Abstract
The human body is colonized by abundant and diverse microorganisms, collectively known as the microbiome. The oral cavity has more than 700 species of bacteria and consists of unique microbiome niches on mucosal surfaces, on tooth hard tissue, and in saliva. The homeostatic balance between the oral microbiota and the immune system plays an indispensable role in maintaining the well-being and health status of the human host. Growing evidence has demonstrated that oral microbiota dysbiosis is actively involved in regulating the initiation and progression of an array of autoimmune diseases.
Oral microbiota dysbiosis is driven by multiple factors, such as host genetic factors, dietary habits, stress, smoking, administration of antibiotics, tissue injury and infection. The dysregulation in the oral microbiome plays a crucial role in triggering and promoting autoimmune diseases via several mechanisms, including microbial translocation, molecular mimicry, autoantigen overproduction, and amplification of autoimmune responses by cytokines. Good oral hygiene behaviors, low carbohydrate diets, healthy lifestyles, usage of prebiotics, probiotics or synbiotics, oral microbiota transplantation and nanomedicine-based therapeutics are promising avenues for maintaining a balanced oral microbiome and treating oral microbiota-mediated autoimmune diseases. Thus, a comprehensive understanding of the relationship between oral microbiota dysbiosis and autoimmune diseases is critical for providing novel insights into the development of oral microbiota-based therapeutic approaches for combating these refractory diseases.
Similar content being viewed by others
Background
The human microbiota, which consists of approximately 10–100 trillion symbiotic microorganisms, defines the sum of all microbes forming communities present in any habitat, including the skin, oral cavity, respiratory system, gastrointestinal tract, and urogenital tract [1]. Normally, the relationship between oral microbes and the host is dynamic and homeostatic. Due to the coevolution between the host and microbiota, the microbiota contributes to adjusting and maintaining human health [2]. On the other hand, components of the innate and adaptive immune subsystems interact and work together to eliminate harmful pathogens and selectively promote commensal tolerance [3]. However, any genetic or environmental change in the bilateral interaction between the microbiota and immune system can lead to pathological conditions such as infection, inflammation and autoimmune diseases [4, 5].
The oral cavity harbors the second largest and most diverse microbiota after the gastrointestinal tract, with 770 opportunistic, commensal, and pathogenic bacterial species [6]. Many unfavorable factors, such as smoking, administration of antibiotics and infections can significantly affect the composition and structure of the oral microbiota, resulting in oral diseases and systemic diseases [7]. Under the conditions of oral microbiota dysbiosis, certain commensal microorganisms in the oral cavity are transformed into pathogenic microorganisms, which are highly virulent and able to escape elimination by the immune system [8,9,10]. Accumulating evidence has revealed that oral microbiota dysbiosis might play a critical role in modulating the initiation and progression of a wide array of autoimmune diseases [11,12,13,14] (Fig. 1). For example, alterations in oral microbiota composition and structure were observed in mice with ligature-induced periodontitis, which may trigger increased secretion of the local gingival Th17 cells [15]. Porphyromonas gingivalis can transform arginine to citrulline in proteins via arginine gingipains and peptidylarginine deiminases (PADs) [16]. The immune system recognizes citrullinated proteins as autoimmune antigens and produces anti-citrullinated peptide antibodies, which subsequently activate the complement system and lead to Rheumatoid arthritis (RA) [17]. Aggregatibacter actinomycetemcomitans (Aa) is the key bacterium triggering autoimmune responses in RA, and it is a known periodontal pathogen that can secrete the pore-forming toxin leukotoxin-A (LtxA). LtxA induces the production of citrullinated antigen by abnormally activating citrullinating enzymes in neutrophils [18]. In addition, varying amounts of antibodies against different oral bacteria were detected in serum samples from patients with systemic lupus erythematosus (SLE). Higher antibody titers against Aa, P. gingivalis, Treponema denticola, and Capnocytophaga. ochracea were found in SLE patients positive for anti-dsDNA antibodies. In particular, antibodies against Aa were significantly associated with SLE disease activity [19].
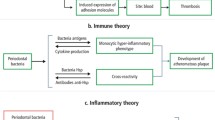
The bidirectional role of oral microbiota in the organism: eubiosis and dysbiosis. The eubiosis of oral microbiota contribute to homeostasis maintaining. Within microbiota dysbiosis, the homeostasis between the host and commensal is destroyed, accompany by abnormal immune response and leading to IBD, RA, SLE, SS, AS, MS, AILDs, IgAN. IBD, Inflammatory bowel disease; RA, Rheumatoid arthritis; SLE, Systemic lupus erythematosus; SS, Sjogren syndrome; AS, Ankylosing spondylitis; MS, Multiple sclerosis; AILDs, Autoimmune liver diseases; IgAN, Immunoglobulin A nephropathy
Oral microbiome is readily accessible and easily sampled, thereby dissecting the composition and function of oral microorganisms is crucial for elucidating the underlying molecular mechanisms for the development of autoimmune diseases. The current review comprehensively covers the pathological changes affecting the bilateral cross-talks between oral microbiota and immune system, which might result in the initiation and progression of autoimmune diseases. We also discussed the major molecular mechanisms that oral microbiota dysbiosis affects autoimmune diseases (microbial translocation, molecular mimicry, autoantigen overproduction and amplification of autoimmunity by cytokines). The potential therapeutic strategies for balancing the oral microbiota are also focused on, which may provide evidence for the precision management and improvement of the efficacy of oral microbiota-mediated autoimmune diseases.
The bilateral cross-talks between human microbiota and immune system
The microbiota plays a decisive role in the induction, training and functions of the host immune system. Early microbial exposure to infants is essential for regulating and influencing the formation of the immune system [20]. The bacterial components from the intestinal tract can be transported to the lactating mammary gland, which plays a crucial role in shaping the early-life gut microbiota and stimulating the immune system in infants [21]. Lactobacilli produce indole-3-aldehyde, and this microbial metabolite stimulates the production of IL-22 and IL17 by activating the aryl hydrocarbon receptor, contributing to the balance of the intestinal mucosal immune system [22]. Likewise, the symbiotic gut microbiota is beneficial for the maturation of gut-associated lymphoid tissues [23]. Severe inflammatory bowel disease (IBD) was observed in mice deficient in Peyer’s patches and lymph nodes [24]. Interestingly, retarded development of lymphoid follicles was observed in the small intestine of germ-free mice [25]. Similarly, germ-free or germ-depleted mice showed smaller secondary lymphoid tissue with lower cellularity [23]. These findings consistently indicate that the microbiota plays a vital role in the establishment and maintenance of the harmonious relationship between the host immune system and microbes, and abnormal changes in the microbiota composition might significantly affect the normal functions of the immune system, leading to immune-mediated diseases [26].
Conversely, the immune system also plays a fundamental role in establishing a sustainable and harmonious host-microbial relationship. The immune response to pathogenic microorganisms is the leading cause of host-mediated tissue damage, which might create a microenvironment that is beneficial for microbiota dysbiosis and invasion by pathogenic microbes. For example, the inflammatory response promotes the growth of aerobic bacteria and significantly decreases the survival of resident colonic bacteria, resulting in IBD [27]. Differences in the secretory function of Paneth cells also affect microbial composition. For instance, a significant distinction in intestinal microbiota composition was observed between human defensin 5 transgenic mice and wild-type mice [28].
Microbiota dysbiosis and its driving factors
Microbiota dysbiosis, an imbalance in the composition and function of the microbiome, has a prominent impact on the initiation and progression of many systemic diseases, such as immunological disorders, cardiovascular diseases, diabetes, and respiratory diseases [29]. Dysbiosis is driven by many factors, such as host genetic factors, dietary habits, stress, smoking, administration of antibiotics, tissue injury and infection (Fig. 2) [3]. The increased consumption of carbohydrate-rich food and industrially processed flour and sugar are the two mainly dietary changes [30]. Dietary shifts contribute a significant role in shaping microbiota profiles [31]. A high-fat, high-sugar diet increased the release of dopamine, a neurotransmitter regulating pleasure, which recruit neural mechanisms driving food wanting [32, 33]. It is becoming increasingly clear that obesity is the risk factor for multiple autoimmune diseases, including RA, IBD and type 1 diabetes (T1D) [34]. It was reported that diet is a key factor in shifts of commensal balance as some oral pathogens were preferentially enriched in hunter-gatherers when compared with traditional farmers [35]. A diet high in fat and sugar can cause gut microbiota dysbiosis, leading to a variety of disorders such as obesity and non-alcoholic fatty liver disease. Sen et al. observed that rats treated with high-fat/high-sugar diet or low-fat/high-sugar diet exhibited an imbalance of gut microbiota with the characteristics of lower overall bacterial diversity and higher Firmicutes/Bacteriodetes ratio [36]. Moreover, Bifidobacterium spp. was important for maintaining gut/intestinal barrier function, and they were found to be absent in the gut microbiota of mice fed a high-fat diet [37]. Compared with high-fat diet, reduced level of plasma TNF-α and inhibition of systematic inflammation were found in the rats treated with low-fat diet [38]. Furthermore, the alterations of oral microbiota and recruitment of neutrophils were found in high-fat diet induced obesity mice model [39]. High-sugar diet may disturb the hepatic lipid metabolism by changing the structure of gut microbial community to cause hyperlipidemia and even non-alcoholic fatty liver disease [40]. Intake of probiotics, Grifola frondosa polysaccharides, healthy diet or polysaccharide peptides are proactive measures that contribute to the amelioration of microbiota dysbiosis caused by high-fat or high-sucrose diet [41,42,43,44]. Excessive administration of antibiotics disturbed the balance of the skin microbiota and retards wound healing by decreasing the expression of RegIIIγ and IL-17 [45]. In addition, overuse of antibiotics also disrupts the gut microbial balance, weakening the body's resistance to pathogenic microorganisms and increasing drug resistance [46]. The number and alpha diversity of gut microbiota in rats treated with antibiotics were significantly lower than the untreated groups [47]. Upon tissue injury, damaged cells release inflammatory chemical signals that facilitate the expansion of the pathogenic microbiota [48]. Moreover, it was reported that P. gingivalis induces microbial dysbiosis by affecting the normal function of the complement system [49]. The P. gingivalis activated Notch-1 signaling in oral epithelial cells to promote the expression of PLA2-IIA that was associated with oral dysbiosis [50].
Microbiota dysbiosis and autoimmune diseases
To date, more than 80 autoimmune diseases has been identified, affecting a wide range of human body parts [51]. A healthy immune system effectively distinguishes self from non-self as well as harmless non-self from dangerous non-self [52]. Autoimmune diseases are characterized by immune response to self-antigens, including aberrant generation of autoantibody-producing B cells, autoreactive T cells, and proinflammatory cytokines [53]. The immune responses are induced by genetic and/or environmental factors acting on the immune system. The level of miR-146a was markedly elevated in THP-1 cells stimulated by IL-1β. The stimulated expression of miR-146a enhanced the tolerance of THP-1 cells to IL-1β, LPS, peptidoglycan, Pam and flagellin [54]. In addition, miR-146a might be involved in autoimmune diseases via the regulation of cytokines induced by 2D2 CD4 T cells. For example, increased IL-6 and IL-21 levels endowed the differentation of Th17 cells in 2D2 CD4 T cells in miR-146a–deficient mice, which induced more severe T-cell-mediated experimental autoimmune encephalomyelitis [55]. The production of inflammatory cytokines in P.gingivalis lipopolysaccharide-stimulated B cells was inhibited by miR-146a [56]. In addition, the expression of miRNA-146a-5p unregulated by Resolvin D1 depleted the levels of connective tissue growth factor, which might ameliorate the severity of RA [57, 58]. Dysbiosis of the oral microbiota (such as microbial diversity reduction and the high level of genus Capnocytophaga) has been found in patients with loss of STAT3 function [59]. Oral microbiota dyregulation has been increasingly considered as a potential risk factor for causing autoimmune diseases, even though the underlying mechanisms remain elusive. At the molecular level, molecular mimicry, autoantigen overproduction, and amplification of autoimmunity by cytokines represent the potential mechanisms for oral microbiota mediated autoimmune disorders. At the cellular level, the translocated oral microbiota directly interacts with immune and tissue cells and contributes to the pathogenesis of systemic autoimmunity [60]. Autoimmune diseases are typically defined by dyregulation of adapitive immune system. In contrast, autoinflammatory diseases result from an aberrant innate response, characterized by fewer T and B cells involvement or autoantibod production [61]. Polymorphonuclear neutrophils expressing major histocompatibility complex (MHC) class II antigens or the T-cell costimulatory molecules CD80 or CD86 in patients with chronic inflammatory diseases induced proliferation of T cells [62, 63]. Importantly, IL-1β, a robust pro-inflammatory cytokine, is secreted by macrophages and activate IL-1 receptor signaling to trigger the defferentiation of Th17 cells [64]. These innate immune cells including neutrophils and macrophages are critical in activating adaptive immune system by antigen presenting [65]. In addition, aberrant inflammatory response is closely correlated with the development autoimmune diseases, such IBD, RA and T1D [66, 67]. Collectively, the innate immune system is the first barrier against pathogen infection and tissue damage in host immune denfense, and subsequently palys a vital role in activating adaptive immunity [68].
Under harmonious host-microbial relationships, the microbiota provides vitamins, protects mucosal immunity and prevents colonization by pathogenic microorganisms, while the host offers an ecological niche and survival substances for the microbiota. Under dysbiosis, the mutually beneficial relationship between the host and microbes is disrupted, leaving the host more vulnerable to attacks by exogenous and endogenous pathogenic microorganisms. The host immune system in turn triggers a series of immune response processes, which might further aggravate microbiota dysbiosis.
Toll-like receptors (TLRs), which are germline-encoded pattern recognition receptors (PRRs), recognize pathogenic microorganisms and activate host signaling pathways to induce the production of cytokines and other inflammatory mediators [69]. In addition, TLRs promote the maturation of antigen-presenting cells (APCs), such as dendritic cells and macrophages, and induce the differentiation of T and B lymphocytes into effector cells, thereby regulating adaptive immunity [70,71,72,73]. Therefore, TLRs play an indispensable role in maintaining host-microbe homeostasis [74]. However, excessive activation of TLRs might lead to uncontrolled production of cytokines and inflammatory mediators, promoting the initiation and development of autoimmune diseases. The dying microbes in the dysfunctional flora release active substances such as lipids, nucleic acids, and proteins that induce the expression of TLRs, exacerbating the severity of autoimmune diseases [75]. Xu et al. found that P. gingivalis inhibited the host-protective TLR2-MyD88 pathway by proteasomal degradation of MyD88 and concurrently activated the host-detrimental TLR2-Mal-PI3K pathway, which destroyed immune function by blocking cellular phagocytosis [76].
Accumulating evidence has demonstrated that microbiota dysbiosis is the critical factor for engendering autoimmune diseases [77]. For instance, the in vivo results showed that intestinal microbial imbalance promoted the initiation of autoimmune pancreatitis by enhancing the production of IFN-α and IL-33 released by plasmacytoid dendritic cells [78]. In addition, intestinal microbiota dysbiosis caused by constipation increased the number of Th17 cells and Teff17 cells in the brain and spinal cord as well as promoted the secretion of cytokines in serum, which deteriorated experimental autoimmune encephalomyelitis in mice [79]. Collectively, gaining deep insights into the relationship between microbiota dysbiosis and autoimmune diseases is crucial for reducing the occurrence of autoimmune diseases and improving prognosis.
The oral cavity is exposed to the external environment and anatomically connected with the gastrointestinal tract. Thus, oral bacteria escaping the inhibition of stomach acid can spread through the gastrointestinal tract and are closely correlated with various systemic diseases, such as IBD and RA [80, 81]. Zhang et al. found that the oral Campylobacter species were detected in the intestinal tissues and stool samples [82]. A significant overlap between the fecal and oral microbiota has been reported, with approximately 45% similarity in bacterial taxa. These findings indicate that it is common for the ectopic colonization of oral bacteria in gastrointestinal tract [83]. Thus, the oral microbiota may play an important role in regulating the composition and structure of intestinal microbiota and have a great influence on the development of autoimmune diseases.
Administration of P. gingivalis significantly altered the gut microbiota in the mice. At the phylum level, Deferribacteres was increased and Bacteroidetes was decreased. Increased Deferribacteriaceae, Gemellaceae, and Clostridiaceae and decreased Paraprevotellaceae and Mogibacteriaceae were observed at the family level. Increased Coriobacteriaceae, Gemellaceae, and Clostridiaceae and decreased Prevotellaceae, Mogibacteriaceae, Dorea, Butyricicoccus, and Bilophila were found at the genus level [84]. The composition of the gut microbiota was disturbed in mice even receiving a single administration of P. gingivalis, with elevated levels of phylum Bacteroidetes and serum endotoxin and a decreased level of phylum Firmicutes [85]. It was reported that inflammation was promoted by gut microbiota dysbiosis via chemokine C–C chemokine ligand 5, which was important for recruiting lymphocytes in the intestine [86]. The endotoxemia induced chronic inflammation and insulin resistance, which contributed to the development of T1D [87]. Intestinal oxidative stress, barrier damage, inflammation and systemic autoimmune responses were common in MRL/lpr mice (a SLE animal model) with aberrant gut microbiome [88]. Oxidative stress promoted the development of autoimmune diseases including autoimmune hepatitis (AIH) and SLE by inducing apoptosis and inflammation [89]. Gut barrier damage in MRL/lpr mice was evidenced by increased levels of fecal albumin and IgA and a decreased level of gut tight junction protein (TJP) ZO-2 [88]. Also, intestinal permeability was increased due to downregulation of TJP-1 and occluding [85]. Thus, the gut microbiota dysbiosis leads to increased intestinal permeability, which in turn promotes the translocation of pathogens and other harmful microbial molecules to bloodstream and even distant organs. In addition, gut microbiota dysbiosis was observed in C57BL6/J mice administrated with salivary samples from periodontitis patients, with an increased abundance of Porphyromonadaceae and Fusobacterium. In addition, the levels of pro-inflammatory cytokines and chemokines such as IL-1β, IL-6 and colony-stimulating factor 1 were elevated, while the expression of anti-inflammatory cytokines such as IL-10 were decreased in colon tissue [90]. Apart from gut microbiota dysbiosis, the increase of TH17 cells proportions among mesenteric lymphocytes and IL-17 in sera were found in mice orally administration of P. gingivalis, followed by aggravation of collagen-induced arthritis [91]. P. gingivalis promotes the production of RA-related cytokines and activation of monocytes through TLR pathways [92]. Intestinal CD4 T cells and inflammation were indirectly induced due to the change in the gut microbiome by P. gingivalis [93]. The levels of TLR4 and CD14 were significantly higher in the livers of MRL/lpr mice [88]. Thus, oral bacteria promote gut microbiota dysbiosis by translocation and subsequently aggravating autoimmune diseases, which highlights the importance of the oral-gut-microbiota axis in the pathogenesis of autoimmune diseases [94].
The major pathways by which oral microbiota dysbiosis affects autoimmune diseases
Microbial translocation
The main possible pathways by which oral pathogenic bacteria reach distant tissues and organs to trigger autoimmune diseases are the hematogenous and enteral routes. Invasive oral treatments, oral mucosal ulcers, injuries, and periodontitis provide convenient entry points for oral pathogenic microorganisms to invade the circulatory system. Subsequently, the oral bacteria and their soluble bacterial products can reach distant organs via the bloodstream [95, 96]. For example, a mouse model of dextran sodium sulfate (DSS)-induced colitis was used to investigate the relationship between Streptococcus mutans and colitis. The results demonstrated that S. mutans may increase the severity of colitis, as it promoted the secretion of IFN-γ when entering the bloodstream [97]. In addition, in a murine model of colorectal cancer, intravenously injected Fusobacterium nucleatum colonized tumor tissues [98]. In addition, the bacteria detected in the oral cavity of the mouse periodontitis model were also found in isolated liver and spleen cells in vitro [99]. Due to the hematogenous dissemination of P. gingivalis, more severe cartilage destruction and higher expression of IL-17 in joints were found in mice orally treated with a mixture of P. gingivalis, T. denticola, and Tannerella forsythia [100].
In addition to the hematogenous route, the enteral route is a major route for oral bacterial dissemination. Increased intestinal permeability is associated with the pathogenesis of many autoimmune diseases, such as type I diabetes, multiple sclerosis (MS), and SLE [101]. An enormous number of oral resident bacteria are swallowed into the digestive tract per day, accompanied by 1.5 L of saliva [102]. Oral microorganisms are ordinarily damaged by stomach acids, so they fail to pass through the stomach to the gut. However, some oral pathogens, such as P. gingivalis, have the ability to withstand stomach acidity and colonize the intestine [80]. The gut permeability and the level of blood endotoxin were significantly increased in mice that were orally administered P. gingivalis [103]. Generally, a healthy gut microbiota or gut barrier can confer resistance to ectopic colonization by oral bacteria. However, bacteria from the oral cavity can easily invade the gut when the intestinal microbiota is disrupted due to the administration of antibiotics. Klebsiella strains, as oral-derived intestinal pathogenic bacteria, possess the ability to resist a variety of antibiotics, including ampicillin (Amp) and tylosin (Tyl). For instance, compared with antibiotic-naïve mice, mice treated with Amp or Ty1 had a perturbed gut microbial profile, which was beneficial for colonization by Kp-2H7. Klebsiella strains strongly induce the accumulation of intestinal TH1 cells, leading to exacerbation of inflammatory diseases through abnormal activation of the intestinal immune system [104]. To investigate the effect of oral microorganisms on intestinal epithelial barrier dysfunction, Nattramilarasu et al. cultured colonic epithelial cell monolayers (HT-29/B6-GR/MR) and M1-macrophage-like THP-1 cells together in vitro. After infection with Campylobacter concisus, THP-1 cells promoted the release of cytokines (TNF-α, IL-1β, and IL-6) and consequently increased the permeability of the intestinal epithelium [105]. Oral pathobionts such as Klebsiella and Enterobacter derived from ligature-induced periodontitis and DSS-treated mice stimulated inflammasome production in colonic mononuclear phagocytes, followed by increased inflammation in the colonic mucosa. Additionally, Klebsiella and Enterobacter activated oral pathobiont-reactive Th17 cells, which had a tendency to migrate toward the inflamed gut [106]. Thus, colonization by oral-associated pathogens in the intestinal system through the enteral route plays a crucial role in the exacerbation of autoimmune diseases [6].
Molecular mimicry
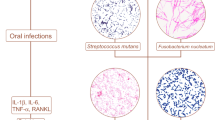
“Molecular mimicry” was coined by R.Damian in 1964. It is demonstrated that more than 30 viral proteomes, especially the proteomes of human T-lymphotropic virus 1, Rubella virus, and hepatitis C virus, have a substantial pentapeptide overlap with the human proteome [107]. These observations strongly confirmed the pathogenesis of immune diseases might be due to the sharing of amino acid sequences between microbiome and the host. As a rich source of antigen, the oral cavity harbors one of the most diverse and abundant microbial communities. Molecular mimicry is involved in oral microbiota-mediated chronic infection in localized and remote organ autoimmunity. Periodontopathic bacteria (P.gingivalis and Aa) express the GroEL-like protein, which is a form of HSP60 family [108, 109]. Of note, higher levels of antibodies against human HSP60 were also observed in the sera of periodontitis patients [110]. It was reported that Streptococcal M protein exhibits structural similarities with cardiac myosin, which underlies a potential pathogenic mechanism of chronic rheumatic heart disease [111]. Increasing evidence has shown that dysregulation of the oral microbiome induces autoimmune diseases through molecular mimicry [112]. For instance, the epitopes of P. denticola and Bacillus cereus have a high degree of similarity with the self-autoantigen Ro 60KDa, which is often detected in SLE and Sjögren’s syndrome (SS). In addition, P. gingivalis, Streptococcus spp., and F. nucleatum have similar epitopes as the self-autoantigen pyruvate dehydrogenase complex E2 (PDC-E2), which is the possible mechanism underlying the initiation and progression of primary biliary cirrhosis (PBC) [113].
Autoantigen overproduction
Cytokines regulate some proteolytic enzymes to hydrolyze extracellular proteins, generating remnant epitopes. The remnant epitopes serve as autoantigens [114]. Immune cells recognize autoantigens and produce autoantibodies. Thus, remnant epitopes are the key factors for the pathogenesis of autoimmune diseases [115]. Neutrophils were recruited and activated by IL-8/CXCL8 to secrete collagenase/ matrix metalloproteinases-8 (MMP-8) and gelatinase B/MMP-9. Gelatinase B in turn potentiated IL-8 to enhance activation of neutrophils and then increase the number of neutrophils and lymphocytes in the joint. Cartilage collagen type II was degraded to multiple protein peptides by MMP-8, which subsequently induced the activation and proliferation of autoimmune T cells [116]. Autoantibodies produced by nonspecific activation of low-affinity autoreactive B cells were triggered by infection. Soulas et al. showed that the levels of rheumatoid factors were higher in Tg mice infected by Borrelia burgdorferi [117]. Of note, the microbiota produces large amounts of proteolytic enzymes, contributing to autoantigen overproduction. For instance, the supernatants of P. gingivalis effectively cleave fibrinogen, fibronectin and type I collagen, which may promote the production of autoantibodies [118]. In addition to remnant epitopes, enzymatic antigen modification is critical in the generation of autoantigens. For instance, P. gingivalis gives rise to the production of bacterial PADs, which catalyzes the citrullination of proteins [119]. Proteins harboring citrullinated epitopes can be recognized by anti-citrullinated protein antibodies (ACPAs), which is the common pathological mechanism of RA [120, 121]. Furthermore, Aa produces pore-forming toxin LtxA to induce neutrophil hypercitrullination in patients with chronic periodontitis, and citrullinated autoantigens are the main immune targets in RA [18].
Amplification of autoimmunity by cytokines
A large body of evidence has revealed that cytokines are actively involved in the initiation and progression of autoimmune diseases [122]. Thus, the proinflammatory microenvironment modulated by the microorganisms is correlated with autoimmune responses [123]. Infection of microorganisms promoted the apoptosis of intestinal epithelial cells, and subsequently elicited the presentation of self-antigens by MHC-II molecules and the generation of autoreactive Th17 cells, leading to the occurrence of auto-inflammation and the production of autoantibodies [124]. It was reported that the level of IL-17 was significantly increased in RA, MS, and IBD [125, 126]. P. gingivalis promotes the production of RA-related cytokines and monocyte activation through TLR pathways. Similarly, F. nucleatum enhances the expression of TNF-α, IFN-gamma, IL-1β, IL-6, and IL-17, which further exacerbates IBD [127]. In addition, C. concisus promotes the secretion of proinflammatory cytokines from THP-1 cells, such as TNF-α, IL-1β, and IL-6, which increase the antigen permeability of the colonic epithelial barrier [105]. It should be pointed out that periodontitis fosters a proinflammatory microenvironment, and the levels of IL-1β, IL-4, IL-10, and IL-17A were significantly higher in diseased sites than healthy sites in the same oral cavity from periodontitis patients [128]. IL-1β stimulated T cells to promote autoimmunity by increasing the production of IL-17 [129]. Amplification of Th17 cells and the production of their effector cytokines (IL-17A, IL-17F, IL-21, CCL20) can be blocked by intravenous immunoglobulin therapy [130]. The major pathways by which oral microbiota dysbiosis affects autoimmune diseases have been summarized in Fig. 3. In addition, the effects of oral microbiota on autoimmune diseases have been summarized in Table 1.
The critical mechanism of oral microorganisms in autoimmune diseases. The major pathways of oral microorganisms-autoimmune diseases axis are shown among the microbiota translocation, molecular mimicry, autoantigen overproduction and amplification of autoimmunity by cytokine. Top left: communication between oral microbiota and the host involves the hematogenous and enteral routes. Under the adverse conditions such Ulcers and periodontal destruction, oral microbiome, such as P.g, S.mutans, and other periodontal pathogens arrive at the distant organs, subsequently increasing the release of cytokines and affecting intestinal barrier integrity. Change in the oral microbiota composition can also increase the response of molecular mimicry, autoantigen overproduction and amplification of autoimmunity by cytokine. Top right: the antigen epitopes of P. denticola, B. cereus, P.gingivalis, F.nucleatum, and Streptococcus increase. Bottom left: The production of residual and citrullinated epitope increase due to the hydrolysis of enzymes form microorganisms. The epitopes might be preferentially captured by APC which presented antigen epitopes to T cells, leading to the production of inflammatory factors. Subsequently, B cell response is activated to produce antibodies. Bottom right: Pathogenic microorganisms including P. gingivalis, F. nucleatum and C. concisus promote the release of TH1/TH17, TNFα, IFNα, Ilβ, IL-6, IL-17. p.g, P.gingivalis; k.s, Klebsiella strains; c.c, C. concisus; APC, Antigen presenting cell; TLR, Toll-like receptor; RA, Rheumatoid arthritis; IBD, Inflammatory bowel disease; Ro60, Anti-dsDNA, Anti-double stranded DNA antibody; Ro60, Ro ribonucleoprotein 60 KDa; PDC-E2, Pyruvate dehydrogenase complex E2
The relationship between the oral microbiota and autoimmune disease
The oral microbiota and IBD
Ulcerative colitis (UC) and Crohn's disease (CD) represent two common types of IBD with chronic and recurrent characteristics [131]. Genetic predisposition, environmental exposure, microbiota dysbiosis and ethnic background are the major contributors to the initiation and progression of IBD [132].
Recently, there has been growing evidence that dysbiosis of the oral microbiome plays a key role in triggering and exacerbating IBDs. A significant increase in the abundance of Saccharibacteria (TM7), Absconditabacteria (SR1), Leptotrichia, Prevotella, Bulleidia, and Atopobium was found in saliva specimens from patients with IBD [133]. Streptococcus and Enterobacteriaceae species were enriched, while Lachnospiraceae and Prevotella were depleted, in saliva samples from patients with UC. Veillonellaceae was abundant, while significant depletion of Neisseriaceae and Haemophilus were observed, in saliva samples from patients with CD [134]. Differences in the sample size and sequencing technologies might be the reasons for the contradictory findings regarding the level of Prevotella in the above two studies. The salivary microbiota profile of CD patients with periodontitis was significantly different from that of those with periodontitis alone and healthy individuals, characterized by an increase in the abundance of Proteobacteria and Firmicutes [135]. It is well established that increased abundance of Proteobacteria often leads to an imbalanced gut microbiota [136]. In addition, approximately 30 abundant bacterial taxa, including g.Prevotella, f.Prevotellaceae, and p.Bacteroidetes, were more abundant in saliva samples from patients with active CD than in samples from control subjects or those in the remission phase [137]. Streptococcus salivarius was consistently detected in saliva and fecal samples from patients with CD by whole-genome shotgun sequencing, which provides compelling evidence for the occurrence of ectopic gut colonization [138, 139]. It should be noted that the majority of Campylobacter species detected in the intestinal tissues and stool samples from patients with IBD were human oral Campylobacter species [82]. The detection rate of Campylobacter species, especially C. concisus, is significantly higher in colonic biopsy samples of UC patients than in control samples [140]. In general, C. concisus is rarely observed in healthy intestines [141]. The pathogenicity of C. concisus in the intestine manifests as destruction of the colonic epithelial barrier, which results in the enhancement of antigen permeability and further facilitates the damage caused by other enteric bacterial species [105, 142].
Mechanistically, the salivary microbiota from patients with periodontitis not only significantly changes the structure of gut microbes but also exacerbates colitis by reducing the level of unsaturated fatty acids and enhancing arachidonic acid metabolism [143]. Klebsiella strains isolated from the salivary microbiota with multidrug resistance were capable of colonizing the colon and increasing the numbers of proinflammatory TH1 cells, which subsequently aggravated the progression of IBD [104]. In addition, PADs, an enzyme or virulence factor specific to P. gingivalis, promotes the production of Th17 cells and IL-17, causing aggravation of UC [144]. It has been demonstrated that P. gingivalis dramatically increases the severity of colitis by breaking down TJPs to increase the permeability of the intestinal epithelium [145]. F. nucleatum is one of the resident bacteria and opportunistic pathogens in the human oral cavity [83, 146, 147]. It is closely associated with IBD progression, acting by damaging intestinal epithelial integrity and increasing intestinal mucosa permeability [127]. More severe epithelial cell damage was found in mice simultaneously treated with DSS and F. nucleatum [148]. Su et al. revealed that F. nucleatum exacerbated UC by promoting intestinal epithelial cell autophagy. In addition, F. nucleatum activates both the endoplasmic reticulum stress and IL-17F/NF-kappaB pathways, leading to the destruction of the intestinal mucosal barrier [149, 150]. Moreover, F. nucleatum regulates proinflammatory M1 macrophages via the AKT2 signaling pathway to exacerbate UC [151].
The oral microbiota and RA
RA is a chronic autoimmune disease with a complex multifactorial etiology. The typical clinical manifestations of RA are symmetrical inflammation and pain in the small joints of the hands and feet. The concordance rate of RA in twins is extremely low, indicating the significance of environmental risk factors for the initiation and development of RA [152].
It has been demonstrated that P. gingivalis is closely correlated with the occurrence and development of RA, and the detailed molecular mechanisms of P. gingivalis for triggering and promoting autoimmune diseases have been summarized in Fig. 4. The correlation analysis revealed that the prevalence of P. gingivalis was significantly higher in tongue biofilms from non-remission RA patients than in those from remission patients [153]. The expression of P. gingivalis was significantly increased in individuals positive for anti-cyclic citrullinated peptide compared with controls [154]. P. gingivalis triggered the initiation of periodontitis and joint inflammation in rats [155]. In addition, collagen-induced arthritis mice injected with P. gingivalis developed more severe synovial inflammation and bone destruction in joints [156]. Similarly, collagen-induced arthritis mice that were orally administered P. gingivalis suffered from more severe arthritis. Interestingly, P. gingivalis affects the progression of RA by directly migrating into joints and inducing the secretion of ACPAs. The fimbriae function of P. gingivalis and the assistance of dendritic cells, macrophages, and neutrophils might contribute to its migration. collagen-induced arthritis mice injected with anti-FimA antibody had less severe arthritis, as anti-FimA antibody inhibited the attachment and aggregation of P. gingivalis [157]. A reduction in periodontal inflammation and anti-P. gingivalis IgG levels was observed in patients with PD after periodontal treatment. However, inflammation and the level of ACPAs changed little in the joint due to ectopic colonization by P. gingivalis [158]. The development of RA is also affected by streptococcal species. Moentadj et al. found that Streptococcus species were abundant in the oral cavity of RA patients. Following the administration of streptococcal cell walls (SCWs) from Streptococcus. parasalivarius strains, ZAP-70-mutant SKG mice developed chronic arthritis [159]. A case‒control study reported that Treponema was greatly enriched in the PD and RA groups compared with the healthy controls by analyzing subgingival plaques [160]. In addition, early-stage RA patients and at-risk individuals had higher levels of Prevotella and Veillonella in saliva samples [161]. Moreover, the levels of Streptococcus parasanguinis and Actinomyces meyeri were higher in subgingival plaque samples of RA patients than in non-RA individuals [162]. The currently available animal models for dissecting the interactions between oral microbiota dysbiosis and autoimmune diseases (IBD and RA) are summarized in Table 2.
Graphical description of targeting the possible mechanisms of P. gingivalis for triggering autoimmune diseases. P. gingivalis is a Gram-negative anaerobic bacillus, mainly locating in the oral cavity. Bottom right: However, due to barrier damage and the function of withstanding stomach acidity, distant translocation of P. gingivalis into joints and enteral tissue by hematogenous dissemination and enteral route has been found in animal models. After successful colonization and survival, the higher level of endotoxin in blood and IL-17 as well as ACPAs in joints could be found in mice treated with P. gingivalis or mixed oral pathogenic microorganisms containing P. gingivalis. Bottom left: Moreover, P. gingivalis possesses immunosuppressive activities by activating the host-detrimental TLR2-Mal-PI3K pathway to blocked cellular phagocytosis. Also, P. gingivalis inhibited the host-protective TLR2-MyD88, which may further weaken the protective function of the immune system. Top middle: In addition, affecting the normal function of the complement system by P. gingivalis conducive to oral microbiota. Top left: PADs from P. gingivalis could catalyze the citrullination of proteins, which promotes the production of Th17 cells as well as IL-17 and exacerbates UC. Top right: P. gingivalis have similar epitopes as the self-autoantigen PDC-E2, which is the possible mechanism underlying the initiation and progression of primary biliary cirrhosis. ACPAs, anti-citrullinated protein antibodies; PADs, peptidylarginine deiminases; PDC-E2, pyruvate dehydrogenase complex E2
The oral microbiota and SLE
SLE is a type of chronic autoimmune connective-tissue disease of unknown etiology that affects multiple organs and leads to tissue damage and extensive inflammation. SLE is more common in women, with a global widespread prevalence of 0.02–0.24% [163]. Notably, genetic makeup, environment, sex, hormones, and the hypothalamic‒pituitary‒adrenal axis play essential roles in increasing the susceptibility to SLE [164].
Recently, a growing body of evidence has indicated the close correlation between the oral microbiota and SLE [165, 166]. Pessoa et al. revealed that 14 unique subgingival bacteria were markedly enriched in SLE patients compared to controls. In addition, the abundances of T. denticola and T. forsythia were increased in SLE-active periodontal sites compared with SLE-inactive sites and sites in healthy controls [167]. The microbial diversity was increased in the saliva samples from SLE patients, as shown by 16S ribosomal RNA gene sequencing. Veillonella, Streptococcus, and Prevotella were the predominant bacteria detected in saliva samples [168]. However, Li et al. discovered that the oral microbiota diversity from mucosal swabs was reduced in SLE patients. The levels of Sphingomonadaceae, Halomonadaceae, and Xanthomonadaceae were dramatically decreased in SLE patients [12]. The inconsistent microbial diversity may be due to the differences in the number of samples.
Notably, periodontitis might activate immune responses by maintaining high expression of TLR2 and TLR4, resulting in aggravation of the occurrence and progression of SLE. Potential cross-recognition was observed between human self Ro60 and homologous peptides from the oral microbiota, which illustrated the potential ability of bacterial antigens to induce an autoimmune response [169]. The expression of TLRs was significantly reduced after periodontal treatments, and the symptoms of SLE were subsequently improved due to the inhibition of immune responses [170]. Additionally, higher levels of IL-6, IL-17A and IL-33 were detected in the saliva of SLE patients with chronic periodontitis. These inflammatory factors contribute to the progression of SLE by inducing the recruitment, activation and differentiation of immune cells to increase the autoimmune response and promote tissue injury [171,172,173]. Interestingly, the abundance of the beneficial bacteria of Bacteroides was negatively linked to SLE disease activity, indicating that manipulating the composition of the oral microbiota might be an effective strategy for treating SLE [168].
The oral microbiota and T1D
T1D is a T-cell mediated autoimmune disease characterized by insulin deficiency due to the destruction of pancreatic β-cells. T1D is the most common type of diabetes in children and adolescence [174]. Accumulating evidence has demonstrated that genetic predisposition and environmental factors (dietary factors, infections, air pollution and microbial composition) affect the pathogenesis of T1D [175,176,177,178]. Lower microbial diversity and higher abundance of opportunistic pathogens were found in the saliva samples from children with new-onset T1D compared to those detected in the saliva samples from healthy children or children with T1D in the chronic phase receiving insulin treatment [179]. The Phyla Actinobacteria and Firmicutes, including Streptococcus spp., Actinomyces spp. and Rothia spp were the predominant bacteria detected in oral swabs of T1D patients [180]. In addition, Moskovitz et al. revealed that Streptococcus was increased in the unstimulated saliva samples from T1D patients by using 16S rRNA sequencing [181]. Aggregatibacter was found only in patients with T1D, while it was absent in the healthy controls and patients with periodontal diseases [182]. Capnocytophaga sputigena and C. ochracea were the more common detected bacterial in subgingival samples from patients with T1D [183]. Noteworthy, a bidirectional relationship was found between periodontitis and T1D. Poor glycemic control deteriorates periodontal disease by aggravating gingival inflammation, increasing probing pocket depth, and accumulating dental plaque. In addition, glycosylated hemoglobin (HbA1c) was positively correlated with periodontal disease index [11, 184]. Importantly, the levels of HbA1c and prostaglandin E2 were significantly reduced in T1D patients after periodontal treatment [185].
Microbial communities residing in oral and intestinal niches may synergistically promote the progression of T1D, highlighting the important role of oral-gut microbiota axis. Metaproteomics revealed that the microbial taxa (Alistipes and Faecalibacterium prausnitzii) contributed to maintain the function of the mucous barrier, microvilli adhesion, and exocrine pancreas were depleted in new-onset T1D [186]. The gut microbial composition in the mice administered with salivary microbiota from a periodontitis patient was significantly different from that in health-associated oral microbiota-administered (HAO) mice. Bifidobacterium, Peptostreptococcus, Atopobium, Mannheimia, Campylobacter, Olsenella, Lactobacillus, and Acidpropionibacterium were abundant in periodontitis-associated microbiota-administered (PAO) mice, while Cutibacterium, Streptococcus, Actinomyces, Faecalibaculum, Granulicatella, Gemella, Alloprevotella, and Staphylococcus were enriched in HAO mice [187]. In addition, oral bacteria led to dysbiosis of intestinal microbiota. Elevated levels of Bacteroides and Staphylococcus and reduced level of Lactobacillus spp may result in a decrease in Th17 cells and an increase in the M1/M2 macrophage ratio [188]. Bacteroides fragilis (BF) tends to aggravate T1D under the condition of increased gut permeability [189]. With the assistance of oral microorganisms such as P. gingivalis or F. nucleatum that increase the intestinal mucosa permeability, BF-like gut microorganisms promote the progression of T1D [127, 145]. Importantly, oral microbiota changes the systemic metabolism by modulating gut microbiota. A higher abundance of 2-hydroxyisobutyric acid and hydroxybenzoic acid was found in PAO mice than in HAO mice [187]. Interestingly, patients with autoimmune diseases had a higher level of 2-hydroxyisobutyric acid and hydroxybenzoic acid [190].
The oral microbiota and SS
SS, including primary SS and secondary SS, is a chronic autoimmune disease. It is mainly characterized by oral and ocular dryness due to damage to salivary and lacrimal glands with the infiltration of lymphocytes [191]. SS is a heterogeneous and multifactorial disease and is typically more common in middle-aged and older women than in men [192]. A recent study showed that Prevotella melaninogenica from the mouthwash samples of SS patients could invade ductal cells. Meanwhile, it increased the expression levels of MHC molecules, including CD80 and IFN-γ [193]. The pathogenesis of SS may be interpreted by the microbial polypeptide activation hypothesis. Peptides from the oral microbiota activated Sjogren’s syndrome antigen A /Ro60- reactive T cells, which induced autoimmune responses in SS. In particular, one of the most effective activators was peptide-P106, which is from von Willebrand factor type A domain protein (vWFA) of C. ochracea [194]. Compared with the healthy controls, both pSS patients and non-SS sicca patients exhibited a perturbed salivary microbiome profile. In addition, significant differences in the salivary microbiota were found between pSS and non-SS sicca patients [195]. It is worth noting that there is no statistically significant difference in the main oral microorganisms between xerostomia patients and the normal population [196].
An increase in Bacteroidetes and Actinobacteria abundance and a decrease in Proteobacteria abundance were found in saliva samples of pSS patients by high-throughput sequencing [197]. Veillonella was approximately four times more abundant in saliva samples from patients with pSS than in healthy subjects [198]. Compared to healthy controls, a significant increase in the relative abundance of Bifidobacterium, Dialister, and Lactobacillus was observed in the salivary samples of pSS patients, while the expression of Leptotrichia was markedly decreased [199]. In the supragingival samples of SS, the abundance of Veillonella parvula, F. nucleatum, ss vincenti, and Propionibacterium acnes was significantly increased. In particular, the level of V. parvula was higher in subgingival and supragingival samples from SS patients than in healthy controls [200]. In tongue microbiome samples, De Paiva et al. discovered that the relative abundance of Streptococcus was higher in mice with SS, while that of Leptotrichia, Fusobacterium, Bergeyella, Peptococcus, and Butyrivibrio was decreased [201]. A lower oral microbial richness was observed in a mouse model of SS [201]. However, no significant differences in bacterial richness and diversity were found between pSS patients and healthy controls [197]. The inconsistent results for oral microbiota richness in SS need further evaluation.
The oral microbiota and ankylosing spondylitis (AS)
AS, a common form of axial spondyloarthritis, is an immune-mediated inflammatory spondyloarthropathy characterized by inflammation of the sacroiliac joints and spinal attachment points. There may be no clinical symptoms in the early stage, but bone fibrosis and calcification appear in the advanced stages of AS. To date, it is generally accepted that AS is influenced by multiple factors, including genetic background, immune reaction, and microbial infection. The exact pathogenic mechanism of AS has not yet been established [202, 203]. Arthritis was developed in the rats under the conventional feeding condition but not in those maintained in the sterile condition. In addition, intramural injection of the bacterial cell wall polymers resulted in peripheral arthritis and the inflammatory responses were blocked by the addition of metronidazole [204]. Wen et al. compared the gut microbial composition in AS patients and healthy controls with quantitative metagenomics analysis. An increased abundance of P. melaninogenica, Prevotella copri, and Prevotella sp. C561 and a decreased level of Bacteroides spp were found in AS patients [205]. An unbalanced gut microbiota was observed in patients with AS, showing decreased quantity of bacteria and a significant increase in Bifidobacterium species [206]. Notably, the levels of Veillonellaceae, Brucella spp., and C. concisus were significantly higher in saliva samples from patients with AS, while the abundance of Streptococcus was decreased [207]. Stoll et al. reported that the levels of salivary Rothia mucilaginosa and plaque-derived Fusobacterium were significantly increased in patients with spondyloarthritis [208], indicating AS is potentially an oral microbiome-driven disease.
The oral microbiota and MS
MS is a chronic immune-mediated, inflammatory neurological disease that widely damages the brain and spinal cord, giving rise to demyelination and axonal and neuronal loss. The exact cause of MS has not been completely elucidated to date. Numerous studies have shown that the gut microbiome plays a critical role in the pathogenesis of MS [209]. However, the correlation between the oral microbiota and MS is still unknown. Compared with healthy controls, an increased abundance of bacteroidetes and proteobacteria, and a reduction in the levels of firmicutes and actinobacteria were found in the saliva specimens collected from monozygotic female twins with MS. At the genus level, prevotella, rothia, and haemophilus were depleted while streptococcus and actinomyces were over-represented in saliva specimens from MS patients [210]. Another study investigated that the salivary microbiome of MS is characterized increased abundance of the genera Staphylococcus, Actinomyces, Fusobacterium, Bacteroides, Porphyromonas, Prevotella, Veillonella, and Propionibacterium and uncultivable bacteria [211]. Lipid 654, a known product from P. gingivalis and functioned as a TLR2 ligand, potentially promotes inflammatory responses [212]. Therefore, we speculate that the detection of oral microbiota and its products may possibly have positive effect to identify the pathogenesis of MS and formulate preventive and therapeutic measures.
The oral microbiota and autoimmune liver diseases (AILDs)
AILDs are chronic inflammatory liver conditions that include AIH, PBC, and primary sclerosing cholangitis (PSC) [213]. AIH, a chronic immune-mediated and progressive condition, is characterized by elevated serum transaminases, hypergamma-globulinemia, and positive autoantibodies. The importance of gut microbiota in shaping and inducing the host immune response as well as promoting the development of AIH has been studied extensively [77, 214]. Abnormal gut microbiota composition in patients with AIH has been reported by Wei et al. The expression of obligate anaerobes was decreased, while Veillonella was increased in fecal samples from AIH patients. In addition, Veillonella dispar was positively associated with serum level of aspartate aminotransferase and the severity of hepatic inflammation. Moreover, abnormal lipopolysaccharide biosynthesis and amino acid metabolism were found in the fecal samples from AIH patients [215]. However, as oral microbiota plays a crucial role in shaping the composition and structure of gut microbiota, the role of oral microbiome in modulating the pathogenesis of AIH needs further investigations. In fact, the oral microbiome diversity was markedly increased in saliva samples from the AIH group compared with the healthy control group. The levels of Streptococcus, Veillonella, and Leptotrichia were significantly elevated in AIH patients [216]. In addition, AIH was characterized by an increase in Veillonella and a decrease in Streptococcus in the saliva samples. A positive correlation was found between the abundance of Veillonella and proinflammatory cytokine levels [217].
PBC is a chronic cholestatic liver disease that occurs predominant in women and is characterized by damage to small intrahepatic bile ducts. PBC may be driven by microbial imbalance, which leads to liver inflammation towards fibrosis and subsequent cirrhosis. A decreased abundance of autochthonous families and an increased abundance of Enterobacteriaceae and Enterococcaceae were observed in the saliva samples from cirrhotic patients [218]. Furthermore, Abe et al. showed that the levels of Eubacterium and Veillonella were increased in saliva samples from PBC patients, while that of Fusobacterium was decreased [219]. By using 16S rDNA sequencing, Bacteroidetes, Campylobacter, Prevotella, and Veillonella were found to be enriched in the saliva samples of PBC patients, while Enterococcaceae, Granulicatella, Rothia, and Streptococcus were depleted. In addition, saliva supernatants from PBC patients induced the production of inflammatory mediators, and the levels of inflammatory cytokines were significantly decreased following effective tooth brushing, indicating that a dysregulated oral microbiota may stimulate inflammatory responses to promote the development of PBC [220]. Of note, the oral microorganisms such as Streptococcaceae were found to be significantly increased in small intestinal lining in cirrhotic mice [221]. Due to the unique anatomical of hepato-enteric portal vein system, bacteria from oral cavity and gastrointestinal tract can translocate into liver tissues to trigger immune responses [222]. The immunosuppressive molecule, IL-35, was found to be lower in peripheral blood mononuclear cells of PBC patients than those of the healthy controls. Importantly, the levels of alkaline phosphatase and IFN-γ were negatively correlated with the level of plasma IL-35 in PBC patients [223]. Moreover, the expression of pro-inflammatory factors including IL-17, IL-6, IFN-γ, TNF-α and IL-10 was significantly higher in serum samples from PBC patients [224]. The bacterium might easily translocate into the portal blood and arrive at liver due to the breakdown of intestinal barrier [37].
PSC is a rare chronic cholestatic disease characterized by progressive inflammation and fibrosis of the intrahepatic and extrahepatic bile ducts. PSC might develop into cirrhosis, portal hypertension, and liver decompensation in the advanced stages [225]. Recently, Lapidot et al. reported that Veillonella, Scardovia, and Streptococcus were greatly enriched, while native bacteria were depleted, in the saliva samples from PSC patients by using 16S rRNA sequencing [226]. Furthermore, Rothia and Haemophilus were less abundant in saliva samples from PSC patients than in healthy controls [227]. By using 16S rRNA sequencing, significant differences in the bacterial community structure were found in the saliva samples from patients with PSC and healthy individuals [228]. Notably, it is estimated that approximately 70% of patients with PSC have IBD, which highlights the involvement of gut-liver axis [229]. It was found that the circulating bacterial translocation markers including lipopolysaccharide-binding protein and soluble CD14 were significantly elevated in the serum samples of patients with PSC [230]. Due to the assistance of impaired intestinal epithelial barrier, we speculated that oral microorganisms and their metabolites reached liver through hepato-enteric portal vein system and subsequently induce activation of immune cells.
The oral microbiota and immunoglobulin a nephropathy (IgAN)
IgAN is an immune-mediated and idiopathic glomerulonephritis characterized by mesangial hyperplasia with IgA deposition. The long-term prognosis for IgAN remains poor, with approximately 30–40% of IgAN patients developing end-stage renal failure within 10–20 years [231, 232]. To date, the immune triggers of IgAN are still not completely understood.
Accumulating evidence has demonstrated that the oral microbiome might be actively involved in the pathogenesis of IgAN. By using high-throughput sequencing technology, a dramatic increase in Granulicatella abundance and a marked reduction in Prevotella and Veillonella abundance were found in the saliva samples of IgAN patients [233]. Piccolo et al. demonstrated that the ratio of Firmicutes to Proteobacteria was significantly reduced, while the Pasteurellaceae family was found to be enriched, in saliva samples of IgAN patients [234]. Compared with healthy controls, the abundance of Neisseria was significantly higher in saliva samples from the IgAN group [235]. In addition, IgAN patients and healthy controls could be differentiated by the abundance Capnocytophaga, Rothia, and Haemophilus in saliva samples. In particular, the abundance of Capnocytophaga was found to be positively correlated with the level of proteinuria [236]. The changes in subgingival microbial structure seem to affect the incidence of IgAN. Proteobacteria and Actinobacteria at the phylum level, Betaproteobacteria, Bacilli, Actinobacteria, Flavobacteriia, and Gammaproteobacteria at the class level, and Bergeyella, Capnocytophaga, Actinomyces, Corynebacterium, Comamonas, Lautropia, and Streptococcus at the genus level were significantly abundant in the subgingival microbiome from CP patients with IgAN compared to those without IgAN. In addition, the bacterial composition in subgingival plaque differed greatly between CP patients with and without IgAN [237].
In particular, Cnm- ( +) S. mutans might be involved in the incidence and progression of IgAN [238]. First, Cnm- ( +) S. mutans was significantly abundant in saliva samples of IgAN patients. In addition, a higher Cnm protein-positive rate in tonsil specimens was observed in patients with IgAN [239]. Recently, Naka et al. reported higher positive staining rates of IgA and C3 in rats treated with Cnm- ( +) S. mutans [240]. More importantly, infection with S. mutans might promote the deposition of IgA1 in mesangial areas in glomeruli by abnormal glycosylation of serine or threonine amino acids of IgA1 [241]. The detailed alterations of oral microbial profile in various autoimmune diseases are summarized in Table 3.
Balancing the oral microbiota for treating autoimmune diseases
Due to the indispensable role of oral microbiota dysbiosis in the initiation and progression of autoimmune diseases, targeting the oral microbiota might be an effective strategy for preventing and treating autoimmune diseases (Fig. 5). Maintenance of the dynamic homeostasis between the oral microbiota and the host immune system contributes to maintaining body health. Avoiding the driving factors for oral microbiota dysbiosis and boosting the host’s immune system are two potential practical approaches for balancing the oral microbiota. Good oral hygiene behaviors, a low carbohydrate diet and healthy lifestyles reduce the oral microbial/bacterial load [242]. In addition, the usage of prebiotics, probiotics, or synbiotics not only reverses oral microbiota dysbiosis but also enhances the resilience of oral microflora [243, 244]. For instance, the prebiotic D-tagatose suppressed the growth of the oral pathogens S. mutans and Streptococcus. gordonii [245]. Interestingly, it has been demonstrated that prebiotics affect gut microbiota composition and enhance the production of beneficial microbial metabolites (short-chain fatty acids, tryptophan and organic acids) for pathogenic bacterial suppression and gut barrier improvement [246]. Targeting microbiota-derived metabolites might also be an effective strategy for treating oral microbiota-mediated autoimmune diseases.
Treatment strategy on balancing oral microbiota for autoimmune diseases. Oral microbiota dysbiosis could lead to development of autoimmune diseases, therefore good oral hygiene behavior, low carbohydrate diet, healthy lifestyles, metabolites (prebiotics, probiotics, synbiotics), OMT, and Nanomedicine-based treatment are viable approaches to reverse dysbiosis and restore eubiosis. Increasing driving factors, such as good oral hygiene behavior, low carbohydrate diet, healthy lifestyles could reduce percentage of oral microbial dysbiosis. Prebiotics, probiotics, synbiotics could inhibit oral pathogenic microorganisms and prevent infections caused by oral pathogens. On the other hand, OMT introduces a new and healthy bacterial community to the recipient with the intent of reversing the established dysbiosis. Moreover, the combined effects of nanomedicine-based therapeutics and OMT could potentially and precisely use to oral microbiota delivery. OMT, oral microbiota transplantation; SCFA, Short-chain fatty acids
Fecal microbiota transplantation (FMT) is an emerging strategy for the treatment of refractory diseases in recent years. For example, this approach has demonstrated promising effects in maintaining remission status in CD and inducing remission in UC [247, 248]. In addition, the gut microbiota could be modulated for the majority of participants by using oral FMT capsules from healthy lean donors [249]. We propose that oral microbiota transplantation (OMT) might be a potential effective strategy for treating oral microbiota-mediated autoimmune diseases, as OMT has shown great promise for curing oral microbiota-mediated diseases. For instance, the microbiota composition in dogs with naturally occurring periodontitis could be regulated by OMT [250]. In addition, OMT alleviated radiotherapy-induced oral mucositis by reshaping the oral bacteria taxonomic composition in mice [251]. Oral bacteria can be effectively encapsulated as a treatment for autoimmune diseases.
Enhancing the synergistic effects of balancing the oral microbiota might be important for achieving better therapeutic outcomes. TNF-α siRNA nanomedicine-targeted macrophages significantly attenuated TNF-α levels and improved IBD in an animal model [252, 253]. Further studies are warranted to determine the combined effects of nanomedicine-based therapeutics and OMT.
Conclusions
It is now well recognized that oral microbiota dysbiosis plays a crucial role in modulating the initiation and development of many autoimmune diseases, and targeting oral microbes might be a promising strategy for ultimately achieving the goal of curing autoimmune diseases. Unfortunately, few clinical trials have evaluated the beneficial effects of microbiota-based therapies on autoimmune diseases. In addition, effective strategies for balancing the oral microbiota to treat autoimmune diseases are urgently needed. Moreover, the underlying molecular mechanisms by which host-microbe interactions in the oral cavity remotely trigger and exacerbate autoimmune diseases remain unclear. Systematic combination of metabolomics, metagenomics, metatranscriptomics, and proteomics might improve our understanding of the complex interactions between the oral microbial ecosystem and immune system, which might provide novel oral microbiota targets for treating autoimmune diseases.
Availability of data and materials
Not applicable.
Abbreviations
- PADs:
-
Peptidylarginine deiminases
- Aa:
-
Aggregatibacter actinomycetemcomitans
- LtxA:
-
Toxin leukotoxin-A
- SLE:
-
Systemic lupus erythematosu
- IBD:
-
Inflammatory bowel disease
- TLRs:
-
Toll-like receptors
- PRRs:
-
Pattern recognition receptors
- APCs:
-
Antigen-presenting cells
- DSS:
-
Dextran sodium sulfate
- MS:
-
Multiple sclerosis
- Amp:
-
Ampicillin
- Tyl:
-
Tylosin
- SS:
-
Sjögren’s syndrome
- PDC-E:
-
Pyruvate dehydrogenase complex E2
- PBC:
-
Primary biliary cirrhosis
- ACPAs:
-
Anti-citrullinated protein antibodies
- UC:
-
Ulcerative colitis
- CD:
-
Crohn's disease
- RA:
-
Rheumatoid arthritis
- SCWs:
-
Streptococcal cell walls
- vWFA:
-
Von willebrand factor type a domain protein
- AS:
-
Ankylosing spondylitis
- AILDs:
-
Autoimmune liver diseases
- AIH:
-
Autoimmune hepatitis
- PSC:
-
Primary sclerosing cholangitis
- IgAN:
-
Immunoglobulin A nephropathy
- FMT:
-
Fecal microbiota transplantation
- OMT:
-
Oral microbiota transplantation
- T1D:
-
Type I diabetes
- BF:
-
Bacteroides fragilis
- PAO:
-
Periodontitis-associated microbiota-administered
- HAO:
-
Health-associated oral microbiota-administered
- MHC:
-
Major histocompatibility complex
- HbA1c:
-
Glycosylated hemoglobin
- TJP:
-
Tight junction protein
References
De Giani A, Zampolli J, Di Gennaro P. Recent trends on biosurfactants with antimicrobial activity produced by bacteria associated with human health: different perspectives on their properties, challenges, and potential applications. Front Microbiol. 2021;12: 655150.
Thomas S, Izard J, Walsh E, Batich K, Chongsathidkiet P, Clarke G, et al. The host microbiome regulates and maintains human health: a primer and perspective for non-microbiologists. Cancer Res. 2017;77:1783–812.
Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol. 2017;17:219–32.
Kamdar K, Khakpour S, Chen J, Leone V, Brulc J, Mangatu T, et al. Genetic and metabolic signals during acute enteric bacterial infection Alter the microbiota and drive progression to chronic inflammatory disease. Cell Host Microbe. 2016;19:21–31.
Bretin A, Lucas C, Larabi A, Dalmasso G, Billard E, Barnich N, et al. AIEC infection triggers modification of gut microbiota composition in genetically predisposed mice, contributing to intestinal inflammation. Sci Rep. 2018;8:12301.
Kitamoto S, Nagao-Kitamoto H, Hein R, Schmidt TM, Kamada N. The bacterial connection between the oral cavity and the gut diseases. J Dent Res. 2020;99:1021–9.
Gao L, Xu T, Huang G, Jiang S, Gu Y, Chen F. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell. 2018;9:488–500.
Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. 2018;16:745–59.
Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol. 2014;16:1024–33.
Coats SR, Jones JW, Do CT, Braham PH, Bainbridge BW, To TT, et al. Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4’-phosphatase activities. Cell Microbiol. 2009;11:1587–99.
Jensen ED, Selway CA, Allen G, Bednarz J, Weyrich LS, Gue S, et al. Early markers of periodontal disease and altered oral microbiota are associated with glycemic control in children with type 1 diabetes. Pediatr Diabetes. 2021;22:474–81.
Li BZ, Zhou HY, Guo B, Chen WJ, Tao JH, Cao NW, et al. Dysbiosis of oral microbiota is associated with systemic lupus erythematosus. Arch Oral Biol. 2020;113: 104708.
Dabdoub SM, Ganesan SM, Kumar PS. Comparative metagenomics reveals taxonomically idiosyncratic yet functionally congruent communities in periodontitis. Sci Rep. 2016;6:38993.
Bruserud O, Siddiqui H, Marthinussen MC, Chen T, Jonsson R, Oftedal BE, et al. Oral microbiota in autoimmune polyendocrine syndrome type 1. J Oral Microbiol. 2018;10:1442986.
Dutzan N, Kajikawa T, Abusleme L, Greenwell-Wild T, Zuazo CE, Ikeuchi T, et al. A dysbiotic microbiome triggers TH17 cells to mediate oral mucosal immunopathology in mice and humans. Sci Transl Med. 2018;10:463.
Curran AM, Naik P, Giles JT, Darrah E. PAD enzymes in rheumatoid arthritis: pathogenic effectors and autoimmune targets. Nat Rev Rheumatol. 2020;16:301–15.
Trouw LA, Haisma EM, Levarht EW, van der Woude D, Ioan-Facsinay A, Daha MR, et al. Anti-cyclic citrullinated peptide antibodies from rheumatoid arthritis patients activate complement via both the classical and alternative pathways. Arthritis Rheum. 2009;60:1923–31.
Konig MF, Abusleme L, Reinholdt J, Palmer RJ, Teles RP, Sampson K, et al. Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci Transl Med. 2016;8:369176.
Silverman GJ, Azzouz DF, Alekseyenko AV. Systemic Lupus Erythematosus and dysbiosis in the microbiome: cause or effect or both? Curr Opin Immunol. 2019;61:80–5.
Toscano M, De Grandi R, Grossi E, Drago L. Role of the human breast milk-associated microbiota on the newborns’ immune system: a mini review. Front Microbiol. 2017;8:2100.
Perez PF, Dore J, Leclerc M, Levenez F, Benyacoub J, Serrant P, et al. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics. 2007;119:e724-732.
Sun M, Ma N, He T, Johnston LJ, Ma X. Tryptophan (Trp) modulates gut homeostasis via aryl hydrocarbon receptor (AhR). Crit Rev Food Sci Nutr. 2020;60:1760–8.
Maynard CL, Elson CO, Hatton RD, Weaver CT. Reciprocal interactions of the intestinal microbiota and immune system. Nature. 2012;489:231–41.
Spahn TW, Herbst H, Rennert PD, Lugering N, Maaser C, Kraft M, et al. Induction of colitis in mice deficient of Peyer’s patches and mesenteric lymph nodes is associated with increased disease severity and formation of colonic lymphoid patches. Am J Pathol. 2002;161:2273–82.
Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–73.
Gebrayel P, Nicco C, Al Khodor S, Bilinski J, Caselli E, Comelli EM, et al. Microbiota medicine: towards clinical revolution. J Transl Med. 2022;20:111.
Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204.
Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422:522–6.
Sudhakara P, Gupta A, Bhardwaj A, Wilson A. Oral dysbiotic communities and their implications in systemic diseases. Dent J. 2018;6(2):10.
Adler CJ, Dobney K, Weyrich LS, Kaidonis J, Walker AW, Haak W, et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and industrial revolutions. Nat Genet. 2013;4:450–5.
Requena T, Martinez-Cuesta MC, Pelaez C. Diet and microbiota linked in health and disease. Food Funct. 2018;9:688–704.
Fritz BM, Munoz B, Yin F, Bauchle C, Atwood BK. A high-fat, high-sugar “western” diet alters dorsal striatal glutamate, opioid, and dopamine transmission in mice. Neuroscience. 2018;372:1–15.
Berridge KC, Ho CY, Richard JM, DiFeliceantonio AG. The tempted brain eats: pleasure and desire circuits in obesity and eating disorders. Brain Res. 2010;1350:43–64.
Manzel A, Muller DN, Hafler DA, Erdman SE, Linker RA, Kleinewietfeld M. Role of “Western diet” in inflammatory autoimmune diseases. Curr Allergy Asthma Rep. 2014;14:404.
Lassalle F, Spagnoletti M, Fumagalli M, Shaw L, Dyble M, Walker C, et al. Oral microbiomes from hunter-gatherers and traditional farmers reveal shifts in commensal balance and pathogen load linked to diet. Mol Ecol. 2018;27:182–95.
Sen T, Cawthon CR, Ihde BT, Hajnal A, DiLorenzo PM, de La Serre CB, et al. Diet-driven microbiota dysbiosis is associated with vagal remodeling and obesity. Physiol Behav. 2017;173:305–17.
Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010;4:232–41.
Lee HC, Yu SC, Lo YC, Lin IH, Tung TH, Huang SY. A high linoleic acid diet exacerbates metabolic responses and gut microbiota dysbiosis in obese rats with diabetes mellitus. Food Funct. 2019;10:786–98.
Chaves IM, Zicker MC, Laranjeira AO, Silveira ALM, Aguiar DC, Barrioni BR, et al. Dysbiotic oral microbiota contributes to alveolar bone loss associated with obesity in mice. J Appl Oral Sci. 2022;30: e20220238.
Sun S, Araki Y, Hanzawa F, Umeki M, Kojima T, Nishimura N, et al. High sucrose diet-induced dysbiosis of gut microbiota promotes fatty liver and hyperlipidemia in rats. J Nutr Biochem. 2021;93: 108621.
Kong C, Gao R, Yan X, Huang L, Qin H. Probiotics improve gut microbiota dysbiosis in obese mice fed a high-fat or high-sucrose diet. Nutrition. 2019;60:175–84.
Lv X-C, Guo W-L, Li L, Yu X-D, Liu B. Polysaccharide peptides from Ganoderma lucidum ameliorate lipid metabolic disorders and gut microbiota dysbiosis in high-fat diet-fed rats. J Funct Foods. 2019;57:48–58.
Haro C, Garcia-Carpintero S, Rangel-Zuniga OA, Alcala-Diaz JF, Landa BB, Clemente JC, et al. Consumption of two healthy dietary patterns restored microbiota dysbiosis in obese patients with metabolic dysfunction. Mol Nutr Food Res. 2017;61:12.
Li L, Guo WL, Zhang W, Xu JX, Qian M, Bai WD, et al. Grifola frondosa polysaccharides ameliorate lipid metabolic disorders and gut microbiota dysbiosis in high-fat diet fed rats. Food Funct. 2019;10:2560–72.
Zhang M, Jiang Z, Li D, Jiang D, Wu Y, Ren H, et al. Oral antibiotic treatment induces skin microbiota dysbiosis and influences wound healing. Microb Ecol. 2015;69:415–21.
Lange K, Buerger M, Stallmach A, Bruns T. Effects of antibiotics on gut microbiota. Dig Dis. 2016;34:260–8.
Wu H, Ma Y, Peng X, Qiu W, Kong L, Ren B, et al. Antibiotic-induced dysbiosis of the rat oral and gut microbiota and resistance to Salmonella. Arch Oral Biol. 2020;114: 104730.
Hajishengallis G. The inflammophilic character of the periodontitis-associated microbiota. Mol Oral Microbiol. 2014;29:248–57.
Olsen I, Lambris JD, Hajishengallis G. Porphyromonas gingivalis disturbs host-commensal homeostasis by changing complement function. J Oral Microbiol. 2017;9:1340085.
Al-Attar A, Alimova Y, Kirakodu S, Kozal A, Novak MJ, Stromberg AJ, et al. Activation of notch-1 in oral epithelial cells by P. gingivalis triggers the expression of the antimicrobial protein PLA(2)-IIA. Mucosal Immunol. 2018;11:1047–59.
Selgrade MK, Cooper GS, Germolec DR, Heindel JJ. Linking environmental agents and autoimmune disease: an agenda for future research. Environ Health Perspect. 1999;107(Suppl 5):811–3.
Nicholson LB. The immune system. Essays Biochem. 2016;60:275–301.
Wahren-Herlenius M, Dorner T. Immunopathogenic mechanisms of systemic autoimmune disease. Lancet. 2013;382:819–31.
Nahid MA, Satoh M, Chan EK. Interleukin 1beta-responsive MicroRNA-146a Is critical for the cytokine-induced tolerance and cross-tolerance to toll-like receptor ligands. J Innate Immun. 2015;7:428–40.
Li B, Wang X, Choi IY, Wang YC, Liu S, Pham AT, et al. miR-146a modulates autoreactive Th17 cell differentiation and regulates organ-specific autoimmunity. J Clin Invest. 2017;127:3702–16.
Jiang S, Hu Y, Deng S, Deng J, Yu X, Huang G, et al. miR-146a regulates inflammatory cytokine production in Porphyromonas gingivalis lipopolysaccharide-stimulated B cells by targeting IRAK1 but not TRAF6. Biochim Biophys Acta Mol Basis Dis. 2018;1864:925–33.
Sun W, Ma J, Zhao H, Xiao C, Zhong H, Ling H, et al. Resolvin D1 suppresses pannus formation via decreasing connective tissue growth factor caused by upregulation of miRNA-146a-5p in rheumatoid arthritis. Arthritis Res Ther. 2020;22:61.
Ding S, Duan H, Fang F, Shen H, Xiao W. CTGF promotes articular damage by increased proliferation of fibroblast-like synoviocytes in rheumatoid arthritis. Scand J Rheumatol. 2016;45:282–7.
Abusleme L, Diaz PI, Freeman AF, Greenwell-Wild T, Brenchley L, Desai JV, et al. Human defects in STAT3 promote oral mucosal fungal and bacterial dysbiosis. JCI Insight. 2018. https://doi.org/10.1172/jci.insight.1220.
Suarez LJ, Garzon H, Arboleda S, Rodriguez A. Oral dysbiosis and autoimmunity: from local periodontal responses to an imbalanced systemic immunity. A Rev Front Immunol. 2020;11: 591255.
Ruff WE, Greiling TM, Kriegel MA. Host-microbiota interactions in immune-mediated diseases. Nat Rev Microbiol. 2020;18:521–38.
Radsak M, Iking-Konert C, Stegmaier S, Andrassy K, Hansch GM. Polymorphonuclear neutrophils as accessory cells for T-cell activation: major histocompatibility complex class II restricted antigen-dependent induction of T-cell proliferation. Immunology. 2000;101:521–30.
Fanger NA, Liu C, Guyre PM, Wardwell K, O’Neil J, Guo TL, et al. Activation of human T cells by major histocompatability complex class II expressing neutrophils: proliferation in the presence of superantigen, but not tetanus toxoid. Blood. 1997;89:4128–35.
Ben-Sasson SZ, Wang K, Cohen J, Paul WE. IL-1beta strikingly enhances antigen-driven CD4 and CD8 T-cell responses. Cold Spring Harb Symp Quant Biol. 2013;78:117–24.
Navegantes KC, de Souza GR, Pereira PAT, Czaikoski PG, Azevedo CHM, Monteiro MC. Immune modulation of some autoimmune diseases: the critical role of macrophages and neutrophils in the innate and adaptive immunity. J Transl Med. 2017;15:36.
Chen Z, Bozec A, Ramming A, Schett G. Anti-inflammatory and immune-regulatory cytokines in rheumatoid arthritis. Nat Rev Rheumatol. 2019;15:9–17.
Nikoopour E, Schwartz JA, Singh B. Therapeutic benefits of regulating inflammation in autoimmunity. Inflamm Allergy Drug Targets. 2008;7:203–10.
Waldner H. The role of innate immune responses in autoimmune disease development. Autoimmun Rev. 2009;8:400–4.
Satoh T, Akira S. Toll-like receptor signaling and its inducible proteins. Microbiol Spectr. 2016. https://doi.org/10.1128/microbiolspec.MCHD-0040-2016.
Pasare C, Medzhitov R. Toll-like receptors: linking innate and adaptive immunity. Adv Exp Med Biol. 2005;560:11–8.
Cabeza-Cabrerizo M, Cardoso A, Minutti CM. Pereira da costa m, reis ESC. Dendritic Cells Revisited Annu Rev Immunol. 2021;39:131–66.
Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801.
Janeway CA Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216.
Aluri J, Cooper MA, Schuettpelz LG. Toll-Like receptor signaling in the establishment and function of the immune system. Cells. 2021. https://doi.org/10.3390/cells10061374.
Toor D, Wsson MK, Kumar P, Karthikeyan G, Kaushik NK, Goel C, et al. Dysbiosis disrupts gut immune homeostasis and promotes gastric diseases. Int J Mol Sci. 2019. https://doi.org/10.3390/ijms20102432.
Maekawa T, Krauss JL, Abe T, Jotwani R, Triantafilou M, Triantafilou K, et al. Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe. 2014;15:768–78.
Xu H, Liu M, Cao J, Li X, Fan D, Xia Y, et al. The dynamic interplay between the gut microbiota and autoimmune diseases. J Immunol Res. 2019;2019:7546047.
Yoshikawa T, Watanabe T, Kamata K, Hara A, Minaga K, Kudo M. Intestinal dysbiosis and autoimmune pancreatitis. Front Immunol. 2021;12: 621532.
Lin X, Liu Y, Ma L, Ma X, Shen L, Ma X, et al. Constipation induced gut microbiota dysbiosis exacerbates experimental autoimmune encephalomyelitis in C57BL/6 mice. J Transl Med. 2021;19:317.
Mo S, Ru H, Huang M, Cheng L, Mo X, Yan L. Oral-intestinal microbiota in colorectal cancer: inflammation and immunosuppression. J Inflamm Res. 2022;15:747–59.
Lu M, Xuan S, Wang Z. Oral microbiota: a new view of body health. Food Sci Human Wellness. 2019;8:8–15.
Zhang L. Oral Campylobacter species: Initiators of a subgroup of inflammatory bowel disease? World J Gastroenterol. 2015;21:9239–44.
Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42.
Kato T, Yamazaki K, Nakajima M, Date Y, Kikuchi J, Hase K, et al. Oral administration of porphyromonas gingivalis alters the gut microbiome and serum metabolome. Msphere. 2018. https://doi.org/10.1128/mSphere.00460-18.
Nakajima M, Arimatsu K, Kato T, Matsuda Y, Minagawa T, Takahashi N, et al. Oral Administration of P gingivalis induces dysbiosis of gut microbiota and impaired barrier function leading to dissemination of enterobacteria to the liver. PLoS One. 2015;10:e0134234.
Hu B, Elinav E, Huber S, Strowig T, Hao L, Hafemann A, et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc Natl Acad Sci USA. 2013;110:9862–7.
Mehta NN, McGillicuddy FC, Anderson PD, Hinkle CC, Shah R, Pruscino L, et al. Experimental endotoxemia induces adipose inflammation and insulin resistance in humans. Diabetes. 2010;59:172–81.
Wang H, Wang G, Banerjee N, Liang Y, Du X, Boor PJ, et al. Aberrant gut microbiome contributes to intestinal oxidative stress, barrier dysfunction, inflammation and systemic autoimmune responses in MRL/lpr mice. Front Immunol. 2021;12: 651191.
Wang H, Wang G, Liang Y, Du X, Boor PJ, Sun J, et al. Redox regulation of hepatic NLRP3 inflammasome activation and immune dysregulation in trichloroethene-mediated autoimmunity. Free Radic Biol Med. 2019;143:223–31.
Bao J, Li L, Zhang Y, Wang M, Chen F, Ge S, et al. Periodontitis may induce gut microbiota dysbiosis via salivary microbiota. Int J Oral Sci. 2022;14:32.
Sato K, Takahashi N, Kato T, Matsuda Y, Yokoji M, Yamada M, et al. Aggravation of collagen-induced arthritis by orally administered Porphyromonas gingivalis through modulation of the gut microbiota and gut immune system. Sci Rep. 2017;7:6955.
Barksby HE, Nile CJ, Jaedicke KM, Taylor JJ, Preshaw PM. Differential expression of immunoregulatory genes in monocytes in response to Porphyromonas gingivalis and Escherichia coli lipopolysaccharide. Clin Exp Immunol. 2009;156:479–87.
Sohn J, Li L, Zhang L, Settem RP, Honma K, Sharma A, et al. Porphyromonas gingivalis indirectly elicits intestinal inflammation by altering the gut microbiota and disrupting epithelial barrier function through IL9-producing CD4(+) T cells. Mol Oral Microbiol. 2021. https://doi.org/10.1111/omi.12359.
Kinashi Y, Hase K. Partners in leaky gut syndrome: intestinal dysbiosis and autoimmunity. Front Immunol. 2021;12: 673708.
Ashare A, Stanford C, Hancock P, Stark D, Lilli K, Birrer E, et al. Chronic liver disease impairs bacterial clearance in a human model of induced bacteremia. Clin Transl Sci. 2009;2:199–205.
Hirschfeld J, Kawai T. Oral inflammation and bacteremia: implications for chronic and acute systemic diseases involving major organs. Cardiovasc Hematol Disord Drug Targets. 2015;15:70–84.
Kojima A, Nomura R, Naka S, Okawa R, Ooshima T, Nakano K. Aggravation of inflammatory bowel diseases by oral streptococci. Oral Dis. 2014;20:359–66.
Abed J, Emgård JE, Zamir G, Faroja M, Almogy G, Grenov A, et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed gal-GalNAc. Cell Host Microbe. 2016;20:215–25.
Tsukasaki M, Komatsu N, Nagashima K, Nitta T, Pluemsakunthai W, Shukunami C, et al. Host defense against oral microbiota by bone-damaging T cells. Nat Commun. 2018;9:701.
Chukkapalli S, Rivera-Kweh M, Gehlot P, Velsko I, Bhattacharyya I, Calise SJ, et al. Periodontal bacterial colonization in synovial tissues exacerbates collagen-induced arthritis in B10 RIII mice. Arthritis Res Ther. 2016;18:161.
Mishra A, Makharia GK. Techniques of functional and motility test: how to perform and interpret intestinal permeability. J Neurogastroenterol Motil. 2012;18:443–7.
Humphrey SP, Williamson RT. A review of saliva: normal composition, flow, and function. J Prosthet Dent. 2001;85:162–9.
Arimatsu K, Yamada H, Miyazawa H, Minagawa T, Nakajima M, Ryder MI, et al. Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci Rep. 2014;4:4828.
Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, et al. Ectopic colonization of oral bacteria in the intestine drives T(H)1 cell induction and inflammation. Science. 2017;358:359–65.
Nattramilarasu PK, Lobo de Sa FD, Schulzke JD, Bucker R. Immune-mediated aggravation of the campylobacter concisus-induced epithelial barrier dysfunction. Int J Mol Sci. 2021. https://doi.org/10.3390/ijms22042043.
Kitamoto S, Nagao-Kitamoto H, Jiao Y, Gillilland MG 3rd, Hayashi A, Imai J, et al. The Intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell. 2020;182:447-462.e414.
Kanduc D, Stufano A, Lucchese G, Kusalik A. Massive peptide sharing between viral and human proteomes. Peptides. 2008;29:1755–66.
Hotokezaka H, Hayashida H, Ohara N, Nomaguchi H, Kobayashi K, Yamada T. Cloning and sequencing of the groESL homologue from Porphyromonas gingivalis. Biochim Biophys Acta. 1994;1219:175–8.
Nakano Y, Inai Y, Yamashita Y, Nagaoka S, Kusuzaki-Nagira T, Nishihara T, et al. Molecular and immunological characterization of a 64-kDa protein of Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 1995;10:151–9.
Tabeta K, Yamazaki K, Hotokezaka H, Yoshie H, Hara K. Elevated humoral immune response to heat shock protein 60 (hsp60) family in periodontitis patients. Clin Exp Immunol. 2000;120:285–93.
Guilherme L, Kalil J, Cunningham M. Molecular mimicry in the autoimmune pathogenesis of rheumatic heart disease. Autoimmunity. 2006;39:31–9.
Cusick MF, Libbey JE, Fujinami RS. Molecular mimicry as a mechanism of autoimmune disease. Clin Rev Allergy Immunol. 2012;42:102–11.
Obando-Pereda G. Trojans in oral environments: evidence of molecular mimicry in oral immunity. Oral Microbiol Periodontitis. 2018. https://doi.org/10.5772/intechopen.75747.
Goajv D. Cytokine-regulated proteases in autoimmune diseases. Immunol Today. 1993;15:103–7.
Opdenakker G, Abu El-Asrar A, Van Damme J. Remnant epitopes generating autoimmunity: from model to useful paradigm. Trends Immunol. 2020;41:367–78.
Opdenakker G, Dillen C, Fiten P, Martens E, Van Aelst I, Van den Steen PE, et al. Remnant epitopes, autoimmunity and glycosylation. Biochim Biophys Acta. 2006;1760:610–5.
Soulas P, Woods A, Jaulhac B, Knapp AM, Pasquali JL, Martin T, et al. Autoantigen, innate immunity, and T cells cooperate to break B cell tolerance during bacterial infection. J Clin Invest. 2005;115:2257–67.
Sofat N, Wait R, Robertson SD, Baines DL, Baker EH. Interaction between extracellular matrix molecules and microbial pathogens: evidence for the missing link in autoimmunity with rheumatoid arthritis as a disease model. Front Microbiol. 2014;5:783.
Wegner N, Wait R, Sroka A, Eick S, Nguyen KA, Lundberg K, et al. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and alpha-enolase: implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 2010;62:2662–72.
Rombouts Y, Ewing E, van de Stadt LA, Selman MH, Trouw LA, Deelder AM, et al. Anti-citrullinated protein antibodies acquire a pro-inflammatory Fc glycosylation phenotype prior to the onset of rheumatoid arthritis. Ann Rheum Dis. 2015;74:234–41.
Hecht C, Englbrecht M, Rech J, Schmidt S, Araujo E, Engelke K, et al. Additive effect of anti-citrullinated protein antibodies and rheumatoid factor on bone erosions in patients with RA. Ann Rheum Dis. 2015;74:2151–6.
Liu C, Chu D, Kalantar-Zadeh K, George J, Young HA, Liu G. Cytokines: from clinical significance to quantification. Adv Sci (Weinh). 2021;8: e2004433.
Chervonsky AV. Microbiota and autoimmunity. Cold Spring Harb Perspect Biol. 2013;5: a007294.
Campisi L, Barbet G, Ding Y, Esplugues E, Flavell RA, Blander JM. Apoptosis in response to microbial infection induces autoreactive TH17 cells. Nat Immunol. 2016;17:1084–92.
Maddur MS, Miossec P, Kaveri SV, Bayry J. Th17 cells: biology, pathogenesis of autoimmune and inflammatory diseases, and therapeutic strategies. Am J Pathol. 2012;181:8–18.
McGeachy MJ, Cua DJ, Gaffen SL. The IL-17 family of cytokines in health and disease. Immunity. 2019;50:892–906.
Liu H, Hong XL, Sun TT, Huang XW, Wang JL, Xiong H. Fusobacterium nucleatum exacerbates colitis by damaging epithelial barriers and inducing aberrant inflammation. J Dig Dis. 2020;21:385–98.
Esparbes P, Legrand A, Bandiaky ON, Cheraud-Carpentier M, Martin H, Montassier E, et al. Subgingival microbiota and cytokines profile changes in patients with periodontitis: a pilot study comparing healthy and diseased sites in the same oral cavities. Microorganisms. 2021. https://doi.org/10.3390/microorganisms9112364.
Mills KHG. IL-17 and IL-17-producing cells in protection versus pathology. Nat Rev Immunol. 2022. https://doi.org/10.1038/s41577-022-00746-9.
Maddur MS, Vani J, Hegde P, Lacroix-Desmazes S, Kaveri SV, Bayry J. Inhibition of differentiation, amplification, and function of human TH17 cells by intravenous immunoglobulin. J Allergy Clin Immunol. 2011;127(823–830):e821-827.
Qi Y, Wu HM, Yang Z, Zhou YF, Jin L, Yang MF, et al. New insights into the role of oral microbiota dysbiosis in the pathogenesis of inflammatory bowel disease. Dig Dis Sci. 2021. https://doi.org/10.1007/s10620-021-06837-2.
Lee SH, Kwon JE, Cho ML. Immunological pathogenesis of inflammatory bowel disease. Intest Res. 2018;16:26–42.
Qi Y, Zang SQ, Wei J, Yu HC, Yang Z, Wu HM, et al. High-throughput sequencing provides insights into oral microbiota dysbiosis in association with inflammatory bowel disease. Genomics. 2021;113:664–76.
Xun Z, Zhang Q, Xu T, Chen N, Chen F. dysbiosis and ecotypes of the salivary microbiome associated with inflammatory bowel diseases and the assistance in diagnosis of diseases using oral bacterial profiles. Front Microbiol. 2018;9:1136.
Sun B, Liu B, Gao X, Xing K, Xie L, Guo T. Metagenomic analysis of saliva reveals disease-associated microbiotas in patients with periodontitis and crohn’s disease-associated periodontitis. Front Cell Infect Microbiol. 2021;11: 719411.
Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503.
Zhang T, Kayani MUR, Hong L, Zhang C, Zhong J, Wang Z, et al. Dynamics of the salivary microbiome during different phases of crohn’s disease. Front Cell Infect Microbiol. 2020;10: 544704.
Hu S, Png E, Gowans M, Ong DEH, de Sessions PF, Song J, et al. Ectopic gut colonization: a metagenomic study of the oral and gut microbiome in Crohn’s disease. Gut Pathog. 2021;13:13.
Goel S, Evans-Johnson JA, Georgieva NI, Boysen G. Exposure profiling of reactive compounds in complex mixtures. Toxicology. 2013;313:145–50.
Mukhopadhya I, Thomson JM, Hansen R, Berry SH, El-Omar EM, Hold GL. Detection of Campylobacter concisus and other Campylobacter species in colonic biopsies from adults with ulcerative colitis. PLoS ONE. 2011;6: e21490.
Nielsen HL, Ejlertsen T, Engberg J, Nielsen H. High incidence of Campylobacter concisus in gastroenteritis in north Jutland, Denmark: a population-based study. Clin Microbiol Infect. 2013;19:445–50.
Mahendran V, Liu F, Riordan SM, Grimm MC, Tanaka MM, Zhang L. Examination of the effects of Campylobacter concisus zonula occludens toxin on intestinal epithelial cells and macrophages. Gut Pathog. 2016;8:18.
Qian J, Lu J, Huang Y, Wang M, Chen B, Bao J, et al. Periodontitis salivary microbiota worsens colitis. J Dent Res. 2021. https://doi.org/10.1177/00220345211049781.
Zhao X, Liu J, Zhang C, Yu N, Lu Z, Zhang S, et al. Porphyromonas gingivalis exacerbates ulcerative colitis via Porphyromonas gingivalis peptidylarginine deiminase. Int J Oral Sci. 2021;13:31.
Tsuzuno T, Takahashi N, Yamada-Hara M, Yokoji-Takeuchi M, Sulijaya B, Aoki-Nonaka Y, et al. Ingestion of Porphyromonas gingivalis exacerbates colitis via intestinal epithelial barrier disruption in mice. J Periodontal Res. 2021;56:275–88.
Gauthier S, Tetu A, Himaya E, Morand M, Chandad F, Rallu F, et al. The origin of Fusobacterium nucleatum involved in intra-amniotic infection and preterm birth. J Matern Fetal Neonatal Med. 2011;24:1329–32.
Feng X, Zhang L, Xu L, Meng H, Lu R, Chen Z, et al. Detection of eight periodontal microorganisms and distribution of Porphyromonas gingivalis fimA genotypes in Chinese patients with aggressive periodontitis. J Periodontol. 2014;85:150–9.
Su W, Chen Y, Cao P, Chen Y, Guo Y, Wang S, et al. Fusobacterium nucleatum promotes the development of ulcerative colitis by inducing the autophagic cell death of intestinal epithelial. Front Cell Infect Microbiol. 2020;10: 594806.
Cao P, Chen Y, Guo X, Chen Y, Su W, Zhan N, et al. Fusobacterium nucleatum activates endoplasmic reticulum stress to promote crohn’s disease development via the upregulation of card3 expression. Front Pharmacol. 2020;11:106.
Chen Y, Chen Y, Cao P, Su W, Zhan N, Dong W. Fusobacterium nucleatum facilitates ulcerative colitis through activating IL-17F signaling to NF-kappaB via the upregulation of CARD3 expression. J Pathol. 2020;250:170–82.
Liu L, Liang L, Liang H, Wang M, Lu B, Xue M, et al. Fusobacterium nucleatum aggravates the progression of colitis by regulating m1 macrophage polarization via AKT2 pathway. Front Immunol. 2019;10:1324.
Silman AJ, MacGregor AJ, Thomson W, Holligan S, Carthy D, Farhan A, et al. Twin concordance rates for rheumatoid arthritis: results from a nationwide study. Br J Rheumatol. 1993;32:903–7.
Ceccarelli F, Orru G, Pilloni A, Bartosiewicz I, Perricone C, Martino E, et al. Porphyromonas gingivalis in the tongue biofilm is associated with clinical outcome in rheumatoid arthritis patients. Clin Exp Immunol. 2018;194:244–52.
Cheng Z, Do T, Mankia K, Meade J, Hunt L, Clerehugh V, et al. Dysbiosis in the oral microbiomes of anti-CCP positive individuals at risk of developing rheumatoid arthritis. Ann Rheum Dis. 2021;80:162–8.
Courbon G, Rinaudo-Gaujous M, Blasco-Baque V, Auger I, Caire R, Mijola L, et al. Porphyromonas gingivalis experimentally induces periodontis and an anti-CCP2-associated arthritis in the rat. Ann Rheum Dis. 2019;78:594–9.
Jung H, Jung SM, Rim YA, Park N, Nam Y, Lee J, et al. Arthritic role of Porphyromonas gingivalis in collagen-induced arthritis mice. PLoS ONE. 2017;12: e0188698.
Jeong SH, Nam Y, Jung H, Kim J, Rim YA, Park N, et al. Interrupting oral infection of Porphyromonas gingivalis with anti-FimA antibody attenuates bacterial dissemination to the arthritic joint and improves experimental arthritis. Exp Mol Med. 2018;50: e460.
Davison E, Johnston W, Piela K, Rosier BT, Paterson M, Mira A, et al. The subgingival plaque microbiome, systemic antibodies against bacteria and citrullinated proteins following periodontal therapy. Pathogens. 2021. https://doi.org/10.3390/pathogens10020193.
Moentadj R, Wang Y, Bowerman K, Rehaume L, Nel H, et al. Streptococcus species enriched in the oral cavity of patients with RA are a source of peptidoglycan-polysaccharide polymers that can induce arthritis in mice. Ann Rheum Dis. 2021;80:573–81.
Liu X, Tian K, Ma X, Wang S, Luo C, Du Q. Analysis of subgingival microbiome of periodontal disease and rheumatoid arthritis in Chinese: a case-control study. Saudi J Biol Sci. 2020;27:1835–42.
Kroese JM, Brandt BW, Buijs MJ, Crielaard W, Lobbezoo F, Loos BG, et al. Differences in the oral microbiome in patients with early rheumatoid arthritis and individuals at risk of rheumatoid arthritis compared to healthy individuals. Arthritis Rheumatol. 2021;73:1986–93.
Lehenaff R, Tamashiro R, Nascimento MM, Lee K, Jenkins R, Whitlock J, et al. Subgingival microbiome of deep and shallow periodontal sites in patients with rheumatoid arthritis: a pilot study. BMC Oral Health. 2021;21:248.
Lisnevskaia L, Murphy G, Isenberg D. Systemic lupus erythematosus. Lancet. 2014;384:1878–88.
Mok CC. Pathogenesis of systemic lupus erythematosus. J Clin Pathol. 2003;56:481–90.
De Luca F, Shoenfeld Y. The microbiome in autoimmune diseases. Clin Exp Immunol. 2019;195:74–85.
Nikitakis NG, Papaioannou W, Sakkas LI, Kousvelari E. The autoimmunity-oral microbiome connection. Oral Dis. 2017;23:828–39.
Pessoa L, Aleti G, Choudhury S, Nguyen D, Yaskell T, Zhang Y, et al. host-microbial interactions in syastemic Lupus Erythematosus and periodontitis. Front Immunol. 2019;10:2602.
Liu F, Ren T, Li X, Zhai Q, Xu X, Zhang N, et al. Distinct microbiomes of gut and saliva in patients with systemic Lupus Erythematous and clinical associations. Front Immunol. 2021;12: 626217.
Clancy RM, Marion MC, Ainsworth HC, Blaser MJ, Chang M, Howard TD, et al. Salivary dysbiosis and the clinical spectrum in anti-Ro positive mothers of children with neonatal lupus. J Autoimmun. 2020;107: 102354.
Marques CP, Maor Y, de Andrade MS, Rodrigues VP, Benatti BB. Possible evidence of systemic lupus erythematosus and periodontal disease association mediated by Toll-like receptors 2 and 4. Clin Exp Immunol. 2016;183:187–92.
Li D, Guo B, Wu H, Tan L, Chang C, Lu Q. Interleukin-17 in systemic lupus erythematosus: a comprehensive review. Autoimmunity. 2015;48:353–61.
Cho KA, Suh JW, Sohn JH, Park JW, Lee H, Kang JL, et al. IL-33 induces Th17-mediated airway inflammation via mast cells in ovalbumin-challenged mice. Am J Physiol Lung Cell Mol Physiol. 2012;302:L429-440.
Naka T, Nishimoto N, Kishimoto T. The paradigm of IL-6: from basic science to medicine. Arthritis Res. 2002;4(Suppl 3):S233-242.
Norris JM, Johnson RK, Stene LC. Type 1 diabetes-early life origins and changing epidemiology. Lancet Diabetes Endocrinol. 2020;8:226–38.
Paschou SA, Papadopoulou-Marketou N, Chrousos GP, Kanaka-Gantenbein C. On type 1 diabetes mellitus pathogenesis. Endocr Connect. 2018;7:R38–46.
Michalska M, Zorena K, Waz P, Bartoszewicz M, Brandt-Varma A, Slezak D, et al. Gaseous pollutants and particulate matter (PM) in ambient air and the number of new cases of type 1 diabetes in children and adolescents in the pomeranian voivodeship. Poland Biomed Res Int. 2020;2020:1648264.
Sofi MH, Gudi R, Karumuthil-Melethil S, Perez N, Johnson BM, Vasu C. pH of drinking water influences the composition of gut microbiome and type 1 diabetes incidence. Diabetes. 2014;63:632–44.
Rewers M, Ludvigsson J. Environmental risk factors for type 1 diabetes. Lancet. 2016;387:2340–8.
Yuan X, Wu J, Chen R, Chen Z, Su Z, Ni J, et al. Characterization of the oral microbiome of children with type 1 diabetes in the acute and chronic phases. J Oral Microbiol. 2022;14:2094048.
de Groot PF, Belzer C, Aydin O, Levin E, Levels JH, Aalvink S, et al. Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PLoS ONE. 2017;12: e0188475.
Moskovitz M, Nassar M, Moriel N, Cher A, Faibis S, Ram D, et al. Characterization of the oral microbiome among children with type 1 diabetes compared with healthy children. Front Microbiol. 2021;12: 756808.
Chakraborty P, Ghosh S. 2409-PUB: assessment of oral microbiota in adolescents with type 1 diabetes mellitus and periodontal disease. Diabetes. 2019. https://doi.org/10.2337/db19-2409-PUB.
Duque C, Joao MF, Camargo GA, Teixeira GS, Machado TS, Azevedo RS, et al. Microbiological, lipid and immunological profiles in children with gingivitis and type 1 diabetes mellitus. J Appl Oral Sci. 2017;25:217–26.
Jindal A, Parihar AS, Sood M, Singh P, Singh N. Relationship between severity of periodontal disease and control of diabetes (glycated hemoglobin) in patients with type 1 diabetes mellitus. J Int Oral Health. 2015;7:17–20.
Rapone B, Ferrara E, Corsalini M, Converti I, Grassi FR, Santacroce L, et al. The effect of gaseous ozone therapy in conjunction with periodontal treatment on glycated hemoglobin level in subjects with type 2 diabetes mellitus: an unmasked randomized controlled trial. Int J Environ Res Public Health. 2020. https://doi.org/10.3390/ijerph17155467.
Gavin PG, Mullaney JA, Loo D, Cao KL, Gottlieb PA, Hill MM, et al. Intestinal metaproteomics reveals host-microbiota interactions in subjects at risk for type 1 diabetes. Diabetes Care. 2018;41:2178–86.
Yamazaki K, Miyauchi E, Kato T, Sato K, Suda W, Tsuzuno T, et al. Dysbiotic human oral microbiota alters systemic metabolism via modulation of gut microbiota in germ-free mice. J Oral Microbiol. 2022;14:2110194.
Kobayashi R, Ogawa Y, Hashizume-Takizawa T, Kurita-Ochiai T. Oral bacteria affect the gut microbiome and intestinal immunity. Pathog Dis. 2020. https://doi.org/10.1093/femspd/ftaa024.
Sofi MH, Johnson BM, Gudi RR, Jolly A, Gaudreau MC, Vasu C. Polysaccharide a-dependent opposing effects of mucosal and systemic exposures to human gut commensal Bacteroides fragilis in type 1 diabetes. Diabetes. 2019;68:1975–89.
Tsoukalas D, Fragoulakis V, Papakonstantinou E, Antonaki M, Vozikis A, Tsatsakis A, et al. Prediction of autoimmune diseases by targeted metabolomic assay of urinary organic acids. Metabolites. 2020. https://doi.org/10.3390/metabo10120502.
Negrini S, Emmi G, Greco M, Borro M, Sardanelli F, Murdaca G, et al. Sjogren’s syndrome: a systemic autoimmune disease. Clin Exp Med. 2022;22:9–25.
Tsigalou C, Stavropoulou E, Bezirtzoglou E. Current insights in microbiome shifts in Sjogren’s syndrome and possible therapeutic interventions. Front Immunol. 2018;9:1106.
Alam J, Lee A, Lee J, Kwon DI, Park HK, Park JH, et al. Dysbiotic oral microbiota and infected salivary glands in Sjogren’s syndrome. PLoS ONE. 2020;15: e0230667.
Szymula A, Rosenthal J, Szczerba BM, Bagavant H, Fu SM, Deshmukh US. T cell epitope mimicry between Sjogren’s syndrome antigen a (SSA)/Ro60 and oral, gut, skin and vaginal bacteria. Clin Immunol. 2014;152:1–9.
Rusthen S, Kristoffersen AK, Young A, Galtung HK, Petrovski BE, Palm O, et al. Dysbiotic salivary microbiota in dry mouth and primary Sjogren’s syndrome patients. PLoS ONE. 2019;14: e0218319.
Weng CT, Huang SL, Yang HW, Kao CC, Wei CC, Huang YF. Oral microbiota in xerostomia patients—a preliminary study. J Dent Sci. 2022;17:324–30.
Zhou S, Cai Y, Wang M, Yang WD, Duan N. Oral microbial flora of patients with Sicca syndrome. Mol Med Rep. 2018;18:4895–903.
Zhou Z, Ling G, Ding N, Xun Z, Zhu C, Hua H, et al. Molecular analysis of oral microflora in patients with primary Sjogren’s syndrome by using high-throughput sequencing. PeerJ. 2018;6: e5649.
Sharma D, Sandhya P, Vellarikkal SK, Surin AK, Jayarajan R, Verma A, et al. Saliva microbiome in primary Sjogren’s syndrome reveals distinct set of disease-associated microbes. Oral Dis. 2020;26:295–301.
Singh M, Teles F, Uzel NG, Papas A. Characterizing microbiota from sjogren’s syndrome patients. JDR Clin Trans Res. 2021;6:324–32.
de Paiva CS, Jones DB, Stern ME, Bian F, Moore QL, Corbiere S, et al. Altered mucosal microbiome diversity and disease severity in sjogren syndrome. Sci Rep. 2016;6:23561.
Voruganti A, Bowness P. New developments in our understanding of ankylosing spondylitis pathogenesis. Immunology. 2020;161:94–102.
Zhu W, He X, Cheng K, Zhang L, Chen D, Wang X, et al. Ankylosing spondylitis: etiology, pathogenesis, and treatments. Bone Res. 2019;7:22.
Sartor RB, Rath HC, Lichtman SN, van Tol EA. Animal models of intestinal and joint inflammation. Baillieres Clin Rheumatol. 1996;10:55–76.
Wen C, Zheng Z, Shao T, Liu L, Xie Z, Le Chatelier E, et al. Quantitative metagenomics reveals unique gut microbiome biomarkers in ankylosing spondylitis. Genome Biol. 2017;18:142.
Cardoneanu A, Mihai C, Rezus E, Burlui A, Popa I, Cijevschi PC. Gut microbiota changes in inflammatory bowel diseases and ankylosing spondilytis. J Gastrointestin Liver Dis. 2021;30:46–54.
Lv L, Jiang H, Yan R, Xu D, Wang K, Wang Q, et al. The salivary microbiota, cytokines, and metabolome in patients with ankylosing spondylitis are altered and more proinflammatory than those in healthy controls. Msystems. 2021;6: e0117320.
Stoll ML, Wang J, Kau CH, Pierce MK, Morrow CD, Geurs NC. Pro-inflammatory oral microbiota in juvenile Spondyloarthritis: a pilot study. Children. 2022. https://doi.org/10.3390/children9111764.
Farshbafnadi M, Agah E, Rezaei N. The second brain: The connection between gut microbiota composition and multiple sclerosis. J Neuroimmunol. 2021;360: 577700.
Boullerne AI, Adami GR, Schwartz JL, Skias D, Maienschein-Cline M, Green SJ, et al. Deep DNA metagenomic sequencing reveals oral microbiome divergence between monozygotic twins discordant for multiple sclerosis severity. J Neuroimmunol. 2020;343: 577237.
Zangeneh Z, Abdi-Ali A, Khamooshian K, Alvandi A, Abiri R. Bacterial variation in the oral microbiota in multiple sclerosis patients. PLoS ONE. 2021;16: e0260384.
Clark RB, Cervantes JL, Maciejewski MW, Farrokhi V, Nemati R, Yao X, et al. Serine lipids of Porphyromonas gingivalis are human and mouse Toll-like receptor 2 ligands. Infect Immun. 2013;81:3479–89.
Bottcher K, Rombouts K, Saffioti F, Roccarina D, Rosselli M, Hall A, et al. MAIT cells are chronically activated in patients with autoimmune liver disease and promote profibrogenic hepatic stellate cell activation. Hepatology. 2018;68:172–86.
Zhang X, Chen BD, Zhao LD, Li H. The gut microbiota: emerging evidence in autoimmune diseases. Trends Mol Med. 2020;26:862–73.
Wei Y, Li Y, Yan L, Sun C, Miao Q, Wang Q, et al. Alterations of gut microbiome in autoimmune hepatitis. Gut. 2020;69:569–77.
Rao B, Lou J, Lu H, Liang H, Li J, Zhou H, et al. Oral microbiome characteristics in patients with autoimmune hepatitis. Front Cell Infect Microbiol. 2021;11: 656674.
Abe K, Fujita M, Hayashi M, Okai K, Takahashi A, Ohira H. Gut and oral microbiota in autoimmune liver disease. Fukushima J Med Sci. 2020;65:71–5.
Bajaj JS, Betrapally NS, Hylemon PB, Heuman DM, Daita K, White MB, et al. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology. 2015;62:1260–71.
Abe K, Takahashi A, Fujita M, Imaizumi H, Hayashi M, Okai K, et al. Dysbiosis of oral microbiota and its association with salivary immunological biomarkers in autoimmune liver disease. PLoS ONE. 2018;13: e0198757.
Lv L, Jiang H, Chen X, Wang Q, Wang K, Ye J, et al. The salivary microbiota of patients with primary biliary cholangitis is distinctive and pathogenic. Front Immunol. 2021;12: 713647.
Kang DJ, Betrapally NS, Ghosh SA, Sartor RB, Hylemon PB, Gillevet PM, et al. Gut microbiota drive the development of neuroinflammatory response in cirrhosis in mice. Hepatology. 2016;64:1232–48.
Aoyama T, Paik YH, Seki E. Toll-like receptor signaling and liver fibrosis. Gastroenterol Res Pract. 2010. https://doi.org/10.1155/2010/192543.
Li T, Huang Y, Liu P, Liu Y, Guo J, Zhang W, et al. Lower plasma levels of IL-35 in patients with primary biliary cirrhosis. Tohoku J Exp Med. 2018;244:123–31.
Sun Q, Wang Q, Feng N, Meng Y, Li B, Luo D, et al. The expression and clinical significance of serum IL-17 in patients with primary biliary cirrhosis. Ann Transl Med. 2019;7:389.
Lazaridis KN, LaRusso NF. Primary sclerosing cholangitis. N Engl J Med. 2016;375:1161–70.
Lapidot Y, Amir A, Ben-Simon S, Veitsman E, Cohen-Ezra O, Davidov Y, et al. Alterations of the salivary and fecal microbiome in patients with primary sclerosing cholangitis. Hepatol Int. 2021;15:191–201.
Iwasawa K, Suda W, Tsunoda T, Oikawa-Kawamoto M, Umetsu S, Takayasu L, et al. Dysbiosis of the salivary microbiota in pediatric-onset primary sclerosing cholangitis and its potential as a biomarker. Sci Rep. 2018;8:5480.
Liwinski T, Zenouzi R, John C, Ehlken H, Ruhlemann MC, Bang C, et al. Alterations of the bile microbiome in primary sclerosing cholangitis. Gut. 2020;69:665–72.
Weismuller TJ, Trivedi PJ, Bergquist A, Imam M, Lenzen H, Ponsioen CY, et al. Patient age, sex, and inflammatory bowel disease phenotype associate with course of primary sclerosing cholangitis. Gastroenterology. 2017;152(1975–1984): e1978.
Dhillon AK, Kummen M, Troseid M, Akra S, Liaskou E, Moum B, et al. Circulating markers of gut barrier function associated with disease severity in primary sclerosing cholangitis. Liver Int. 2019;39:371–81.
Lv J, Zhang H, Zhou Y, Li G, Zou W, Wang H. Natural history of immunoglobulin a nephropathy and predictive factors of prognosis: a long-term follow up of 204 cases in China. Nephrology. 2008;13:242–6.
Chen D, Zhang Z, Yang Y, Hong Q, Li W, Zhuo L. High-throughput sequencing analysis of genes encoding the B-lymphocyte receptor heavy-chain CDR3 in renal and peripheral blood of IgA nephropathy. 2019. Biosci Rep. https://doi.org/10.1042/BSR20190482.
Luan S, Zhang S, Zhong H, Zhang Y, Wei X, Lin R, et al. Salivary microbial analysis of Chinese patients with immunoglobulin a nephropathy. Mol Med Rep. 2019;20:2219–26.
Piccolo M, De Angelis M, Lauriero G, Montemurno E, Di Cagno R, Gesualdo L, et al. Salivary microbiota associated with immunoglobulin a nephropathy. Microb Ecol. 2015;70:557–65.
Khasnobish A, Takayasu L, Watanabe KI, Nguyen TTT, Arakawa K, Hotta O, et al. Dysbiosis in the salivary microbiome associated with iga nephropathy-a japanese cohort study. Microbes Environ. 2021. https://doi.org/10.1264/jsme2.ME21006.
He JW, Zhou XJ, Hou P, Wang YN, Gan T, Li Y, et al. Potential roles of oral microbiota in the pathogenesis of immunoglobin a nephropathy. Front Cell Infect Microbiol. 2021;11: 652837.
Cao Y, Qiao M, Tian Z, Yu Y, Xu B, Lao W, et al. Comparative analyses of subgingival microbiome in chronic periodontitis patients with and without IgA nephropathy by high throughput 16S rRNA sequencing. Cell Physiol Biochem. 2018;47:774–83.
Misaki T, Naka S, Kuroda K, Nomura R, Shiooka T, Naito Y, et al. Distribution of Streptococcus mutans strains with collagen-binding proteins in the oral cavity of IgA nephropathy patients. Clin Exp Nephrol. 2015;19:844–50.
Ito S, Misaki T, Naka S, Wato K, Nagasawa Y, Nomura R, et al. Specific strains of Streptococcus mutans, a pathogen of dental caries, in the tonsils, are associated with IgA nephropathy. Sci Rep. 2019;9:20130.
Naka S, Wato K, Misaki T, Ito S, Matsuoka D, Nagasawa Y, et al. Streptococcus mutans induces IgA nephropathy-like glomerulonephritis in rats with severe dental caries. Sci Rep. 2021;11:5784.
Nagasawa Y, Misaki T, Ito S, Naka S, Wato K, Nomura R, et al. Title IgA nephropathy and oral bacterial species related to dental caries and periodontitis. Int J Mol Sci. 2022;23:725.
Jia G, Zhi A, Lai PFH, Wang G, Xia Y, Xiong Z, et al. The oral microbiota - a mechanistic role for systemic diseases. Br Dent J. 2018;224:447–55.
Khor B, Snow M, Herrman E, Ray N, Mansukhani K, Patel KA, et al. Interconnections between the oral and gut microbiomes: reversal of microbial dysbiosis and the balance between systemic health and disease. Microorganisms. 2021;9:496.
Rosier BT, Marsh PD, Mira A. resilience of the oral microbiota in health: mechanisms that prevent dysbiosis. J Dent Res. 2018;97:371–80.
Mayumi S, Kuboniwa M, Sakanaka A, Hashino E, Ishikawa A, Ijima Y, et al. potential of prebiotic d-tagatose for prevention of oral disease. Front Cell Infect Microbiol. 2021;11: 767944.
Peredo-Lovillo A, Romero-Luna HE, Jimenez-Fernandez M. Health promoting microbial metabolites produced by gut microbiota after prebiotics metabolism. Food Res Int. 2020;136: 109473.
Costello SP, Hughes PA, Waters O, Bryant RV, Vincent AD, Blatchford P, et al. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA. 2019;321:156–64.
Huang C, Huang Z, Ding L, Fu Y, Fan J, Mei Q, et al. Fecal microbiota transplantation versus glucocorticoids for the induction of remission in mild to moderate ulcerative colitis. J Transl Med. 2022;20:354.
Paramsothy S, Nielsen S, Kamm MA, Deshpande NP, Faith JJ, Clemente JC, et al. Specific bacteria and metabolites associated with response to fecal microbiota transplantation in patients with ulcerative colitis. Gastroenterology. 2019;156(1440–1454): e1442.
Beikler T, Bunte K, Chan Y, Weiher B, Selbach S, Peters U, et al. Oral microbiota transplant in dogs with naturally occurring periodontitis. J Dent Res. 2021;100:764–70.
Xiao H, Fan Y, Li Y, Dong J, Zhang S, Wang B, et al. Oral microbiota transplantation fights against head and neck radiotherapy-induced oral mucositis in mice. Comput Struct Biotechnol J. 2021;19:5898–910.
Xiao B, Laroui H, Ayyadurai S, Viennois E, Charania MA, Zhang Y, et al. Mannosylated bioreducible nanoparticle-mediated macrophage-specific TNF-alpha RNA interference for IBD therapy. Biomaterials. 2013;34:7471–82.
Liu J, Wan M, Lyon CJ, Hu TY. Nanomedicine therapies modulating macrophage dysfunction: a potential strategy to attenuate cytokine storms in severe infections. Theranostics. 2020;10:9591–600.
Acknowledgements
Not applicable.
Funding
This work was supported by National Natural Science Foundation of China (81901006), Guangdong Basic and Applied Basic Research Foundation (2020A1515110051), Scientific Research Talent Cultivation Project of Stomatological Hospital, Southern Medical University (RC202005) and Science Research Cultivation Program of Stomatological Hospital, Southern Medical University (PY2020002), Clinical Research Startup Program of Southern Medical University by High-level University Construction Funding of Guangdong Provincial Department of Education (LC2017PY001).
Author information
Authors and Affiliations
Contributions
XH and XH were involved in conceptualization, writing of the original draft, constructing figures and the tables, reviewing and editing. XZ and LC were involved in visualization, reviewing and editing. YH, JZ, ZM and YL were involved in writing of the original draft and investigation. SH was involved in reviewing and funding acquisition. All authors contributed to the article and approved the submitted version. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors have declared no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Huang, X., Huang, X., Huang, Y. et al. The oral microbiome in autoimmune diseases: friend or foe?. J Transl Med 21, 211 (2023). https://doi.org/10.1186/s12967-023-03995-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-023-03995-x