Abstract
Inflammatory bowel disease (IBD) is a chronic gastrointestinal condition characterized by severe gut inflammation, commonly presenting as Crohn’s disease, ulcerative colitis or categorized as IBD- unclassified. While various treatments have demonstrated efficacy in adult IBD patients, the advent of anti-TNF therapies has significantly revolutionized treatment outcomes and clinical management. These therapies have played a pivotal role in achieving clinical and endoscopic remission, promoting mucosal healing, averting disease progression, and diminishing the necessity for surgery. Nevertheless, not all patients exhibit positive responses to these therapies, and some may experience a loss of responsiveness over time. This review aims to present a comprehensive examination of predictive biomarkers for monitoring the therapeutic response to anti-TNF therapy in IBD patients. It will explore their limitations and clinical utilities, paving the way for a more personalized and effective therapeutic approach.
Similar content being viewed by others
Introduction
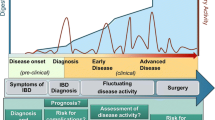
Inflammatory bowel disease (IBD) is a chronic, recurrent inflammatory disorder of the gastrointestinal (GI) tract, resulting in a wide range of clinical symptoms [1]. The global incidence of IBD is increasing, affecting even younger individuals and children, possibly due to various factors such as urbanization, westernization, dietary changes, increased antimicrobial exposure, and alterations in the host-microbial balance [2]. IBD is characterized by functional damage to the GI tract and intestinal epithelium, requiring lifelong medication [1]. Based on clinical symptoms, endoscopy imaging, and histology, IBD is broadly categorized into three major subtypes, Ulcerative Colitis (UC), which primarily affects the colon and Crohn’s Disease (CD) which affects multiple sites within the GI tract [3]. This third subtype, where histology assessments do not categorize patients into either Ulcerative Colitis (UC) or Crohn’s Disease (CD), is referred to as ‘‘Inflammatory Bowel Disease, type unclassified’’ or ‘‘Undetermined’’ (IBD-U) [2]. Diagnosing patients with IBD-U is challenging due to the ambiguous nature of their symptoms. They may display characteristics of both UC and CD, or present with atypical symptoms that do not fit into either category [4]. The disease follows a pattern of remission and relapse and often leads to complications and the need for surgical intervention in a majority of cases [5].
Advances in our understanding of the underlying immunopathogenic mechanisms of IBD have paved the way for the development of targeted therapies. The initial class of biological therapies approved for the treatment of IBD patients focused on inhibiting the pro-inflammatory cytokine tumor necrosis factor (TNF) in 1998 [6]. This biologic has revolutionized IBD treatment, markedly enhancing response rates and increasing the chances of remission among patients, and exerting a significant influence on the existing therapeutic algorithms [7]. Anti-TNF-alpha therapies, such as Infliximab and Adalimumab, have proven effective for moderate to severe CD cases [8]. Infliximab, the first biologic used for IBD treatment, is a genetically engineered chimeric immunoglobulin (Ig)G1 anti-human TNF agent [9]. It can activate complement and target cells expressing membrane-bound TNF-alpha, effectively downregulating inflammatory mechanisms throughout the mucosal layer [10, 11].
Anti-TNF therapies are generally well-tolerated and landmark studies like the ACCENT I and II trials have contributed significantly to our understanding of their effectiveness [12, 13], however, not all patients respond well to these biologics [14,15,16]. In addition, these agents carry risks of infection and malignancy, contributing to treatment costs [17]. Therefore, effective management of patients requires careful consideration of essential factors, including the potential for immunogenicity, the safety profile of treatments, and the optimal therapeutic duration. Over the past decade, numerous studies have identified a range of predictive biomarkers that hold the promise of delivering personalized and effective treatments for patients [8, 18]. This review will provide a comprehensive summary of findings derived from various studies in predicting response to anti-TNF therapies. Additionally, the review will highlight the limitations and clinical utilities of these predictive biomarkers in monitoring optimized patient outcomes and delivering personalized care.
Loss of response to anti-TNF therapy
Over the last two decades, anti-TNF-alpha therapies have emerged as highly promising therapies for autoimmune and inflammatory conditions such as IBD, rheumatoid arthritis, psoriasis, and ankylosing spondylitis. However, a significant challenge lies in the fact that not all patients respond favorably to these therapies [19]. Approximately 30% of IBD patients do not respond to anti-TNF therapy (primary non-responders), and nearly half of those who initially benefit from these drugs experience a loss of clinical benefits within the first year, necessitating dose escalation or treatment alteration, referred to as “secondary loss of response” [20, 21]. Therefore predicting response to IBD therapy is crucial to avoid severe IBD-associated complications such as surgery and hospitalizations. Indeed these predictive biomarkers are instrumental in administering the appropriate treatment to the right patient at the right time. Moreover, considering that a significant number of IBD patients either develop intolerance or experience a loss of response to treatment over time, the ability to predict treatment response opens avenues for more personalized treatment options [22]. Recent studies have unveiled a wealth of potential markers, encompassing both genetic and non-genetic factors, that show promise as indicators for predicting responses to anti-TNF therapies.
Predictive genetic markers
Some genetic markers were shown to have a correlation with predictive primary response to the biological treatment in IBD patients. For example, a favorable clinical response was observed to be positively correlated with polymorphisms in genes like FCGR3A, TLR4, TNFRSF1A, IFNG, IL6, and IL1B. Conversely, variants of TLR2 and TLR9 showed a negative correlation with treatment response and are categorized as primary non-responder [23]. Although, most genetic predictive markers are related to cytokines/chemokines or their receptors and immunoglobulin receptors, including TNF gene, TNFRSF1B gene, ATG16L1, and ATG16L2 gene, apoptosis genes, NOD2/CARD15 genes, IL23R and IL12 genes and Fc receptors related genes [24,25,26]. For example, genetic variants in TNF or TNFRSF1B gene and NFkB gene, may influence the level of TNF-alpha production or affect the binding affinity of TNF-alpha blockers to TNF-alpha receptor and, in turn, affect the primary response to anti-TNF (Infliximab, adalimumab, and golimumab) therapy in CD patients [27,28,29] (Table 1), or UC patients [30] (Table 2). Another polymorphism in NOD2/CARD15 gene has been linked to CD and may influence the response to anti-TNF-alpha therapy in individuals with this mutation, while polymorphisms in the IL23 receptor have been linked to the response to Infliximab treatment in UC patients [31]. In addition, when examining the Fas ligand gene, a CC genotype related to apoptosis has been positively associated with non-primary responsiveness to Infliximab. In contrast, individuals with a TC or TT genotype have been predictive of a positive response to anti-TNF therapy [27]. Moreover, investigations into the FCGR3A and ATG16L1 gene polymorphisms have revealed their impact on response to anti-TNF treatment. [25, 26, 32]. IBD patients with ATG16L1 T/T and C/T genotypes have demonstrated significantly higher CRP levels and a more favorable response to Adalimumab compared to those with the C/C genotype [24, 33]. Similarly, some specific genetic variations in genes like TNF-β and TNFRSF1B (rs1061624_A‐rs3397_T), in conjunction with a minor allele (A) polymorphism of the TNF gene (rs1800629), have been linked to the prediction of non-responsiveness to anti-TNF (Infliximab) therapy among patients with CD [27,28,29]. Conversely, a heterozygous genotype of IL12B—10,993 G > C (rs3212217) has shown a positive correlation with non-responsiveness to anti-TNF therapy in patients with UC [30, 34]. Hence, patients with these genetic profile are categorized as primary non-responders to anti-TNF therapy.
Fecal markers
Although, fecal calprotectin and lactoferrin are commonly considered surrogate markers for assessing luminal disease activity [35]. These markers have been suggested as potential indicators of how individuals with CD [36] and UC [37] patients might respond to anti-TNF therapy [38]. However, the results from various studies present a mixed picture. In some instances, higher calprotectin levels were associated with a better response [39], while in others, the relationship was the opposite [40, 41]. Additionally, certain studies have failed to confirm any of these associations [42,43,44]. In summary, it appears that fecal calprotectin levels alone may not reliably predict an individual patient's response to anti-TNF therapy. While analyzing metabolic network reconstruction and metabolic profiles in fecal samples might offer insights into which patients with IBD are more likely to achieve clinical remission with anti-TNF therapy [45].
Immune markers
Monitoring changes in blood or mucosal parameters has proven valuable in evaluating the effectiveness of anti-TNF therapy. Effective anti-TNF therapy results in a decrease in TNF-α and interferon (IFN)-γ levels, signifying reduced inflammation at the mucosal level [41, 46]. In addition, Th17 signature cytokines, including IL-17A, IL-17B, IL-17D, and IL-17F, have also shown potential as candidate biomarkers for assessing the efficacy of anti-TNF therapy in patients with IBD [38]. They can modulate the expression of various cytokines and chemokines associated with autoimmunity and immune functions. In non-responders compared to responders, genes in the chemokine signaling pathway are down-regulated. Normally, high mucosal expression of IL7R, IL-6, sTNFR2, IL-1, IL-10, IL-8, and oncostatin M (OSM) signaling before treatment is associated with primary non-response to anti-TNF therapy in CD and UC patients [47,48,49,50,51]. Although these immune biomarkers might be considered as biomarker candidates for the evaluation of anti-TNF therapeutic efficacy in IBD patients, however, the clinical utilities remain a challenge, and further validation studies are required to confirm their potential as a biomarker. Currently, to the best of our knowledge no comparative studies assess the efficacy of anti-TNF treatments in IBD patients with normalized versus high TNF levels. Moreover, treatment decisions normally rely on disease activity evaluations based on clinical symptoms and endoscopic observations. It remains uncertain whether IBD patients with normalized mucosal TNF levels can benefit from anti-TNF therapy.
Protein markers
Proteomics can provide valuable insights into treatment responses to anti-TNF therapy, disease mechanisms, and individualized patient care; hence proteomics profiling is rapidly being explored to identify predictive biomarkers for monitoring or therapeutic response [52, 53]. For example, recent studies have identified multiple proteins markers (ANG1, ANG2, CRP, CEACAM1, SAA1, EMMPRIN, TGFA, MMP1, MMP2, MMP3, MMP9, IL-6, IL-7, sCD40L, PF4, complement C4-B, apolipoprotein A-I, apolipoprotein E, serotransferrin, beta-2-glycoprotein 1 and clusterin and VCAM1) in CD patients responding to Infliximab [53, 54]. Although proteomics has great potential for evaluating therapeutic responses, individual responses to therapy can be influenced by multiple factors beyond proteomics, such as genetics, lifestyle, microbiome, and environmental factors. Hence proteomics markers alone are not enough to predict the efficacy of biologics, an integration of proteomics data with other omics data as well as clinical information will provide better predictive universal markers for therapeutic response in IBD patients.
Microbial biomarkers
Although the etiology of IBD remains exclusive, the complex interaction of the gut microbial communities with immune cells can influence the disease severity and susceptibility to immune therapy in IBD patients [2]. The relationship between the gut microbiome and anti-TNF therapy is complex and not fully understood, however, research on the role of the microbiome in the context of anti-TNF-alpha therapy is continuously evolving. The gut microbiome, which consists of trillions of microorganisms residing in the gastrointestinal tract, plays a crucial role in immune modulation [55,56,57,58], intestinal inflammation [59], and response to immunomodulatory therapies [2, 60, 61], including anti-TNF-alpha drugs [62]. Emerging studies have suggested that certain microbial markers may be associated with treatment outcomes and response to anti-TNF-alpha therapy [62, 63]. For example; diverse microbes are generally associated with a healthier gut and patients with a more diverse gut microbiome respond better to anti-TNF-alpha therapy, while the presence of certain microbial species or groups of bacteria are associated with either a positive or negative response to anti-TNF-alpha therapy [63]. For example, a higher abundance of Faecalibacterium prausnitzii, Ruminococcus bromii, Bifidobacterium ssp., Clostridium colinum, Eubacterium rectale, and lower levels of Streptococcus mitis have have been linked to a better treatment response to anti-TNF therapy in IBD patients [64,65,66]. Moreover, increased levels of butyrate‐producing species (such as Roseburia inulinivorans and Burkholderiales) and a higher level of branched-chain amino acids are shown to be positively correlated with the clinical response to Vedolizumab [67]. In contrast, short-chain fatty acids producing bacteria such as Lachnospiraceae and Ruminococcaceae families, have been found to be associated with primary non-responders to the anti-TNF-α therapies [68]. In addition, the patients with gut microbial dysbiosis [69] or with additional fibrostenotic disease often exhibit no-response or poor response to anti-TNF therapy [70,71,72,73]. The balance between the two dominant microbial groups mainly Prevotella and Bacteroides in the gut has also been explored in relation to anti-TNF therapeutic response in patients [74, 75]. Higher levels of Prevotella relative to Bacteroides have been associated with better response to biologics [76]. Interestingly, a recent study by Caenepeel et al. (2024) investigated various combinations of clinical and microbial data to predict the efficacy of TNF-alpha (infliximab, adalimumab, and golimumab) in patients with both CD and UC [77]. Their model, integrating clinical parameters, stool features (moisture and calprotectin), and identification of microbial dysbiosis, achieved a 73.9% accuracy rate in predicting treatment outcomes with different biologicals. Notably, the study unveiled the significant microbiota-modulating effect of anti-TNF-α therapy, while vedolizumab appeared less effective in patients with dysbiotic microbiota. In addition, abundance of Fusobacterium also correlated with fecal calprotectin concentrations and postoperative relapse in patients with CD [78].
In addition to changes in the bacteriome, dysbiosis in the mycobiome also plays a crucial role in the pathophysiology of IBD and may influences responses to therapy. For instance, an increase in Basidiomycota and a decrease in Ascomycota, especially Saccharomyces cerevisiae, are associated with disease activity and biologic response [79]. Shifting Basidiomycota to Ascomycota ratios are observed during remission, hence indicating a potential marker for fungal dysbiosis [80]. Notably, the abundance of Candida albicans in IBD is reduced in patients who are primary responders to anti-TNF-alpha therapies [80]. While viruses are more abundant in the microbiome compared to bacteria, there is limited exploration of the inflammatory bowel disease (IBD) virome. Despite the bacteriome being a more accurate reflection of IBD activity, specific viruses such as those from the Retroviridae family are associated with CD [81]. However, their precise connection to the response to anti-TNF-alpha therapies remains unclear [82]. Although finding microbial biomarkers for clinical response to biological therapies seems to be a promising target, considering the diversity of changes in different populations and lack of statistical power among studies, it is important to note that research in this area is ongoing, and findings may not yet have fully translated into routine clinical practice.
Anti-drug-antibodies
In some patients, anti-TNF therapy can stimulate the immune system to produce neutralizing anti-drug antibodies (ADAs) [83, 84]. This process is known as immunogenicity, which can be modulated by various factors and is a crucial consideration in IBD treatment especially with biological therapies. For example, certain genetic factors have been associated with an increased risk of ADA formation in CD patients undergoing anti-TNF therapy. These factors include HLA-DQA1*05, HLA-DRB1 alleles, and polymorphisms at the FCGR3A locus, which encodes the IgG Fc receptor IIIa [85, 86]. Monoclonal antibodies, particularly chimeric antibodies like Infliximab, can stimulate the production of ADA [87], which can lead to treatment inefficacy or secondary loss of response (Fig. 1), therefore, monitoring of serum ADAs and anti-TNF levels in case of > 3 μg/ml Infliximab therapy in IBD patients is an important parameter to optimum treatment response in IBD patients [88]. Although the presence of ADAs in patients is not permanent, it may disappear within a year after discontinuing the anti-TNF therapy [89]. In addition, some studies have also reported a link between poor efficacy of Infliximab therapy and anti-OmpC (Escherichia coli outer membrane porin) antibodies as well as anti-neutrophil cytoplasmic antibodies (pANCA) [90,91,92]. However, testing for pANCA positivity to predict non-response to Infliximab therapy showed a sensitivity of only 25% and a specificity of 85% resulting in the pANCA testing not commonly being employed in routine clinical practice for predicting therapy response [93].
Others
Loss of responsiveness to anti-TNF therapies in IBD might also be due to increased activity of matrix metalloproteinases (MMPs), which break down anti-TNF antibodies [94], and heightened clearance of TNF-anti-TNF antibody complexes through Fc receptor-mediated endocytosis, leading to proteolytic degradation [77] and contributing to secondary loss of response in IBD patients [95].
MicroRNAs (miRNAs) are small non-coding RNAs, and play an important role in pro-inflammatory cytokine production and the inflammatory processes in IBD patients [96]. In recent years, circulating and fecal miRNAs have emerged as potential novel biomarkers for predicting therapeutic responses in IBD patients [97]. For example, Batra et al. demonstrated significant changes in the expression of seven miRNAs after treatment in responders compared to non-responders, within a small cohort of pediatric IBD patients receiving anti-TNF therapies [98]. However, another study focusing on miRNA polymorphisms and their association with anti-TNF treatment response in CD did not find any correlations between identified miRNA polymorphisms (miR-146 rs2910164, miR-196a rs11614913, miR-221 rs113054794, and miR-224 rs188519172) and patients' responses to anti-TNF mAbs in CD patients [99]. Therefore, further studies are required to fully investigate and validate the utility of miRNAs as predictive markers for treatment outcomes in IBD.
Previous surgery or exposure to anti-TNF therapy in IBD patients has been associated with an increased risk of both primary non-responders and secondary loss of response in subsequent anti-TNF therapies [100,101,102]. The effectiveness of a second anti-TNF treatment appears to be influenced by the reason for switching, with higher remission rates observed in patients who discontinued anti-TNF therapy due to intolerance (61%) compared to those with secondary (45%) or primary failure (30%) [103]. Additionally, concomitant immunomodulator therapy, as shown in the SONIC trial for CD patients [104] and the SUCCESS trial for UC patients [105], has been demonstrated to enhance corticosteroid-free remission rates when combined with infliximab. Although, these data suggest that non-genetic, environmental factors can influence the response to anti-TNF therapies in IBD patients, they do not pinpoint any specific markers or concrete indicators.
Future directions
In summary, the discovery of anti-TNF therapies has resulted in remarkable changes in the treatment approach of IBD, shifting from gradual attainment of symptomatic clinical remission to achieving sustained remission. This transformation has enabled the realization of short- and long-term clinical and endoscopic remission, a goal once considered unattainable [53, 106]. As a result, anti-TNF therapies occupy a central position within the current IBD treatment paradigm, and they can be further enhanced through strategies such as continuous monitoring of disease progression and/or response to therapy [107].
Although the use of laboratory biomarkers shows potential for enhancing the evaluation and customization of anti-TNF therapy for IBD, the search of predictive biomarkers is still in its nascent phase. Significant challenges remain to be addressed.. Many of the identified biomarkers primarily indicate a generalized inflammation and lack specificity for IBD. In reality, the response to this therapy is influenced by a variety of factors, including disease-related characteristics, biochemical markers, genetics, microbial composition, metabolic factors, and local mucosal conditions. As a consequence, this could be the underlying reason why none of the identified biomarkers have been incorporated into routine clinical practice as definitive tools for enabling personalized treatment approaches. The recent model reported by Caenepeel et al. which integrated a combination of clinical data, stool characteristics (microbial load, moisture content, and calprotectin concentration), and fecal microbiota, enabled the prediction of treatment response to biological therapies [77]. However, it still classified more than half of IBD patients as refractory to all anti-inflammatory therapies studied [77]. To address this, future efforts should focus on robustly validating biomarkers through well-designed studies involving a significant number of IBD patients.
Nevertheless, despite their pivotal role in the current IBD treatment paradigm, certain aspects of anti-TNF use still pose unanswered questions. These include considerations about the timing, dosing, monitoring, and addressing issues related to loss of treatment response, such as identifying risk factors, biomarkers, mechanistic insights, and devising strategies for both prevention and management. More efforts should be focused on leveraging advanced multi-Omics and computational techniques to assess gut microbiota and immune signatures of IBD patients before and during treatment. These approaches can help pinpoint dysbiotic microbiota and mucosal signatures, enabling tailored treatment plans involving microbial consortia and anti-inflammatory therapies. Several promising novel treatment strategies are in clinical trials for IBD. The ultimate goal should be to establish a comprehensive profile including genetics, cytokines, inflammatory markers, and microbiota at the time of diagnosis, allowing clinicians to tailor specific treatments to individual patients—a true precision medicine approach for IBD. Integration of multi-omic data may offer insights into treatment response patterns, yet adoption in clinical practice is hindered by resource scarcity and lack of standardized methodologies. We feel IBD organizations and policy mackers can play a crucial role in formulating clear methodologies, secure funding for inovative multi-omic approaches into research as well as for easily implementable clinical tools, leading to effective clinical solutions for IBD in the near future.
Data availability
All available data is presented in the submitted work.
References
Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12:205–17.
Kumar M, Garand M, Al KS. Integrating omics for a better understanding of Inflammatory Bowel disease: a step towards personalized medicine. J Transl Med. 2019;17:419.
Everhov AH, Sachs MC, Malmborg P, Nordenvall C, Myrelid P, Khalili H, Elmberg M, Ekbom A, Askling J, Jakobsson G, Halfvarson J, Ludvigsson JF, Olen O. Changes in inflammatory bowel disease subtype during follow-up and over time in 44,302 patients. Scand J Gastroenterol. 2019;54:55–63.
Perler BK, Ungaro R, Baird G, Mallette M, Bright R, Shah S, Shapiro J, Sands BE. Presenting symptoms in inflammatory bowel disease: descriptive analysis of a community-based inception cohort. BMC Gastroenterol. 2019;19:47.
Liverani E, Scaioli E, Digby RJ, Bellanova M, Belluzzi A. How to predict clinical relapse in inflammatory bowel disease patients. World J Gastroenterol. 2016;22:1017–33.
van Deventer SJH. Anti-TNF antibody treatment of Crohn’s disease. Ann Rheum Dis. 1999;58(I114):I120.
Berg DR, Colombel JF, Ungaro R. The role of early biologic therapy in inflammatory bowel disease. Inflamm Bowel Dis. 2019;25:1896–905.
Elhag DA, Kumar M, Saadaoui M, Akobeng AK, Al-Mudahka F, Elawad M, Al KS. Inflammatory Bowel disease treatments and predictive biomarkers of therapeutic response. Int J Mol Sci. 2022;23:6966.
Poggioli G, Laureti S, Campieri M, Pierangeli F, Gionchetti P, Ugolini F, Gentilini L, Bazzi P, Rizzello F, Coscia M. Infliximab in the treatment of Crohn’s disease. Ther Clin Risk Manag. 2007;3:301–8.
Jang DI, Lee AH, Shin HY, Song HR, Park JH, Kang TB, Lee SR, Yang SH. The role of tumor necrosis factor alpha (TNF-α) in autoimmune disease and current TNF-α inhibitors in therapeutics. Int J Mol Sci. 2021;22:2719.
Levin AD, Wildenberg ME, van den Brink GR. Mechanism of action of anti-TNF therapy in inflammatory bowel disease. J Crohns Colitis. 2016;10:989–97.
Sands BE, Anderson FH, Bernstein CN, Chey WY, Feagan BG, Fedorak RN, Kamm MA, Korzenik JR, Lashner BA, Onken JE, Rachmilewitz D, Rutgeerts P, Wild G, Wolf DC, Marsters PA, Travers SB, Blank MA, van Deventer SJ. Infliximab maintenance therapy for fistulizing Crohn’s disease. N Engl J Med. 2004;350:876–85.
Hanauer SB, Feagan BG, Lichtenstein GR, Mayer LF, Schreiber S, Colombel JF, Rachmilewitz D, Wolf DC, Olson A, Bao W, Rutgeerts P. Maintenance infliximab for Crohn’s disease: the ACCENT I randomised trial. Lancet. 2002;359:1541–9.
Logan RF, Skelly MM. Maintenance infliximab delayed loss of response in active Crohn disease. ACP J Club. 2002;137:92.
Roda G, Jharap B, Neeraj N, Colombel JF. Loss of response to anti-TNFs: definition, epidemiology, and management. Clin Transl Gastroenterol. 2016;7: e135.
Gisbert JP, Panés J. Loss of response and requirement of infliximab dose intensification in Crohn’s disease: a review. Am J Gastroenterol. 2009;104:760–7.
Galloway JB, Hyrich KL, Mercer LK, Dixon WG, Fu B, Ustianowski AP, Watson KD, Lunt M, Symmons DP. Anti-TNF therapy is associated with an increased risk of serious infections in patients with rheumatoid arthritis especially in the first 6 months of treatment: updated results from the British Society for Rheumatology biologics register with special emphasis on risks in the elderly. Rheumatology. 2011;50:124–31.
Neurath MF. Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat Immunol. 2019;20:970–9.
Kopylov U, Seidman E. Predicting durable response or resistance to antitumor necrosis factor therapy in inflammatory bowel disease. Therap Adv Gastroenterol. 2016;9:513–26.
Adegbola SO, Sahnan K, Warusavitarne J, Hart A, Tozer P. Anti-TNF therapy in Crohn’s disease. Int J Mol Sci. 2018;19:2244.
Billioud V, Sandborn WJ, Peyrin-Biroulet L. Loss of response and need for adalimumab dose intensification in Crohn’s disease: a systematic review. Am J Gastroenterol. 2011;106:674–84.
Gerich ME, McGovern DP. Towards personalized care in IBD. Nat Rev Gastroenterol Hepatol. 2014;11:287–99.
Bek S, Nielsen JV, Bojesen AB, Franke A, Bank S, Vogel U, Andersen V. Systematic review: genetic biomarkers associated with anti-TNF treatment response in inflammatory bowel diseases. Aliment Pharmacol Ther. 2016;44:554–67.
Koder S, Repnik K, Ferkolj I, Pernat C, Skok P, Weersma RK, Potocnik U. Genetic polymorphism in ATG16L1 gene influences the response to adalimumab in Crohn’s disease patients. Pharmacogenomics. 2015;16:191–204.
Moroi R, Endo K, Kinouchi Y, Shiga H, Kakuta Y, Kuroha M, Kanazawa Y, Shimodaira Y, Horiuchi T, Takahashi S, Shimosegawa T. FCGR3A-158 polymorphism influences the biological response to infliximab in Crohn’s disease through affecting the ADCC activity. Immunogenetics. 2013;65:265–71.
Louis E, El Ghoul Z, Vermeire S, Dall’Ozzo S, Rutgeerts P, Paintaud G, Belaiche J, De Vos M, Van Gossum A, Colombel JF, Watier H. Association between polymorphism in IgG Fc receptor IIIa coding gene and biological response to infliximab in Crohn’s disease. Aliment Pharmacol Ther. 2004;19:511–9.
Netz U, Carter JV, Eichenberger MR, Dryden GW, Pan J, Rai SN, Galandiuk S. Genetic polymorphisms predict response to anti-tumor necrosis factor treatment in Crohn’s disease. World J Gastroenterol. 2017;23:4958–67.
Taylor KD, Plevy SE, Yang H, Landers CJ, Barry MJ, Rotter JI, Targan SR. ANCA pattern and LTA haplotype relationship to clinical responses to anti-TNF antibody treatment in Crohn’s disease. Gastroenterology. 2001;120:1347–55.
Medrano LM, Taxonera C, Marquez A, Barreiro-de Acosta M, Gomez-Garcia M, Gonzalez-Artacho C, Perez-Calle JL, Bermejo F, Lopez-Sanroman A, Martin Arranz MD, Gisbert JP, Mendoza JL, Martin J, Urcelay E, Nunez C. Role of TNFRSF1B polymorphisms in the response of Crohn’s disease patients to infliximab. Hum Immunol. 2014;75:71–5.
Bank S, Andersen PS, Burisch J, Pedersen N, Roug S, Galsgaard J, Turino SY, Brodersen JB, Rashid S, Rasmussen BK, Avlund S, Olesen TB, Hoffmann HJ, Nexo BA, Sode J, Vogel U, Andersen V. Genetically determined high activity of IL-12 and IL-18 in ulcerative colitis and TLR5 in Crohns disease were associated with non-response to anti-TNF therapy. Pharmacogenomics J. 2018;18:87–97.
Jürgens M, Laubender RP, Hartl F, Weidinger M, Seiderer J, Wagner J, Wetzke M, Beigel F, Pfennig S, Stallhofer J, Schnitzler F, Tillack C, Lohse P, Göke B, Glas J, Ochsenkühn T, Brand S. Disease activity, ANCA, and IL23R genotype status determine early response to infliximab in patients with ulcerative colitis. Am J Gastroenterol. 2010;105:1811–9.
Louis EJ, Watier HE, Schreiber S, Hampe J, Taillard F, Olson A, Thorne N, Zhang H, Colombel JF. Polymorphism in IgG Fc receptor gene FCGR3A and response to infliximab in Crohn’s disease: a subanalysis of the ACCENT I study. Pharmacogenet Genom. 2006;16:911–4.
Barber GE, Yajnik V, Khalili H, Giallourakis C, Garber J, Xavier R, Ananthakrishnan AN. Genetic markers predict primary non-response and durable response to anti-tnf biologic therapies in Crohn’s disease. Am J Gastroenterol. 2016;111:1816–22.
Planell N, Masamunt MC, Leal RF, Rodriguez L, Esteller M, Lozano JJ, Ramirez A, Ayrizono MLS, Coy CSR, Alfaro I, Ordas I, Visvanathan S, Ricart E, Guardiola J, Panes J, Salas A. Usefulness of transcriptional blood biomarkers as a non-invasive surrogate marker of mucosal healing and endoscopic response in ulcerative colitis. J Crohns Colitis. 2017;11:1335–46.
Yamamoto T, Shiraki M, Bamba T, Umegae S, Matsumoto K. Faecal calprotectin and lactoferrin as markers for monitoring disease activity and predicting clinical recurrence in patients with Crohn’s disease after ileocolonic resection: a prospective pilot study. United Eur Gastroenterol J. 2013;1:368–74.
Bohra A, Mohamed G, Vasudevan A, Lewis D, Van Langenberg DR, Segal JP. The utility of faecal calprotectin, lactoferrin and other faecal biomarkers in discriminating endoscopic activity in crohn’s disease: a systematic review and meta-analysis. Biomedicines. 2023;11:1408.
Ueno N, Sugiyama Y, Kobayashi Y, Murakami Y, Iwama T, Sasaki T, Kunogi T, Takahashi K, Tanaka K, Ando K, Kashima S, Inaba Y, Moriichi K, Tanabe H, Taruishi M, Saitoh Y, Okumura T, Fujiya M. Fecal calprotectin is a useful biomarker for predicting the clinical outcome of granulocyte and monocyte adsorptive apheresis in ulcerative colitis patients: a prospective observation study. BMC Gastroenterol. 2021;21:316.
Crowe JS, Roberts KJ, Carlton TM, Maggiore L, Cubitt MF, Clare S, Harcourt K, Reckless J, MacDonald TT, Ray KP, Vossenkämper A, West MR. Preclinical development of a novel, orally-administered anti-tumour necrosis factor domain antibody for the treatment of inflammatory Bowel disease. Sci Rep. 2018;8:4941.
Benítez JM, García-Sánchez V. Faecal calprotectin: Management in inflammatory bowel disease. World J Gastrointest Pathophysiol. 2015;6:203–9.
Ho GT, Lee HM, Brydon G, Ting T, Hare N, Drummond H, Shand AG, Bartolo DC, Wilson RG, Dunlop MG, Arnott ID, Satsangi J. Fecal calprotectin predicts the clinical course of acute severe ulcerative colitis. Am J Gastroenterol. 2009;104:673–8.
Cui G, Fan Q, Li Z, Goll R, Florholmen J. Evaluation of anti-TNF therapeutic response in patients with inflammatory bowel disease: current and novel biomarkers. EBioMedicine. 2021;66: 103329.
Hassan EA, Ramadan HK, Ismael AA, Mohamed KF, El-Attar MM, Alhelali I. Noninvasive biomarkers as surrogate predictors of clinical and endoscopic remission after infliximab induction in patients with refractory ulcerative colitis. Saudi J Gastroenterol. 2017;23:238–45.
Dahlén R, Magnusson MK, Bajor A, Lasson A, Ung K-A, Strid H, Öhman L. Global mucosal and serum cytokine profile in patients with ulcerative colitis undergoing anti-TNF therapy. Scand J Gastroenterol. 2015;50:1118–26.
Angelison L, Almer S, Eriksson A, Karling P, Fagerberg U, Halfvarson J, Thörn M, Björk J, Hindorf U, Löfberg R, Bajor A, Hjortswang H, Hammarlund P, Grip O, Torp J, Marsal J, Hertervig E. Long-term outcome of infliximab treatment in chronic active ulcerative colitis: a Swedish multicentre study of 250 patients. Aliment Pharmacol Ther. 2017;45:519–32.
Aden K, Rehman A, Waschina S, Pan WH, Walker A, Lucio M, Nunez AM, Bharti R, Zimmerman J, Bethge J, Schulte B, Schulte D, Franke A, Nikolaus S, Schroeder JO, Vandeputte D, Raes J, Szymczak S, Waetzig GH, Zeuner R, Schmitt-Kopplin P, Kaleta C, Schreiber S, Rosenstiel P. Metabolic functions of gut microbes associate with efficacy of tumor necrosis factor antagonists in patients with inflammatory Bowel diseases. Gastroenterology. 2019;157:1279-1292.e11.
Olsen T, Goll R, Cui G, Christiansen I, Florholmen J. TNF-alpha gene expression in colorectal mucosa as a predictor of remission after induction therapy with infliximab in ulcerative colitis. Cytokine. 2009;46:222–7.
Belarif L, Danger R, Kermarrec L, Nerrière-Daguin V, Pengam S, Durand T, Mary C, Kerdreux E, Gauttier V, Kucik A, Thepenier V, Martin JC, Chang C, Rahman A, Guen NS, Braudeau C, Abidi A, David G, Malard F, Takoudju C, Martinet B, Gérard N, Neveu I, Neunlist M, Coron E, MacDonald TT, Desreumaux P, Mai HL, Le Bas-Bernardet S, Mosnier JF, Merad M, Josien R, Brouard S, Soulillou JP, Blancho G, Bourreille A, Naveilhan P, Vanhove B, Poirier N. IL-7 receptor influences anti-TNF responsiveness and T cell gut homing in inflammatory bowel disease. J Clin Invest. 2019;129:1910–25.
Bertani L, Baglietto L, Antonioli L, Fornai M, Tapete G, Albano E, Ceccarelli L, Mumolo MG, Pellegrini C, Lucenteforte E, de Bortoli N, Bellini M, Marchi S, Blandizzi C, Costa F. Assessment of serum cytokines predicts clinical and endoscopic outcomes to vedolizumab in ulcerative colitis patients. Br J Clin Pharmacol. 2020;86:1296–305.
Billiet T, Cleynen I, Ballet V, Claes K, Princen F, Singh S, Ferrante M, Van Assche G, Gils A, Vermeire S. Evolution of cytokines and inflammatory biomarkers during infliximab induction therapy and the impact of inflammatory burden on primary response in patients with Crohn’s disease. Scand J Gastroenterol. 2017;52:1086–92.
Kim WM, Kaser A, Blumberg RS. A role for oncostatin M in inflammatory bowel disease. Nat Med. 2017;23:535–6.
Garand M, Kumar M, Huang SSY, Al KS. A literature-based approach for curating gene signatures in multifaceted diseases. J Transl Med. 2020;18:279.
Kalla R, Adams AT, Bergemalm D, Vatn S, Kennedy NA, Ricanek P, Lindstrom J, Ocklind A, Hjelm F, Ventham NT, Ho GT, Petren C, Repsilber D, Söderholm J, Pierik M, D’Amato M, Gomollón F, Olbjorn C, Jahnsen J, Vatn MH, Halfvarson J, Satsangi J. Serum proteomic profiling at diagnosis predicts clinical course, and need for intensification of treatment in inflammatory bowel disease. J Crohns Colit. 2020;15:699–708.
Gisbert JP, Chaparro M. Clinical usefulness of proteomics in inflammatory bowel disease: a comprehensive review. J Crohns Colitis. 2019;13:374–84.
D’Haens G, Kelly O, Battat R, Silverberg MS, Laharie D, Louis E, Savarino E, Bodini G, Yarur A, Boland BS, Afif W, Li XJ, Hale M, Ho J, Kondragunta V, Huang B, Kuy C, Okada L, Hester KD, Bray KR, Mimms L, Jain A, Singh S, Collins A, Valasek MA, Sandborn WJ, Vermeire S, Dulai PS. Development and validation of a test to monitor endoscopic activity in patients with Crohn’s disease based on serum levels of proteins. Gastroenterology. 2020;158:515-526.e10.
Kumar M, Singh P, Murugesan S, Vetizou M, McCulloch J, Badger JH, Trinchieri G, Al KS. Microbiome as an Immunological Modifier. Methods Mol Biol. 2020;2055:595–638.
Kumar M, Murugesan S, Singh P, Saadaoui M, Elhag DA, Terranegra A, Kabeer BSA, Marr AK, Kino T, Brummaier T, McGready R, Nosten F, Chaussabel D, Al KS. Vaginal microbiota and cytokine levels predict preterm delivery in asian women. Front Cell Infect Microbiol. 2021;11: 639665.
Kumar MS, Khodor M. Infections and pregnancy: effects on maternal and child health. Front Cell Infect Microbiol. 2022;12:873253.
Chopra C, Bhushan I, Mehta M, Koushal T, Gupta A, Sharma S, Kumar M, Khodor SA, Sharma S. Vaginal microbiome: considerations for reproductive health. Future Microbiol. 2022;17:1501–13.
Augustine T, Kumar M, Al Khodor S, van Panhuys N. Microbial dysbiosis tunes the immune response towards allergic disease outcomes. Clin Rev Allergy Immunol. 2022. https://doi.org/10.1007/s12016-022-08939-9.
Akobeng AK, Singh P, Kumar M, Al KS. Role of the gut microbiota in the pathogenesis of coeliac disease and potential therapeutic implications. Eur J Nutr. 2020;59:3369–90.
Elawad MA, Kumar M, Saadaui M, Elhag D, Akobeng A, Al-Mudahka F, Hendaus M, Al KS. P245 Dynemics of the Immune-transcriptome and microbiome in paediatric inflammatory Bowel disease. J Crohns Colitis. 2022;16:i292–i292.
Ding NS, McDonald JAK, Perdones-Montero A, Rees DN, Adegbola SO, Misra R, Hendy P, Penez L, Marchesi JR, Holmes E, Sarafian MH, Hart AL. Metabonomics and the gut microbiome associated with primary response to anti-TNF therapy in Crohn’s disease. J Crohns Colitis. 2020;14:1090–102.
Sanchis-Artero L, Martínez-Blanch JF, Manresa-Vera S, Cortés-Castell E, Valls-Gandia M, Iborra M, Paredes-Arquiola JM, Boscá-Watts M, Huguet JM, Gil-Borrás R, Rodríguez-Morales J, Cortés-Rizo X. Evaluation of changes in intestinal microbiota in Crohn’s disease patients after anti-TNF alpha treatment. Sci Rep. 2021;11:10016.
Singh N, Rabizadeh S, Jossen J, Pittman N, Check M, Hashemi G, Phan BL, Hyams JS, Dubinsky MC. Multi-center experience of vedolizumab effectiveness in pediatric inflammatory Bowel disease. Inflamm Bowel Dis. 2016;22:2121–6.
Kolho KL, Korpela K, Jaakkola T, Pichai MV, Zoetendal EG, Salonen A, de Vos WM. Fecal microbiota in pediatric inflammatory Bowel disease and its relation to inflammation. Am J Gastroenterol. 2015;110:921–30.
Lee JWJ, Plichta D, Hogstrom L, Borren NZ, Lau H, Gregory SM, Tan W, Khalili H, Clish C, Vlamakis H, Xavier RJ, Ananthakrishnan AN. Multi-omics reveal microbial determinants impacting responses to biologic therapies in inflammatory bowel disease. Cell Host Microbe. 2021;29:1294-1304.e4.
Ananthakrishnan AN, Luo C, Yajnik V, Khalili H, Garber JJ, Stevens BW, Cleland T, Xavier RJ. Gut Microbiome function predicts response to anti-integrin biologic therapy in inflammatory Bowel diseases. Cell Host Microbe. 2017;21(603–610): e3.
Yilmaz B, Juillerat P, Øyås O, Ramon C, Bravo FD, Franc Y, Fournier N, Michetti P, Mueller C, Geuking M, Pittet VEH, Maillard MH, Rogler G, Wiest R, Stelling J, Macpherson AJ. Microbial network disturbances in relapsing refractory Crohn’s disease. Nat Med. 2019;25:323–36.
Jones-Hall YL, Nakatsu CH. The intersection of TNF, IBD and the microbiome. Gut Microbes. 2016;7:58–62.
Colombel JF, Sandborn WJ, Rutgeerts P, Enns R, Hanauer SB, Panaccione R, Schreiber S, Byczkowski D, Li J, Kent JD, Pollack PF. Adalimumab for maintenance of clinical response and remission in patients with Crohn’s disease: the CHARM trial. Gastroenterology. 2007;132:52–65.
Hyams J, Crandall W, Kugathasan S, Griffiths A, Olson A, Johanns J, Liu G, Travers S, Heuschkel R, Markowitz J, Cohen S, Winter H, Veereman-Wauters G, Ferry G, Baldassano R. Induction and maintenance infliximab therapy for the treatment of moderate-to-severe Crohn’s disease in children. Gastroenterology. 2007;132:863–73.
Moran GW, Dubeau MF, Kaplan GG, Yang H, Seow CH, Fedorak RN, Dieleman LA, Barkema HW, Ghosh S, Panaccione R. Phenotypic features of Crohn’s disease associated with failure of medical treatment. Clin Gastroenterol Hepatol. 2014;12(434–42): e1.
Peters CP, Eshuis EJ, Toxopeus FM, Hellemons ME, Jansen JM, D’Haens GR, Fockens P, Stokkers PC, Tuynman HA, van Bodegraven AA, Ponsioen CY, Holland N, GUTc. Adalimumab for Crohn’s disease: long-term sustained benefit in a population-based cohort of 438 patients. J Crohns Colitis. 2014;8:866–75.
Bazin T, Hooks KB, Barnetche T, Truchetet ME, Enaud R, Richez C, Dougados M, Hubert C, Barré A, Nikolski M, Schaeverbeke T. Microbiota composition may predict anti-tnf alpha response in spondyloarthritis patients: an exploratory study. Sci Rep. 2018;8:5446.
Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151:363–74.
Franzin M, Stefančič K, Lucafò M, Decorti G, Stocco G. Microbiota and drug response in inflammatory bowel disease. Pathogens. 2021. https://doi.org/10.3390/pathogens10020211.
Caenepeel C, Falony G, Machiels K, Verstockt B, Goncalves PJ, Ferrante M, Sabino J, Raes J, Vieira-Silva S, Vermeire S. Dysbiosis and associated stool features improve prediction of response to biological therapy in inflammatory bowel disease. Gastroenterology. 2024;166:483–95.
Machiels K, Pozuelo Del Rio M, Martinez-De la Torre A, Xie Z, Pascal Andreu V, Sabino J, Santiago A, Campos D, Wolthuis A, D’Hoore A, De Hertogh G, Ferrante M, Manichanh C, Vermeire S. Early postoperative endoscopic recurrence in Crohn’s disease is characterised by distinct microbiota recolonisation. J Crohns Colitis. 2020;14:1535–46.
Sokol H, Leducq V, Aschard H, Pham HP, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, Cosnes J, Seksik P, Langella P, Skurnik D, Richard ML, Beaugerie L. Fungal microbiota dysbiosis in IBD. Gut. 2017;66:1039–48.
Kowalska-Duplaga K, Krawczyk A, Sroka-Oleksiak A, Salamon D, Wędrychowicz A, Fyderek K, Gosiewski T. Dependence of colonization of the large intestine by Candida on the treatment of Crohn’s disease. Pol J Microbiol. 2019;68:121–6.
Meade S, Liu Chen Kiow J, Massaro C, Kaur G, Squirell E, Bressler B, Lunken G. Gut microbiome-associated predictors as biomarkers of response to advanced therapies in inflammatory bowel disease: a systematic review. Gut Microbes. 2023;15:2287073.
Ungaro F, Massimino L, D’Alessio S, Danese S. The gut virome in inflammatory bowel disease pathogenesis: from metagenomics to novel therapeutic approaches. United European Gastroenterol J. 2019;7:999–1007.
Wolbink GJ, Aarden LA, Dijkmans BA. Dealing with immunogenicity of biologicals: assessment and clinical relevance. Curr Opin Rheumatol. 2009;21:211–5.
West RL, Zelinkova Z, Wolbink GJ, Kuipers EJ, Stokkers PC, van der Woude CJ. Immunogenicity negatively influences the outcome of adalimumab treatment in Crohn’s disease. Aliment Pharmacol Ther. 2008;28:1122–6.
Sazonovs A, Kennedy NA, Moutsianas L, Heap GA, Rice DL, Reppell M, Bewshea CM, Chanchlani N, Walker GJ, Perry MH, McDonald TJ, Lees CW, Cummings JRF, Parkes M, Mansfield JC, Irving PM, Barrett JC, McGovern D, Goodhand JR, Anderson CA, Ahmad T. HLA-DQA1*05 carriage associated with development of anti-drug antibodies to infliximab and adalimumab in patients with crohn’s disease. Gastroenterology. 2020;158:189–99.
Billiet T, Vande Casteele N, Van Stappen T, Princen F, Singh S, Gils A, Ferrante M, Van Assche G, Cleynen I, Vermeire S. Immunogenicity to infliximab is associated with HLA-DRB1. Gut. 2015;64:1344–5.
Vermeire S, Gils A, Accossato P, Lula S, Marren A. Immunogenicity of biologics in inflammatory bowel disease. Therap Adv Gastroenterol. 2018. https://doi.org/10.1177/1756283X17750355.
Bortlik M, Duricova D, Malickova K, Machkova N, Bouzkova E, Hrdlicka L, Komarek A, Lukas M. Infliximab trough levels may predict sustained response to infliximab in patients with Crohn’s disease. J Crohns Colitis. 2013;7:736–43.
Adegbola SO, Sahnan K, Warusavitarne J, Hart A, Tozer P. Anti-TNF therapy in Crohn’s disease. Int J Mol Sci. 2018;19:2244.
Dubinsky MC, Mei L, Friedman M, Dhere T, Haritunians T, Hakonarson H, Kim C, Glessner J, Targan SR, McGovern DP, Taylor KD, Rotter JI. Genome wide association (GWA) predictors of anti-TNFalpha therapeutic responsiveness in pediatric inflammatory bowel disease. Inflamm Bowel Dis. 2010;16:1357–66.
Kevans D, Waterman M, Milgrom R, Xu W, Van Assche G, Silverberg M. Serological markers associated with disease behavior and response to anti-tumor necrosis factor therapy in ulcerative colitis. J Gastroenterol Hepatol. 2015;30:64–70.
Ferrante M, Vermeire S, Katsanos KH, Noman M, Van Assche G, Schnitzler F, Arijs I, De Hertogh G, Hoffman I, Geboes JK, Rutgeerts P. Predictors of early response to infliximab in patients with ulcerative colitis. Inflamm Bowel Dis. 2007;13:123–8.
Nguyen DL, Nguyen ET, Bechtold ML. pANCA positivity predicts lower clinical response to infliximab therapy among patients with IBD. South Med J. 2015;108:139–43.
Biancheri P, Brezski RJ, Di Sabatino A, Greenplate AR, Soring KL, Corazza GR, Kok KB, Rovedatti L, Vossenkämper A, Ahmad N, Snoek SA, Vermeire S, Rutgeerts P, Jordan RE, MacDonald TT. Proteolytic cleavage and loss of function of biologic agents that neutralize tumor necrosis factor in the mucosa of patients with inflammatory bowel disease. Gastroenterology. 2015;149:1564-1574.e3.
Rosen MJ, Minar P, Vinks AA. Review article: applying pharmacokinetics to optimise dosing of anti-TNF biologics in acute severe ulcerative colitis. Aliment Pharmacol Ther. 2015;41:1094–103.
Jung H, Kim JS, Lee KH, Tizaoui K, Terrazzino S, Cargnin S, Smith L, Koyanagi A, Jacob L, Li H, Hong SH, Yon DK, Lee SW, Kim MS, Wasuwanich P, Karnsakul W, Shin JI, Kronbichler A. Roles of microRNAs in inflammatory bowel disease. Int J Biol Sci. 2021;17:2112–23.
James JP, Riis LB, Malham M, Høgdall E, Langholz E, Nielsen BS. MicroRNA biomarkers in IBD—differential diagnosis and prediction of colitis-associated cancer. Int J Mol Sci. 2020;21:7893.
Batra SK, Heier CR, Diaz-Calderon L, Tully CB, Fiorillo AA, van den Anker J, Conklin LS. Serum miRNAs are pharmacodynamic biomarkers associated with therapeutic response in pediatric inflammatory bowel disease. Inflamm Bowel Dis. 2020;26:1597–606.
Papaconstantinou I, Kapizioni C, Legaki E, Xourgia E, Karamanolis G, Gklavas A, Gazouli M. Association of miR-146 rs2910164, miR-196a rs11614913, miR-221 rs113054794 and miR-224 rs188519172 polymorphisms with anti-TNF treatment response in a Greek population with Crohn’s disease. World J Gastroint Pharmacol Ther. 2017;8:193–200.
Billiet T, Papamichael K, de Bruyn M, Verstockt B, Cleynen I, Princen F, Singh S, Ferrante M, Van Assche G, Vermeire S. A matrix-based model predicts primary response to infliximab in Crohn’s disease. J Crohns Colitis. 2015;9:1120–6.
Iborra M, Pérez-Gisbert J, Bosca-Watts MM, López-García A, García-Sánchez V, López-Sanromán A, Hinojosa E, Márquez L, García-López S, Chaparro M, Aceituno M, Calafat M, Guardiola J, Belloc B, Ber Y, Bujanda L, Beltrán B, Rodríguez-Gutiérrez C, Barrio J, Cabriada JL, Rivero M, Camargo R, van Domselaar M, Villoria A, Schuterman HS, Hervás D, Nos P. Effectiveness of adalimumab for the treatment of ulcerative colitis in clinical practice: comparison between anti-tumour necrosis factor-naïve and non-naïve patients. J Gastroenterol. 2017;52:788–99.
Gonczi L, Vegh Z, Golovics PA, Rutka M, Gecse KB, Bor R, Farkas K, Szamosi T, Bene L, Gasztonyi B, Kristóf T, Lakatos L, Miheller P, Palatka K, Papp M, Patai Á, Salamon Á, Tóth GT, Vincze Á, Biro E, Lovasz BD, Kurti Z, Szepes Z, Molnár T, Lakatos PL. Prediction of short—and medium-term efficacy of biosimilar infliximab therapy. do trough levels and antidrug antibody levels or clinical and biochemical markers play the more important role? J Crohns Colitis. 2017;11:697–705.
Gisbert JP, Marín AC, McNicholl AG, Chaparro M. Systematic review with meta-analysis: the efficacy of a second anti-TNF in patients with inflammatory bowel disease whose previous anti-TNF treatment has failed. Aliment Pharmacol Ther. 2015;41:613–23.
Colombel JF, Sandborn WJ, Reinisch W, Mantzaris GJ, Kornbluth A, Rachmilewitz D, Lichtiger S, D’Haens G, Diamond RH, Broussard DL, Tang KL, van der Woude CJ, Rutgeerts P. Infliximab, azathioprine, or combination therapy for Crohn’s disease. N Engl J Med. 2010;362:1383–95.
Panaccione R, Ghosh S, Middleton S, Márquez JR, Scott BB, Flint L, van Hoogstraten HJ, Chen AC, Zheng H, Danese S, Rutgeerts P. Combination therapy with infliximab and azathioprine is superior to monotherapy with either agent in ulcerative colitis. Gastroenterology. 2014;146:392-400.e3.
Billiet T, Rutgeerts P, Ferrante M, Van Assche G, Vermeire S. Targeting TNF-α for the treatment of inflammatory bowel disease. Expert Opin Biol Ther. 2014;14:75–101.
Torres J, Cravo M, Colombel JF. Anti-TNF withdrawal in inflammatory bowel disease. GE Port J Gastroenterol. 2016;23:153–61.
Funding
This work was made possible by grant # NPRP10-0125-170242 received from the Qatar National Research Fund (a member of Qatar Foundation). This research was also co-funded by Sidra Medicine, Qatar, project number SDR100028. The funding bodies played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript. The APC was funded by Research Department, Sidra Medicine, Qatar.
Author information
Authors and Affiliations
Contributions
All authors have made substantial contributions to this manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kumar, M., Murugesan, S., Ibrahim, N. et al. Predictive biomarkers for anti-TNF alpha therapy in IBD patients. J Transl Med 22, 284 (2024). https://doi.org/10.1186/s12967-024-05058-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-024-05058-1