes conventional and spectroscopic methods to obtain both spatial and spectral image information far beyond > 740 nm [18]. Each pixel is assigned a specific vector of radiation values which depends on the chemical material composition of the corresponding localized pixel. This results in large amounts of data, which, however, enables automated tissue recognition through the implementation of machine learning (ML) as an outstanding method that enables researchers to recognize patterns and regularities in increasingly complex data automatically. Modern approaches like deep learning (DL) enlarge origin procedures of ML and move to more complex modelling. As a result, the explicit formulation of rules for high dimensional data can be avoided [19]. DL has become a popular tool in medical data analysis by attaining great achievements in tasks like tissue classification [20, 21] or cancer segmentation [22, 23]. Rapid developments in computer hardware and algorithms have accelerated the success of DL, typically implemented with deep neural networks (DNN) architectures [24, 25]. In medicine DL assists in analysing HSI as a result of the promising non-contact, optical image modality [26,27,28,29]. Since we believe that the differentiation of pathological tissue changes, similar to the assessment of blood parameters, is only possible on the basis of a “healthy” standard, the aim of this study was, for the first time, to create a representative HSI data collection of healthy human fat, muscle and oral mucosa, which will serve as a reference library for the assessment of pathological tissue conditions by processing their spectral characteristics with deep learning methods.
Methods
Tissue samples
In this prospective, non-randomized experimental study, human excess tissue samples were intraoperatively taken and scanned via HSI. The study was approved by the local ethic committee of Rhineland-Palatinate (registration number: 2020–14,952) and was conducted in accordance with the protocol and in compliance with the moral, ethical and scientific principles governing clinical research as set out in the Declaration of Helsinki of 1975 as revised in 1983. The tissue types examined were transverse muscles, fat and mucosa from oral sites (cheek, vestibule, floor of mouth as well as hard and soft palate). These are the tissue structures most frequently exposed during surgical procedures in the head and neck area. Tissue samples that were macroscopically damaged by the use of bipolar and/or monopolar caustics were excluded from the study. To avoid measurement errors due to blood residues, the tissue samples were washed in a 0.9% saline solution before the examination.
Hyperspectral cube processing
Briefly, HSI is based on the assessment of contiguous spectra (i.e. light of different wavelengths) individually re-emitted by molecules, whereby the molecule-specific re-emitted wave spectrum is generated on the basis of the light spectrum of the halogen spotlights initially emitted for examination. The HS cubes were acquired using a TIVITA Tissue system (Diaspective Vision GmbH, Pepelow, Germany), composed with a 120 W halogen illumination source and a radiometrically calibrated 32-bit complementary metal-oxide semiconductor spectrometer capturing images at a resolution of 480 × 640 pixels. Spatial resolution of the CMOS sensor is 22 μm. The hyperspectral cube contains 100 spectral bands, ranging from 500 nm to 1000 nm with a 5 nm sampling interval and illustrates just a tissue sample belongs to one class. In order to provide training data, 316 tissue samples from 174 patients in total were taken, scanned via HSI, inspected histologically and assigned to the examination classes fat, muscle and oral mucosa. The hyperspectral images for the inspected and classified tissue samples were normalized at each wavelength. For accurate and reproducible measurements, the standard measuring distance was 47.5 cm, ensured by two separate indicator lasers (red laser crosshairs and green laser dot) in an overlapped position in which the green laser dot lies in the centre of the red crosshairs (App. 1.) For image analysis, the camera-specific software package (TIVITA™ Suite) was used [14]. After that, overexposure effects were removed and the arithmetic mean of 7 to 8 manually positioned circular regions of interest (ROIs) with a radius of 5 pixels, each distributed across the tissue sample, were calculated to keep the local proximity (Fig. 1). The arithmetic mean corresponded to the recorded wavelength-specific reflectance values of the ROIs per tissue sample. A radius of 5 pixels has proven to be suitable for placing at least five ROIs on the specimens’ surface. ROIs were placed at different positions whereby its number depended to the surface geometry with ROIs along the border and at least one ROI in the specimens’ centre whenever possible to achieve a homogeneous distribution of the measured area (Fig. 2). The ROIs, named numeric patches, are distributed equally over the tissue sample. To further increase the amount of data, each individual ROI per tissue sample was evaluated as an individual sample in the sense of data augmentation (image subdivision). The classification in this study was conducted using a deep neural network built from scratch using H2O Flow (H2O.ai, version 3.32.1.1, for Microsoft Windows, Open source). For running the experiments, a high-performance notebook operating on Windows 10 Professional with 16GB of RAM and an NIVIDA Quadro T1000 GPU was used.
Histology
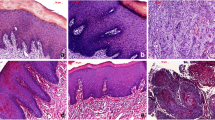
After HSI was performed on the tissue samples, the samples were prepared for histological evaluation. For this purpose, the samples were fixed in formalin, embedded in paraffin, cut in 5 μm steps using a microtome, applied to slides and stained with haematoxylin and eosin (H&E). After slide digitization, they were examined with regard to the tissue composition (i.e. proportion of fat and musculature in the total tissue sample, as well as lack of inflammation). Samples containing other tissue types (e.g. muscle on mucosa specimen), which accounted for > 5% of the examined preparation surface, as well as inflammatory modified mucosa samples, were excluded from the evaluation due to result distortion. Examples of histological sections are shown in Fig. 3.
Statistics
Raw data sets were saved in Excel® sheets (Microsoft Corporation, Redmond, USA) and subsequently transferred into SPSS Statistics® (version 23.0.0.2, MacOS X; SPSS Inc., IBM Corporation, Armonk, NY, USA). Data were expressed as median (MD), mean (M), standard deviation (SD±), minimum (min), maximum (max) and standard error of the mean (SEM). Normal distribution was checked using non-parametric Kolmogorov-Smirnov test (KS test) and results were analysed for statistical significance by the use of analysis of variance (ANOVA (#)), unpaired non-parametric Mann-Whitney U tests = ($) and students’ t-test = (*). P-values of ≤0.05 were termed significant. For a proof of principle study sample size calculation is not practicable, thus this study is in accordance to other published proof of concept works dealing with similar group sizes [30, 31]. Line charts were used for illustration purposes.
Neural network
The processed training data were split in ratio 85:8:7 (training, validation, test), while training patches used to fit the networks weights. In addition, validation patches intended to optimize hyperparameter and test patches used for evaluation purpose of the fitted model. The data split procedure was performed considering leave-patient-out approach. This feed-forward neural network was trained stepwise in mini-batches of 64 numeric patches with a patient-ID based stratified cross-validation paradigm for 4000 epochs using early stopping techniques for plateauing. A uniform adaptive distribution was used to set initial weights. To improve generalisation, a dropout rate of 30% was applied after first and 20% after second and third hidden layer. Each neuron was activated using rectified linear unit (ReLU). Training was performed using balanced classes and an adaptive learning rate for stochastic gradient descent optimization [32] with momentum of 0.99 and a smoothing factor equal to 1 × 10− 8. Furthermore, L1 and L2 regularisation terms of 5 × 10− 3 each for reducing the cross-entropy loss was set up as shown in Fig. 6. Before training, hyperparameter such as neural network architecture, dropout quantity, activation function, learning rate related parameters, regularization terms and batch size were adjusted with systematic grid search technique by taking care of the overfitting gap between training and validation loss. Every epoch, the validation performance was evaluated but shuffling was disabled caused by higher losses. The final softmax-layer predicts inputs with respect to the highest probability of each class. Testing is done after training and validation loss converges equally. As plotted in Fig. 6, the model generalises in a fast manner. The deep neural network performance was evaluated on the optimal checkpoint applied on fully independent test data to calculate accuracy (Acc; \( Accuracy=\frac{TP\ \left( true\ positive\right)+ TN\left( true\ negative\right)}{Total\ No. of\ patches} \)), specificity (Spec; \( Specificity=\frac{TN}{TN+ FP} \)) and sensitivity (Sens; \( Sensitivity=\frac{TP}{TP+ FN} \)).
Results
Ex-vivo fat, muscle and mucosa - spectral signatures
The spectral signature for each class obtained from the hyperspectral imaging processing is plotted in Fig. 4.
When comparing fat (group-1) and muscle (group-2), as well as muscle (group-2) and mucosa (group-3) the mean reflectance values differed significantly at all wavelengths (p < .001). In contrast, the spectral signatures of fat (group-1) and mucosa (group-3) appeared much more similar in the graphical overview (Fig. 4), but also differed significantly at most wavelengths (500 nm to 520 nm, 545 nm to 790 nm, 910 nm to 940 nm and 950 nm to 995 nm (p < .001$*)). Detailed information on mean values, standard deviations and individual significances are available on request.
Neural network
The amount of training data and the distribution among classes are shown in Table 1. As shown in Fig. 5, a light-weight architecture was implemented for the studies.
Evaluation
Classification scores were calculated using values reported in Table 2 based on common equations [20]. Interestingly, a comparatively high classification error between musculature and mucosa was found (error 0.21). Table 3 shows the evaluation results.
Discussion
Although non-invasive examination methods (e.g. brush biopsy or tissue autofluorescence) have been developed to monitor oral potentially malignant disorders (OPMD), histopathological examination still represents the diagnostic gold standard for lesion monitoring. HSI is a non-invasive, non-contact optical wide-field modality that holds the potential to sense tumours in varying depth using visible spectrum (VIS) and near infrared (NIR) light and therefore to improve OPMD monitoring, early oral cancer diagnosis and reduce cancer-related mortality and morbidity [20, 33]. To process the extensive amount of spectra-spatial data cube information efficiently and automatically, the use of DL methods is suitable. Knowledge of the spectral characteristics of its main components (oral mucosa, muscle and fat) is essential for the assessment of complex, pathologically altered oral mucosa. This study presents a method to classify 316 fresh surgical ex-vivo human oral tissue samples’ reflectance values into fat, muscle and mucosa, based on HSI data of a representative number of samples. This light-weight deep learning (DL) approach achieved an overall accuracy score over 87% in an ordinary and time-saving manner, but with commonly used optimization techniques. Together with a lot more patient’s metadata and a hyperspectral database of many samples from different individuals, the clinical use for non-invasive, automated assessment of oral mucosal changes would be a conceivable and tangible approach. Therefore, the data should simultaneously serve as a kind of hyperspectral reference library for future applications such as the in-vivo examination of chronic inflammatory oral diseases, the intraoperative assessment of surgical safety margins or the intraoperative assessment of lymph nodes when deciding to include higher lymph node levels. Using a light-weight 6-layer deep neural network with only 10,445 parameters trained about 4000 epochs, we can distinguish tissue samples in fat, muscle and oral mucosa with a class accuracy over 80% each. Though, the presented solution provided fully independent test data [20] and this study’s training was not affected by overfitting, however the optimisation potential can be increased even further as shown in Fig. 6. A limitation of this approach is the high-dimensional feature space with about 100 (500 to 1000 nm) wavelengths. Further experiments with significantly reduced number of features based on feature selection and dimensionality reduction processes present a more complex problem the research group is currently working on. By gaining more sample data, the generalisation potential of the deep learning approach and the numerical discrepancy between sensitivity and specificity regarding muscle and mucosa as shown in Table 3 could improve. However, the false-positive classification of muscle and mucosa in the test data set (error 0.21) compared to fat versus muscle or mucosa (error 0.10) proved to be relatively high (Table 2). One explanation for this would be the anatomically determined increased contamination of the mucosal tissue with musculature, which was mainly found in the area of the cheeks and soft palate mucosa.
Preliminary results of this proof of concept study demonstrate the ability of deep learning methodology for discriminating between hyperspectral tissue samples. Future studies are going to deal with classification between healthy, dysplastic and cancerous tissue samples based on Convolutional Neural Network (CNN) approaches with non-pre-processed hyperspectral cube data.
Conclusions
The processing of hyperspectral tissue data by a neural network allows the automated classification of tissue samples with increasing model accuracies. Our research deals with the classification of healthy oral fat, muscle and mucosa by using of HS-reflectance values and thus differs to alternative approaches build upon HS-images and CNNs. With an accuracy of > 80% our model in comparison to Halicek et al. [18] lacks accuracy but relies on numeric patches characterises fast training and feed-forward phase. In order to eliminate the accuracy gap, future studies will emphasize convolutional building blocks and image data. Provided that sufficient hyperspectral training data of dys- and anaplastic mucosa samples are available, the combination of hyperspectral imaging and deep learning can thus represent a promising method for a real time non-invasive assessment of oral mucosal changes.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- Acc:
-
Accuracy
- CNN:
-
Convolutional Neural Network
- DL:
-
Deep Learning
- DNN:
-
Deep Neural Networks
- H&E:
-
Haematoxylin and Eosin
- HS:
-
Hyperspectral
- HSI :
-
Hyperspectral Imaging
- OSCC:
-
Oral Squamous Cell Carcinoma
- m:
-
Mean
- Max:
-
Maximum
- MD:
-
Median
- Min:
-
Minimum
- ML:
-
Machine Learning
- NIR:
-
Near Infrared
- OPMD:
-
Oral Potentially Malignant Disorders
- ReLU:
-
Rectified Linear Unit
- ROIs:
-
Regions of Interest
- SD:
-
Standard Deviation
- Sens:
-
Sensitivity
- Spec:
-
Specificity
- VIS:
-
Visible Spectrum
References
Remmerbach TW, Mathes SN, Weidenbach H, Hemprich A, Bocking A. Noninvasive brush biopsy as an innovative tool for early detection of oral carcinomas. Mund Kiefer Gesichtschir. 2004;8:229–36.
Kammerer PW, Koch FP, Santoro M, Babaryka G, Biesterfeld S, Brieger J, et al. Prospective, blinded comparison of cytology and DNA-image cytometry of brush biopsies for early detection of oral malignancy. Oral Oncol. 2013;49(5):420–6. https://doi.org/10.1016/j.oraloncology.2012.12.006.
Kammerer PW, Rahimi-Nedjat RK, Ziebart T, Bemsch A, Walter C, Al-Nawas B, et al. A chemiluminescent light system in combination with toluidine blue to assess suspicious oral lesions-clinical evaluation and review of the literature. Clin Oral Investig. 2015;19(2):459–66. https://doi.org/10.1007/s00784-014-1252-z.
Patton LL, Epstein JB, Kerr AR. Adjunctive techniques for oral cancer examination and lesion diagnosis: a systematic review of the literature. J Am Dent Assoc. 2008;139(7):896–905; quiz 993-894. https://doi.org/10.14219/jada.archive.2008.0276.
Koch FP, Kaemmerer PW, Biesterfeld S, Kunkel M, Wagner W. Effectiveness of autofluorescence to identify suspicious oral lesions--a prospective, blinded clinical trial. Clin Oral Investig. 2011;15(6):975–82. https://doi.org/10.1007/s00784-010-0455-1.
Cicciù M, Cervino G, Fiorillo L, et al. Early Diagnosis on Oral and Potentially Oral Malignant Lesions: A Systematic Review on the VELscope® Fluorescence Method. Dent J (Basel). 2019;7(3):93. https://doi.org/10.3390/dj7030093. Published 2019 Sep 4.
Macey R, Walsh T, Brocklehurst P, Kerr AR, Liu JL, Lingen MW, Ogden GR, Warnakulasuriya S, Scully C. Diagnostic tests for oral cancer and potentially malignant disorders in patients presenting with clinically evident lesions. Cochrane Database Syst Rev. 2021;7:CD010276. https://doi.org/10.1002/14651858.CD010276.pub2.
Ishida T, Kurihara J, Viray FA, Namuco SB, Paringit EC, Perez GJ, et al. A novel approach for vegetation classification using UAV-based hyperspectral imaging. Comput Electron Agric. 2018;144:80–5. https://doi.org/10.1016/j.compag.2017.11.027.
Miljković V, Gajski D: Adaptation of Industrial Hyperspectral Line Scanner for Archaeological Applications. ISPRS - International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences 2016, XLI-B5:343–345.
Pavurala N, Xu X, Krishnaiah YSR. Hyperspectral imaging using near infrared spectroscopy to monitor coat thickness uniformity in the manufacture of a transdermal drug delivery system. Int J Pharm. 2017;523(1):281–90. https://doi.org/10.1016/j.ijpharm.2017.03.022.
de la Ossa MAF, Amigo JM, Garcia-Ruiz C. Detection of residues from explosive manipulation by near infrared hyperspectral imaging: a promising forensic tool. Forensic Sci Int. 2014;242:228–35. https://doi.org/10.1016/j.forsciint.2014.06.023.
Teke M, Deveci HS, Haliloglu O, Gurbuz SZ, Sakarya U: A Short Survey of Hyperspectral Remote Sensing Applications in Agriculture. Proceedings of 6th International Conference on Recent Advances in Space Technologies (Rast 2013) 2013:171–176.
Qin JW, Kim MS, Chao KL, Chan DE, Delwiche SR, Cho BK. Line-scan hyperspectral imaging techniques for food safety and quality applications. Applied Sciences-Basel. 2017;7.
Holmer A, Marotz J, Wahl P, Dau M, Kammerer PW. Hyperspectral imaging in perfusion and wound diagnostics - methods and algorithms for the determination of tissue parameters. Biomed Tech (Berl). 2018;63(5):547–56. https://doi.org/10.1515/bmt-2017-0155.
Thiem D, Frick RW, Goetze E, Gielisch M, Al-Nawas B, Kämmerer PW. Hyperspectral analysis for perioperative perfusion monitoring-a clinical feasibility study on free and pedicled flaps. Clin Oral Investig. 2021;25(3):933–45. https://doi.org/10.1007/s00784-020-03382-6.
Fei B, Lu G, Wang X, Zhang H, Little JV, Patel MR, et al. Label-free reflectance hyperspectral imaging for tumor margin assessment: a pilot study on surgical specimens of cancer patients. J Biomed Opt. 2017;22(08):1–7. https://doi.org/10.1117/1.JBO.22.8.086009.
Maktabi M, Kohler H, Ivanova M, Jansen-Winkeln B, Takoh J, Niebisch S, et al. Tissue classification of oncologic esophageal resectates based on hyperspectral data. Int J Comput Assist Radiol Surg. 2019;14(10):1651–61. https://doi.org/10.1007/s11548-019-02016-x.
Halicek M, Fabelo H, Ortega S, Callico GM, Fei B. In-vivo and ex-vivo tissue analysis through hyperspectral imaging techniques: revealing the invisible features of Cancer. Cancers (Basel). 2019;11(6). https://doi.org/10.3390/cancers11060756.
Plaß B, Zschiesche K, Altinbas T, Karla D, Rau L, Schlüter M: Artificial intelligence as a strategy in engineering geodesy - first steps in the railway environment. ZFV - Journal for Geodesy, Geoinformation and Land Management 2020, 04.
Halicek M, Little JV, Wang X, Chen AY, Fei B. Optical biopsy of head and neck cancer using hyperspectral imaging and convolutional neural networks. J Biomed Opt. 2019;24(03):1–9. https://doi.org/10.1117/1.JBO.24.3.036007.
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. https://doi.org/10.1016/j.media.2017.07.005.
G ST, Biswas M, O GK, Tiwari A, H SS, Turk M, Laird JR, Asare CK, A AA, N NK, et al: A Review on a Deep Learning Perspective in Brain Cancer Classification. Cancers (Basel) 2019, 11.
Petersson H, Gustafsson D, Bergstrom D: Hyperspectral image analysis using deep learning — A review. In 2016 Sixth International Conference on Image Processing Theory, Tools and Applications (IPTA). pp. 1–6; 2016:1–6.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44. https://doi.org/10.1038/nature14539.
Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60(6):84–90. https://doi.org/10.1145/3065386.
Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19(1):221–48. https://doi.org/10.1146/annurev-bioeng-071516-044442.
Lu G, Fei B. Medical hyperspectral imaging: a review. J Biomed Opt. 2014;19(1):10901. https://doi.org/10.1117/1.JBO.19.1.010901.
Gao L, Smith RT. Optical hyperspectral imaging in microscopy and spectroscopy - a review of data acquisition. J Biophotonics. 2015;8(6):441–56. https://doi.org/10.1002/jbio.201400051.
Martin ME, Wabuyele MB, Chen K, Kasili P, Panjehpour M, Phan M, et al. Development of an advanced hyperspectral imaging (HSI) system with applications for cancer detection. Ann Biomed Eng. 2006;34(6):1061–8. https://doi.org/10.1007/s10439-006-9121-9.
Lee SY, Pakela JM, Helton MC, Vishwanath K, Chung YG, Kolodziejski NJ, Stapels CJ, McAdams DR, Fernandez DE, Christian JF, O'Reilly J, Farkas D, Ward BB, Feinberg SE, Mycek M. Compact dual-mode diffuse optical system for blood perfusion monitoring in a porcine model of microvascular tissue flaps. J Biomed Opt. 2017;22(12):121609.
Khan M, Pretty CG, Amies AC, Balmer J, Banna HE, Shaw GM, et al. Proof of concept non-invasive estimation of peripheral venous oxygen saturation. Biomed Eng Online. 2017;16(1):60. https://doi.org/10.1186/s12938-017-0351-x.
Zeiler, M D: ADADELTA: An Adaptive Learning Rate Method. Available online at http://arxiv.org/pdf/1212.5701v1; 2012.
Gimi B, Krol A, Lu G, Qin X, Wang D, Muller S, Zhang H, Chen A, Chen ZG, Fei B. Hyperspectral imaging of neoplastic progression in a mouse model of oral carcinogenesis. In Medical imaging 2016: biomedical applications in molecular, structural, and functional Imaging; 2016.
Acknowledgements
Not applicable.
Funding
Intramural level 1 funding to support young researchers at the University Medical Centre Mainz, Germany. Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
All authors read and approved the final manuscript. Daniel G.E. Thiem (daniel.thiem@uni- mainz.de) takes responsibility for the integrity of the work as a whole, from inception to finished article. Daniel G.E. Thiem substantially contributed to the conception and the design of the study, acquisition, analysis and interpretation of data as well as drafted the manuscript. Paul Römer substantially contributed to acquisition, analysis and interpretation of data. Matthias Gielisch substantially contributed to acquisition, analysis and interpretation of data. Bilal Al-Nawas contributed to data interpretation and substantively revised the manuscript. Martin Schlüter contributed to data interpretation and substantively revised the manuscript. Bastian Plaß substantially contributed to data analysis and data interpretation and substantively revised the manuscript. Peer W. Kämmerer substantially contributed to data interpretation to the conception and the design of the study and substantively revised the manuscript.
Authors’ information
The results of the present study are part of the doctoral thesis of the co-author and student of the University of Mainz, Paul Römer, and as such will be made publicly available as part of the doctorate.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the local ethic committee of Rhineland-Palatinate (registration number: 2020–14952) and was conducted in accordance with the protocol and in compliance with the moral, ethical and scientific principles governing clinical research as set out in the Declaration of Helsinki of 1975 as revised in 1983. Informed consent was obtained from all individual participants included in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Appendix 1.
The figure shows the measurement situation with laser marking.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Thiem, D.G.E., Römer, P., Gielisch, M. et al. Hyperspectral imaging and artificial intelligence to detect oral malignancy – part 1 - automated tissue classification of oral muscle, fat and mucosa using a light-weight 6-layer deep neural network. Head Face Med 17, 38 (2021). https://doi.org/10.1186/s13005-021-00292-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13005-021-00292-0