Abstract
Non-coding RNA has aroused great research interest recently, they play a wide range of biological functions, such as regulating cell cycle, cell proliferation, and intracellular substance metabolism. Piwi-interacting RNAs (piRNAs) are emerging small non-coding RNAs that are 24–31 nucleotides in length. Previous studies on piRNAs were mainly limited to evaluating the binding to the PIWI protein family to play the biological role. However, recent studies have shed more lights on piRNA functions; aberrant piRNAs play unique roles in many human diseases, including diverse lethal cancers. Therefore, understanding the mechanism of piRNAs expression and the specific functional roles of piRNAs in human diseases is crucial for developing its clinical applications. Presently, research on piRNAs mainly focuses on their cancer-specific functions but lacks investigation of their expressions and epigenetic modifications. This review discusses piRNA’s biogenesis and functional roles and the recent progress of functions of piRNA/PIWI protein complexes in human diseases.
Video Abstract
Similar content being viewed by others
Introduction
Human health achieves dynamic balance through various adjustments. The homeostasis of the internal environment is a necessary condition for normal life activities [1,2,3]. Disturbances in the internal environment cause to mutations and disruption of the balance of various genes, leading to a variety of human diseases, including cancer and cardiovascular and cerebrovascular diseases that kill millions yearly [4,5,6,7]. Normal functional activities of cells support the homeostasis of the internal environment, essentially activated by various intracellular signaling pathways and epigenetic modifications [2, 8, 9].
PIWI-interacting RNAs (piRNAs), the least studied class of ncRNAs to date, is an emerging class of ncRNAs ranging in length from 24 to 32 nucleotides and was first identified in Drosophila, mouse, and rat germ cells in 2006 [9,10,11]. piRNA, initially identified as highly expressed in a germ-line cell, maintains genomic integrity [12,13,14]. More in-depth research shows piRNA to be widely expressed in somatic cells and engaged in some fundamental regulations [15, 16], such as cell cycle regulation, proliferation, energy metabolism and immune microenvironment regulation through epigenetic modification and regulation of multiple signaling pathways [17, 18]. The functional roles of piRNA in germ and somatic cells have greatly interested researchers for further studies.
Growing bodies of evidence have revealed the essential functions of piRNA in the occurrence and prognosis of various human diseases and its application prospect in diagnosing and treating related conditions [19,20,21]. In addition, piRNA/piwi complexes have been shown to bind piRNA to piwi proteins and to perform regulatory functions such as transposon silencing, epigenetic modification, and protein regulation [22, 23]. Moreover, piRNA plays a vital role as a diagnostic marker in the early stage of various cancers, serving as critical prognostic indicators after treatment [24,25,26]. At the same time, piRNA/piwi proteins enhance the resistance of tumor cells to currently known chemotherapeutics, indicating the possibility of it being a new therapeutic target that can potentially solve the problem of chemotherapy resistance [27].
This review summarized the latest progress of piRNA biogenesis research. It expounded on the biological functions and clinical value of piRNA in recent years to provide a systematic summary of the potential role of piRNA in human diseases (Fig. 1).
The association of abnormally dysregulated piRNAs/PIWI proteins with human diseases can be broadly divided into two categories. On the one hand, the blue background of piRNA/PIWI proteins represent their important roles in the development of different human cancers. On the other hand, the pink background of piRNA/PIWI proteins represent the correlation in the occurrence and development of human non-cancer diseases
Biosynthesis and basic functions of piRNAs
Homeostasis of an organism primarily depends on homeostasis and regulation of the genome, critically influencing development and balance [28, 29]. The maintenance of this homeostasis is mainly achieved by diverse molecular events, in which RNA silencing plays a significant role in eukaryotes [30, 31]. Argonaute proteins are composed of the AGO and the PIWI family. Among them, PIWI proteins are mainly expressed in germ cells and form specific RNA-induced silencing complex (RISC) with piRNAs, collectively referred to as piRISC [10, 32, 33], which primarily regulate the expression and transposition of transposons in the genome and affect the activities. Here, we mainly review the biogenesis and functional mechanisms of piRNAs (Fig. 2).
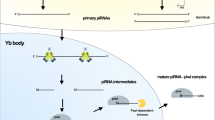
Left: piRNA cluster formation and germ line piRNA biogenesis. Rhino forms a terpolymer complex with Deadlock and Cutoff called RDC, and recognizes H3K9me3 on the piRNA cluster. First, RDC recruits Moonshiner and Trf2 to initiate transcription, and upon completion of transcription, piRNA cluster precursor is formed. Then RDC complex achieves the binding of NXF3-NXT1 complex by recruiting adapter protein Bootlegger (boot). Finally, Nxf3 binds Crm1 to mediate the transfer of piRNA precursor transcription out of the nucleus and localization to Nuage in the cytoplasm. In nuage, the 3 ‘end of the loaded piRNA precursor transcribed on Aub is trimmed by Nibbler, and then the end is subjected to 2’ -O-methylation by the methyltransferase Hen1 to become mature piRNAs. Finally, Krimper realized the transition of mature piRNA. In the mitochondria, Armi transfers the piRNA precursor transcript to the mitochondrial outer membrane, where it is cleaved by Zuc to produce the first piRNA. And then it is transported back to nuage. The rest of the piRNA precursor transcripts are repeatedly bound by PIWI and cleaved by Zuc, generating a string of PIWI-loaded piRNAs that are methylated by Hen1. Right: piRNA cluster formation and ovarian somatic cells piRNA biogenesis. The single-stranded piRNA cluster in ovarian cells contains a promoter itself, whose transcription initiation depends on transcription factor Pol II. The piRNA precursor transcripts form standard mRNA structures in the nucleus by standard splicing, 5 ‘-end closure, and 3’ -end polyadenylation. The Nxf1-Nxt1 complex and nuclear porins Nup54 and Nup58 then mediate the export of these transcripts to the Yb body. In Yb bodies, PIWI binds to the 5 ‘end of the flam precursor transcript and connects to Armi, then translocate to the mitochondrial outer membrane, where zuc cleavage produces a new 3’ end, which is finally subjected by Hen-1 to produce mature piRNA
piRNA cluster: the origin of piRNAs
Large amounts of piRNA, known as “piRNA clusters”, are generated from specific genomic loci, which are identified as genomic regions ranging from few hundred to several thousand kilobases [34, 35]. piRNA clusters are first transcribed by RNA polymerase II (Pol II) into long noncoding transcripts and piRNA precursors in the nucleus and then enter the cytoplasm for further processing and modification [36]. The Drosophila melanogaster model is the earliest and most mature model for piRNA cluster research [37]. Therefore, in this section, we will use the Drosophila model as a template to introduce the piRNA cluster generation in detail from the main production sites, the key enzymes required, and the main processes.
In germ cells, the piRNA cluster is bidirectional, lacking a well-defined promoter region. Hence transcription regulation mostly depends on the histone H3 Lys9 (H3K9) trimethylation mark present in D. Melanogaster female germ cells [38, 39]. Rhino, a heterochromatin protein 1 (HP1) family protein, first binds to this mark and simultaneously recruits Deadlock and Cutoff proteins to form a Rhino-Deadlock -Cutoff (RDC) complex [40, 41]. RDC activates the transcription of piRNA clusters through irregular mechanisms. Specifically, Deadlock recruits the transcription initiation factor Moonshiner, which induces Pol II-mediated transcription by binding to TATA box-binding protein-associated Factor 2 (Trf2) [42, 43]. Cutoff binds the new piRNA precursors to protect them from degradation by blocking splicing and polyadenylation [44, 45]. Thus, the absence of splicing or polyadenylation is one of the most striking features of piRNA precursor transcripts [42, 46]. Finally, the piRNA prerequisite transcript passes through the nuclear pore through the nuclear output adapter complex Nxf3-Nxt1 [47]. The resulting piRNA precursors are then transported to the “Nuage” region in the cytoplasm and further processed into mature piRNAs [48, 49].
Another classic type comprises the unidirectional piRNA cluster with promoters in Drosophila melanogaster ovary cells [50]. The Flamenco (flam) locus is the most representative one, with no H3K9 trimethylation mark but has a specific independent well-defined promoter, also transcribed by Pol II [51,52,53]. In this case, the resulting transcripts are spliced and processed in the nucleus to form characteristic RNA precursors (5’ caps and 3’ poly(A) tails) [54]. The piRNA precursors are then transported to the perinuclear RNA particle “Yb body” for further processing [55]. The classical output complex NXF1-NXT1 is instrumental in the intracellular transport of piRNA precursors from the nucleus to the cytoplasm, necessary for the subsequent localization to the Yb body [56]. The nucleoporins Nup54 and Nup58 specifically transfer flam precursor transcripts from the nucleus and link to the Yb body to participate in the precise localization of piRNA precursor [54].
Biosynthesis of piRNA: two classical pathways
Upon export to the cytoplasm, the piRNA precursor transcripts are localized to ribonucleoprotein particles near the nucleus, such as Yb body and Nuage, and then processed into piRNA with mature functions [57, 58].
In D. melanogaster, the biogenesis of piRNA in germ cells is processed through the ping-pong cycle, mainly in a specific region in the cytoplasm called Nuage and partly in the outer membrane of mitochondria [59]. The transport of piRNA precursor transcripts in this process primarily depends on the exchange of two cytoplasmic PIWI proteins: Aubergine (Aub) and Argonaute 3 (Ago3) [60,61,62].
The ping-pong cycle is initiated by the Aub, recognizing complementary piRNA precursor transcripts and slices from the 5’ end between nucleotides 10 and 11 to generate a new 5’ end [63]. Ago3 then interacts with a 5 ‘-monophosphorylated fragment, cleaved in Nuage by an endonuclease of Aub or Ago3 or by Zucchini (zuc) on the mitochondrial outer membrane to produce a 3’ end [64]. Zuc-dependent cleavage produces piRNAs of the final size, and the 3 ‘to 5’ exonuclease Nibbler, which forms the 3 ‘end of piRNAs by trimming the 3’ end to its final length, is also involved in AGO3-mediated piRNA generation [65, 66]. Methyltransferase Hen1 then methylates the 3’ end with 2’-O- methylation to produce the mature piRNA, which is protected from degradation [67, 68]. In addition, the Aub of the loaded dimethylated piRNA and the Ago3 in the unloaded state are specifically identified and combined by Krimper to ensure a sustainable ping-pong cycle [64]. The following process is mediated by the RNA helicase Armitage (Armi) and is initiated in the mitochondria. First, Aub bound to the 5 ‘-monophosphorylated piRNA precursor mutually and linked to Armi, which transfers the complex to the outer membrane of the mitochondria [69]. Here, it is cleaved by Zuc to produce a new 3’ end. The remaining piRNA precursor transcripts are bound by Piwi at the 5’ end and then continue to be cleaved by Zuc to produce a series of piRNAs ranging in length from 24 to 32 nucleotides that are eventually transferred to the nucleus [70].
The lack of expression of Aub and Ago3 in the somatic cells of Drosophila ovaries causes the piRNA precursor transcripts to be only dependent on the cleavage by Zuc [71]. Flam-piRNA precursor transcripts are first accumulated with protein Yb, then transferred by Armi to the mitochondrial outer membrane, where they are cleaved by Zuc to produce a new 3’ end [72]. Eventually, they are treated by 2’ -O - methylation of Hen1 to produce the mature piRNA.
Cellular functions of PIWI–piRNA complexes
The interaction between piRNAs and PIWI proteins prevents transposable element (TE) activity and regulates the encoded mRNAs through different mechanisms [73]. This section primarily describes the piRNA-mediated transcriptional silencing of TE and the functional role of piRNA/PIWI protein complexes in regulating protein-coding genes.
Silencing of transposable element
Transposon activity must be maintained through integration into germ cell DNA. In contrast, silencing of a transposon leads to inactivation of the protein encoded by the transposon as well as transposon death demise. The PIWI-piRNA complex was initially discovered to silence transposable elements (TEs) and maintain the stability and integrity of the germ cell genome [74, 75]. Different complexes formed by PIWI proteins and piRNAs silence TEs at the transcriptional and post-transcriptional levels, respectively [76]. Cytoplasmic PIWI proteins, such as Aub and Ago3 in Drosophila and MIWI and MILI in mice, primarily slice TE transcripts after transcription via piRNA targeting [77]. Nuclear PIWI proteins, including those in Drosophila PIWI proteins and mouse MIWI2, were incorporated into the nucleus after being loaded with phased piRNAs [78, 79]. Nuclear PIWI proteins bind to nascent RNAs through guiding piRNAs at the site of TE transcription and recruiting cofactors for transcriptional silencing [80].
Regulation of protein-coding genes
In addition to TE silencing, piRNA regulates protein-coding genes during reproductive development; first discovered in Drosophila embryos, where TE-derived piRNA directed Aub to Nanos mRNA to subsequently mediated its deadenylation by recruiting deadenylate complexes [19, 26, 81]. Similarly, Aub loaded with piRNA can also directly degrade maternal mRNA in somatic cells in the early embryos of Drosophila [82]. Aub also interacts with poly(A) binding protein PABP and eIF3 subunits to activate the translation initiation of Nanos mRNA in embryonic reproductive cells, highlighting the crucial role of the evolutionary conservation of PIWI-piRNA complexes in translation activation [60]. These findings reflect the diversity of piRNA-regulated protein-coding genes, but the mechanisms behind these regulations remain to be fully understood.
Roles of piRNA/PIWI complexes in regulating biological processes in human diseases
With increasing research on piRNA/piwi protein, dozens of abnormally expressed complexes are found to be related to the occurrence and development of diseases [15, 83, 84]. The mechanisms underlying their function include activating a pathway or target. These changes alter the initial homeostasis in cells and tissues to cause abnormal regulation of piRNA/PIWI proteins, which include sustained proliferation signaling, immune microenvironment remodeling, angiogenesis induction, apoptotic resistance, activation invasion and metastasis, and cell energy metabolism [85,86,87,88,89]. We classified the reported abnormalities according to their roles in disease and elaborated on their functions to elucidate the mechanisms fueling the disease-causing functions of the piRNA/PIWI proteins (Table 1).
Epigenetic modifications mediated by piRNA/PIWI complex
Existing research suggests that piRNAs primarily mediate epigenetic changes through abnormal methylation and histone modification in somatic cells, eventually causing the diseases [124, 125] (Fig. 3).
Differential methylation modification in different diseases. The top half is the DNA methylation mediated by piRNA and PIWI proteins in different diseases. In this section, the top part are the corresponding disease types, the middle part are the enzymes mediating the methylation modification, and the bottom part are the targets of the ultimate effect. The bottom half is the m6A methylation modification carried out by piRNA/PIWI protein in the diseases. In this section, the bottom part are the corresponding disease types, the middle color blocks are the enzymes whose methylation modification is performed, and the top part are the targets of the final effect.
DNA methylation is critical to epigenetic modification, primarily by silencing transposons and other repeating elements [126, 127]. Four major types of DNA methyltransferases (DNMTS) regulate DNA methylation and its patterns [128]. In this section, we mainly summarize the biological roles of epigenetic modifications mediated by piRNA/PIWI proteins in normal functions and human diseases (Table 2).
piR-823 regulates the methylation of cancer stem cells by regulating DNMT1, DNMT3A, and DNMT3B. Ai et al. demonstrated that silencing of piRNA-823 reduced global DNA methylation induced by myeloid-derived suppressor cells (MDSCs) and reduced the stemness of multiple myeloma stem cells (MMSCs) maintained by MDSC, reducing tumor burden and angiogenesis in vivo [111, 129]. In addition, Yan et al. further explored the specific mechanism of piR-823 in MM, where the expression of piR-823 positively correlated with DNMT3A and DNMT3B. The high expression of DNMTs induces p16INK4A hypermethylation, leading to the loss of many protein-encoding tumor-suppressor genes (TSGs) [111]. piR-823 also induces aberrant DNA methylation through DNMT3B and has a tumorigenic role in ESCC [130]. Ping et al. reported piR-823 to be highly expressed in breast cancer cells and positively correlated with the expression of DNMT1, DNMT3A, and DNMT3B. Further research showed that the upregulation of piR-823 was positively correlated with m5C, the methylation target in the adenomatous polyposis coli (APC) promoter region. Meanwhile, aberrant APC promoter methylation also mediates Wnt signaling pathway activation [131]. Interestingly, piR-823 confer a tumor suppressor role in gastric cancer, these aberrant expression of piRNAs may directly or indirectly suppress tumor suppressor genes, resulting in an abnormal “stem-like” cellular state, inducing tumorigenesis [132].
In leukemia cells, upregulated hsa_piR_011186 binds to DNMT1 and inhibits CDKN2B gene expression by increasing DNA methylation at the CDKN2 promoter, inducing cell cycle progression and reducing cell apoptosis [133]. piR-651 is highly expressed in breast cancer tissues and promotes the expression of oncogenes such as MDM2, CKD4, and CyclinD1. Upregulated piR-651 leads to an increase in DNMT1, which binds to the promoter of Phosphatase and tensin homolog (PTEN), increases its methylation and inhibits its expression [135]. Zhang et al. reported that piR-31,470/PIWIL4 complex induces hypermethylation of glutathione S-transferase pi 1 (GSTP1) by recruiting DNMT1, DNMT3A, and MBD2, resulting in lower expression of GSTP1, which renders the cells more susceptible to oxidative stress, making them more easily acquire the tumorigenic phenotype [134]. piR-63,076 is highly expressed in hypoxic pulmonary arteries, upregulating DNMT1 and DNMT3a, which subsequently increases the methylation of the Acadm promoter and leads to the inhibition of its expression under hypoxia [136].
PIWIL1 is silenced in normal endometrium and is reactivated and carcinogenic in endometrial cancer. In endometrial cancer cells, estrogen E2 induces ERα to bind to the PIWIL1 promoter, leading to hypomethylation of the promoter, which then upregulates PIWIL1 transcription [137]. Studies have also shown that PIWIL1 induces hypermethylation of PTEN by increasing the expression of DNMT1 and inhibiting the tumor suppressor PTEN to promote cancer progression [138]. In addition, silencing PIWIL2 plays a protective role in transient global cerebral ischemia (tGCI) tolerance induced by hypoxic postconditioning (HPC). Zhan et al. found that HPC downregulated PIWIL2 expression in CA1. Low expression of PIWIL2 decreased the expression of apoptosis-related proteins and exerted a neuroprotective effect. CREB2 is a crucial regulator of neuronal plasticity and memory [143, 144]. PIWIL2 damages neurons after cerebral ischemia by regulating DNA methylation in the CpG island of the CREB2 promoter. Moreover, HPC-induced PIWIL2 downregulation following tGCI leads to a significant reduction in DNMT3A level, eliminating the TGCI-induced increase in DNA methylation of CREB2 [139].
Recent studies have also shown that piRNAs contribute to cancer development and progression by regulating abnormal m6A methylation modification [145,146,147]. m6A methylation is the most common and abundant internal post-transcriptional modification [148, 149]. Evidence shows that m6A methylation significantly impacts RNA biosynthesis/catabolism and regulates the pathogenesis of various cancers [150, 151].
Previous studies demonstrated that Wilms’ tumor 1-associating protein (WTAP) (an m6A methyltransferase) is required for m6A methyltransferase activity in vivo [152]. In diffuse large B-cell lymphoma (DLBCL), Han et al. showed that piRNA-30,473 increases WTAP mRNA expression by binding to its 3’UTR, reducing WTAP mRNA degradation and enhancing its stability. The high expression of WTAP further promotes the stable expression of its downstream target HK2. Then, IGF2BP2 binds to the 5 ‘UTR region of HK2 to stabilize its methylated transcripts [140]. piRNA-14633 is highly expressed in cervical cancer, and the total abundance of m6A increases with its overexpression. Further studies showed that piRNA-14633 specifically binds METTL14 in cervical cancer cells and enhances its expression by improving mRNA stability. The high expression of METTL14 promotes cancer progression by promoting m6A methylation of its downstream target, CYP1B1 [141]. Senescent neutrophil-derived exosome piRNA-17560 increases binds to fat mass and obesity-associated protein (FTO), reduces its mRNA decay and enhances its stability in breast cancer. FTO is one of the vital m6A mRNA demethylation enzymes, the upregulation of which further enhances the strength and expression of ZEB1 transcripts by reducing the m6A methylation of mRNA [153, 154]. In addition, m6A reader protein YTHDF2 binds to ZEB1 transcripts to induce post-transcriptional regulation [155, 156]. In conclusion, YTHDF2 and FTO increase the stability and expression of ZEB1 mRNA, leading to drug resistance and high aggressiveness of breast cancer cells [112].
piRNA-mediated methylation also plays a role in normal physiological functions. In osteoblast differentiation, piRNA-36,741 and METTL3 are highly expressed, and their silencing inhibits the normal differentiation of osteoblasts. Mechanistically, piR-36,741 sequesters METTL3 to prevent its binding to BMP2 mRNA. The m6A modification level of BMP2 is then significantly increased, allowing YTHDF2 (m6A reader) to recognize and degrade the m6A methylated BMP2 transcript, resulting in decreased expression and reduction in osteogenic phenotype and matrix mineralization [121]. In addition, Gao initially found that cardiac-hypertrophy-associated piRNA (CHAPIR) deficiency ameliorated cardiac hypertrophy and remodeling. Mechanistically, the CHAPIR-PIWIL4 complex binds to METTL3, reduces its activity to prevent the binding to PARP10 mRNA, and reduces its m6A methylation, thereby avoiding recognition and degradation by YTHDF2. PARP10 then accelerates cardiac hypertrophy by blocking GSK3β activity and weakening NFATC4 phosphorylation [142].
Abnormal cell cycle and resistance to apoptosis
Apoptosis is representative of programmed cell death, the inhibition of which leads to uncontrolled cell division [157, 158]. An abnormal cell cycle further leads to uncontrolled malignant proliferation, causing cancer [159, 160].
Cyclin D1 and CDK4 are essential regulators of the G1 phase of the cell cycle [161, 162]. Silencing piR-823 via an anti-sense sequence inhibits Cyclin D1 and CDK4 expression, blocking the cell cycle by inducing G1 phase arrest and inhibiting G1/S transition [90]. Moreover, piR-017061 upregulation induces apoptosis of pancreatic cells and significantly reduces the tumor volume and weight in vivo. Notably, overexpression of piR-017061 significantly reduced the expression of Bcl-2, simultaneously increasing the cleaved Caspase3 and decreasing p-ERK levels, suggesting the critical role of ERK signaling in apoptosis regulation [91]. As a tumor suppressor, piR-36,712 suppresses the malignant phenotype. Mechanistically, the interaction of piR-36,712 with SEPW1P reduces SEPW1 expression and function, with a subsequent increase in p53 levels and resulting in G1 cell cycle arrest. These results suggest that piR-36,712 increases the expression of p53 to exert its tumor-suppressive effects [92]. Overexpression of piR-004800 promotes proliferation and inhibits apoptosis in multiple myeloma (MM) cells. Mechanistically, the piR-004800 downregulation decreases the expression of anti-apoptotic genes, such as Bcl2 and caspase-3, and increases the expression of related pro-apoptotic proteins BAD, cleaved PARP, and cleaved caspase-3 [93]. After piR-8041 overexpression, glioma cells were arrested at the G0/G1 checkpoint, with decreased entry to the S-phase. However, the specific signaling pathways or proteins regulating the cell cycle have not been discovered and confirmed [94]. PIWIL1 knockdown induces the expression of CDKN1B and CCND2, resulting in upregulation of the cyclin-dependent kinase inhibitor p27, which regulates G1 progression by inhibiting cyclin activity. In addition, PIWIL1 knockdown inhibits MCL1 expression, promoting apoptosis and senescence [95]. PIWIL3 and piR-30,188 complex bind to carcinoma-related OIP5-AS1 to reduce its expression, reversing its inhibitory effect on its downstream tumor suppressor target miR-367-3p. The highly expressed miR-367-3p, in turn, binds and reduces CEBPA expression, inhibiting its binding to the downstream TRAF4 promoter and weakening TRAK4 activation. Finally, it inhibits the malignant growth of glioma and promotes apoptosis [96].
Overexpression of piR-651 promoted G cell cycle progression by inducing cyclin D1 and CDK4, and cells overexpressing piR-651 had a greater ability to form colonies. In vivo experiments showed that overexpression of piR-651 resulted in greater tumor volume and weight [97]. In addition, piR-651 also exerted its anti-apoptotic effect by affecting mitochondria-related proteins, such as increasing caspase-3-p17, Bax, and cleaved-PARP-1 and decreasing Bcl-2 [98]. PIWIL2 promotes cancer progression by inhibiting apoptosis primarily through IκB kinase (IKK)/IκB/NF-κB pathway in esophageal cell squamous carcinoma (ESCC). PIWIL2 binds to IKKα and IKKβ in the cytoplasm, significantly upregulating the total protein and phosphorylation levels of IKKα/β. Phosphorylated IKK induces IκB phosphorylation and NF-κB release for nuclear translocation and apoptosis inhibition. In addition, PIWIL2 competitively inhibits IKKβ binding to TSC1, inactivating mTORC1 and promoting ULK1 phosphorylation and initiating autophagy [99].
Aberrant proliferative signaling pathways
Uncontrolled malignant cell proliferation is one of the most prominent features of cancer, causing tumor growth [163, 164]. Mechanistically, intracellular and extracellular signaling molecules regulate the orderly proliferation and apoptosis of cells to regulate regular operations [165,166,167,168].
Oncogenic piR-31,115 promotes the unrestricted proliferation of ccRCC cells. In contrast, silencing piR-31,115 inhibits cell proliferation and significantly reduces AKT phosphorylation and PI3K expression, potentially promoting proliferation by activating EMT and PI3K/AKT signaling pathways [100]. piR-001773/piR-017184 and PCDH9 mRNA co-exist in the PIWIL4 complex and decrease its expression. In PCa cells, PCDH9 acts as a competitive substrate for PI3K binding and hinders the catalytic PIP3 synthesis reaction. Thus, PCDH9 silencing leads to increased AKT phosphorylation and activated AKT contributes to increased cell proliferation and inhibition of apoptosis [101]. Overexpression of piR-823 activates the Wnt/β-catenin pathway, leading to the malignant proliferation of breast cancer cells [131]. piR-55,490 binds to the 3’UTR of mTOR mRNA and degrades it, preventing the activation of the AKT/mTOR signaling pathway [103].
Invasion and metastasis activation
Tumor cell malignant behaviors, invasion, and metastasis are closely related to the poor prognosis of patients, causing the death of patients with different types of cancer [169,170,171]. Most tumors miss the best treatment opportunities due to early metastasis, and patients with the initially diagnosed metastatic disease have the worst 5-year survival rate compared with individuals with localized disease [172,173,174].
Ding et al. showed that piR-57,125 upregulation specifically inhibits chemokine (C-C motif) ligand 3 (CCL3) expression. Mechanistically, piR-57,125 suppresses invasion and metastasis by inhibiting CCL3 and its downstream AKT/ERK signaling pathway activation [104]. piR-19,166 overexpression significantly hinders the migration of PCa cells, while it’s silencing significantly promotes their migration. Mechanistically, piR-19,166 inhibited the direct downstream target CTTN expression and the matrix metalloproteinases (MMP) signaling pathway, and then reducing the invasion and metastasis of PCa cells [105]. Overexpression of MMPs plays an important role in tumor invasion and metastasis, and MMP-2 has been characterized as the most potential therapy target [175, 176]. The highly expressed piR-39,980 directly reduces the expression of the tumor suppressor gene SERPINB1, relieving its inhibitory effect on the downstream target MMP-2. High MMP-2 levels significantly induce osteosarcoma cell proliferation, invasion, and metastasis [106]. piR-54,265 binds to PIWIL2 and promotes the formation of PIWIL2/STAT3/p-SRC complex. Upregulated piR-54,265 and PIWIL2 increase the expression of downstream targets MMP2 and MMP9 by promoting the activation of the STAT3 signaling pathway, enhancing the invasion and metastasis ability of CRC cells [107]. Knockdown of PIWIL2 reduces the expression of MM9 in prostate cancer cells, thereby reducing invasion and migration ability [108]. The high expression of PIWIL1 significantly enhances the invasion and metastasis of pancreatic ductal adenocarcinoma (PDAC) cells in vitro and in vivo. PIWIL1 activated the APC/ C-Ub pathway, and further drives the proteolysis of adhesion protein Pinin, enhancing PDAC metastasis [109].
The imbalance between hypoxia-induced overexpression of pro-angiogenic factor (VEGF) and anti-angiogenic factors leads to angiogenesis in the tumor microenvironment, further aggravating the proliferation and spread of cancer cells [177,178,179,180]. piR-823 overexpression in MM cells transmits signals to the tumor microenvironment (TME) via extracellular vesicles (EVs) to increase the expression levels of VEGF and IL-6 in endothelial cells, inducing angiogenesis and increasing tumor cells’ invasion and metastasis [110]. As a negative regulator of VEGF expression, overexpression of p16INK4A inhibits VEGF. Overexpressed piR-823 inhibits the expression of p16INK4A through methylation, thus promoting VEGF-mediated angiogenesis [111].
Epithelial-mesenchymal transition (EMT)
Epithelial-mesenchymal transition (EMT) is the morphological transformation associated with the aggressiveness, metastasis, and drug resistance of malignant cells [181,182,183,184]. EMT is characterized by upregulated mesenchymal markers (N- cadherin and vimentin) and down-regulated epithelial markers (E‐ cadherin) [185, 186].
In breast cancer, senile neutrophils produce exosome piR-17,560 in a STAT3-dependent manner, which binds to FTO and increases ZEB1 expression by reducing m6A methylation, promoting chemotherapy resistance and EMT in BC cells [112]. piRNA-31,115 downregulation leads to the upregulation of E-cadherin, vimentin, decreased snail, and inhibition of cell migration in vitro. This indicated that the knockdown of piR-31,115 inhibits EMT and reduces cancer cell invasion [100].
EMT and the induction of stem-like properties of cancer cells are associated with solid tumors. PIWIL1 upregulates Snail mRNA expression and promotes EMT. Endometrial cancer cells with low PIWIL1 expression have upregulated E-cadherin, while Vimentin and N-cadherin were down-regulated. In addition, the expression of Vimentin, CD44, and ALDH1 was significantly reduced in the low-expression group, while that of E-cadherin was higher [113]. In OSCC cells, piR-1037 targeting significantly inhibited cell migration and invasion, with upregulated expression of E-cadherin and decreased levels of the N-cadherin, suggesting that piR-1037 knockdown could inhibit EMT [114].
DNA damage and repair
DNA molecules are the fundamental genetic materials. The structural and functional integrity of DNA is crucial to support normal life activities and stable species traits [187, 188]. Endogenous or exogenous stress causes cells to produce various DNA damage, causing corresponding biological changes, such as carcinogenesis and abnormal cell apoptosis [189,190,191,192].
piR-39,980 causes apoptosis by increasing doxorubicin (DOX) accumulation. Mechanically, co-treatment of piR-39,980 mimics and DOX increased DNA fragmentation and γH2AX accumulation, which were mainly dependent on the regulation of the downstream targets RRM2 and CYP1A2 by piR-39,980. RRM2 is a catalytic subunit of ribonucleotide reductase (RR), which regulates DNA repair [193,194,195]. CYP1A2 is a phase I/II metabolic enzyme whose upregulation regulates resistance mechanisms by inactivating DOX [196, 197]. piR-39,980 inhibits the expression of RRM2 and CYP1A2 to prevent the natural resistance of fibrosarcoma to DOX by inhibiting DNA repair [119]. PIWIL4 can increase intracellular reactive oxygen species (ROS) accumulation by inhibiting the expression of antioxidant enzymes from causing DNA damage. The loss of antioxidant enzymes leads to excessive ROS generation in mitochondria [198,199,200]. Upon reaching a specific threshold, the mitochondrial membrane channel opens to release a large amount of ROS into the cytoplasm, causing cell and DNA damage [201, 202]. Overexpression of PIWIL4 induced G2/M arrest and apoptosis in fibrosarcoma cells, whereas knockdown of PIWIL4 enhanced cell proliferation, metastasis, and invasion [120]. piR-31,470, highly expressed in prostate cancer, forms a complex with PIWIL4 and maintains the hypermethylation of GSTP1, inactivating it and reducing its expression. GSTP1, an antioxidant factor with protective and antitumor functions, plays an important regulatory role in anti-oxidative damage, regulating cellular oxidative stress and promoting cytotoxic metabolism [102, 203, 204]. Epigenetic silencing of GSTP1 leads to exposure to long-term oxidative damage and increased accumulation of potentially mutagenic DNA adducts, ultimately causing prostate cancer [121].
Regulation of metabolism
Mitochondria fulfills cellular energy requirements through ATP synthesis using oxygen to metabolize carbohydrates and fatty acids, which are essential for long-term cell survival [205,206,207,208]. piR-823 could increase the expression of mitochondrial chaperone HSP60 and 70, which could maintain mitochondrial stability and inhibit mitochondrial autophagy, thereby upregulating the quantity to meet the transitional capacity requirements in tumor cells [209,210,211]. The PINK1-Parkin pathway is necessary for mitophagy, leading to apoptosis [212, 213]. PINK1 recruits the E3 ubiquitin ligase, Parkin, from the cytosol to the mitochondria, phosphorylates it at Ser65, and induces mitophagy through ubiquitination [214, 215]. piR-823 promotes proteasome-mediated degradation of PINK1, causing mitophagy to maintain the high-energy metabolic state of tumor cells [115]. Pyruvate carboxylase (PC) is essential for islet glutathione synthesis and promotes its antioxidant capacity [216]. In addition, PC mediates glucose metabolism to meet the increased energy demands to facilitate the rapid proliferation of cancer cells [217,218,219]. piR-hsa-211,106 inhibits PC mRNA from directly interacting with its protein, impairing its normal function. PC knockdown attenuates cell proliferation and colony formation in vitro and suppresses tumor xenograft growth in vivo. In addition, PC inhibition significantly consumes the glutathione (GSH) pool, disturbs GSH homeostasis and reduces the ability to survive oxidative stress [116]. Overexpression of PIWIL1 in hepatocellular carcinoma cells significantly increases the intracellular ATP content to meet the needs of rapid cell proliferation. However, surprisingly, PIWIL1 expression failed to increase HCC cells’ glucose uptake and significantly increased the mitochondrial ROS. HCC cells increased intracellular free fatty acid consumption, leading to PIWIL1-mediated promotion of fatty acid metabolism to speed up energy production to meet the needs of rapid tumor growth [118]. PIWIL2 performs its anti-fibrotic function by reprogramming purine metabolism. PIWIL2 overexpression increases the levels of serine, nucleoside intermediates (AICAR), and nucleoside monophosphates (IMP and GMP), suggesting the probable requirement of purine biosynthesis in PIWIL2-driven remission of RILF. TGF-β1 is a critical factor in the development of radiation-induced pulmonary fibrosis [220,221,222]. Upregulation of PIWIL2 alone inhibits the radiation-induced increase of TGF-β1 and achieves the effect of anti-pulmonary fibrosis. Mycophenolate mofetil (MMF) reverses the inhibitory effect of Nrf2 or PIWIL2 on radiation-induced TGF-β1 expression. These results collectively suggest that PIWIL2 exerts its therapeutic impact on RILF by activating the purine metabolism axis [117].
Immune microenvironment remodeling
HIV-1, as an integrated provirus, is hidden in the cellular genome and evades immune surveillance [223, 224]. The 5’ end of HIV-1 long terminal repeat (LTR) is a strong promoter driving viral gene transcription [225, 226]. PIWIL4 inhibits HIV-1 5’LTR-driven transcription by recruiting SETDB1, HP1α/β/γ, and other repressors [227]. Conversely, the depletion of PIWIL4 reactivates the virus from latent CD4 + T lymphocytes and initiates replication. These results reveal an association between nuclear PIWIL4 and provide new insight into the mechanisms underlying the mandatory latency of HIV [122]. PIWIL1 overexpression accelerates energy production by promoting fatty acid metabolism to meet the needs of rapid tumor growth in HCC cells. This can be achieved by improving the fatty acid β-oxidation (FAO). In addition, PIWIL1-induced metabolic transformation to FAO creates an immunosuppressive microenvironment, promoting cancer progression. Inhibition of FAO suppresses MDSC and activates CD8 + toxic T cells, inhibiting tumor growth. Overexpression of PIWIL1 leads to increased C3 secretion, transmitting the immunosuppressive signals generated by PIWIL1 to its microenvironment. In addition, C3 secretion also activates IL-10 production, inhibiting T-cell immunity. In conclusion, PIWIL1 critically plays a dual role in driving metabolic reprogramming and immune escape in HCC [118]. Macrophages undergo polarization to M1 and M2 types under the stimulation of different stimulus factors, among which M2-polarized macrophages are pro-tumorigenic, known as tumor-associated macrophages (TAM) [228,229,230]. During M2 polarization, DQ551351 expression was downregulated, while DQ551308 expression was upregulated. Further research showed that overexpression of piR-DQ551351 and low expression of piR-DQ551308 could promote the mRNA expression of M1-macrophage marker TNF-α to promote the antitumor activity. The tumor volume in the overexpression group of piR-DQ551351 is significantly smaller than that in the control group [123]. The results of this study may provide new ideas for the M1 polarization of macrophages in the tumor immune microenvironment, thereby inhibiting the immune escape of M2-polarized macrophages.
Association of piRNAs/PIWI proteins with clinical characterization in diverse diseases
The dysregulated piRNA/PIWI proteins play an important role in the occurrence and development of diverse diseases. A growing body of evidence has confirmed the high correlation between piRNA/PIWI proteins and the clinical features of tumors and other non-cancer diseases. Here, we summarize the clinical relevance of piRNA/PIWI proteins in various systemic cancers and non-cancer diseases (Table 3).
Digestive system cancers
Most new cases of oral cancer diagnosed each year worldwide are squamous-cell cancers leading to painful deaths [267, 268]. The expression of PIWIL2 is significantly increased in all oral cancer cell lines. Patients with high PIWIL2 have more lymph node metastasis, poor histological type, clinical stage, and shorter survival [231, 232]. piR-1037 promotes chemotherapy resistance of OSCC cells to cisplatin (CDDP) by promoting invasion and metastasis via EMT, leading to poor prognosis [114].
Esophageal cancer is the sixth leading cause of cancer death worldwide, with esophageal squamous cell carcinoma (ESCC) accounting for about 90% of all esophageal cancers [269,270,271,272]. Su et al. reported that piR-823, highly expressed in ESCC, was positively correlated with pathological features such as lymph node metastasis and tumor size [130]. PIWIL2 is highly expressed in ESCC with a positive correlation with pathological T-stage and a negative correlation with long-term survival rate. In vitro experiments confirmed that it aided malignancy by inhibiting apoptosis [99].
Gastric cancer (GC) is the third leading cause of cancer death worldwide [273, 274]. piR-823 levels in gastric cancer tissues were significantly lower than in para-cancerous tissues. Furthermore, elevated piR-823 levels significantly inhibited tumor growth in vivo in a dose-dependent manner. However, the specific mechanism has not been further studied [132]. In addition, piRNA-1245 expression positively correlated with tumor size and TNM stage and inversely with overall survival in GC patients [233]. Moreover, the upregulated PIWIL1 in gastric cancer samples significantly correlated with tumor node metastasis but inversely with the degree of differentiation. High PIWIL1 mRNA expression is associated with poor prognosis, frequently seen in patients with poor differentiation and high TNM stage [234].
The development of clinical research and molecular targets have increased the treatment options for patients with advanced colorectal cancer [275,276,277]. However, colorectal cancer (CRC) is still the fourth deadliest cancer globally [278,279,280]. The highly expressed piR-020619 and piR-020450 act as oncogenes in CRC. The expression levels of these piRNAs in serum closely correlated to tumor size and early stage. The piR-02061 and piR-02450 detected in serum samples of patients after surgery were significantly lower than those before surgery [235]. Mai et al. demonstrated that the serum level of piR-54,265 in CRC patients is significantly higher than in the control groups, suggesting that its expression significantly increases in patients with advanced and metastatic colorectal cancer. Moreover, the overall survival due to its high expression was significantly shorter than those with low piR-54,655 levels [107]. In addition, the expression of piR-54,265 in stage I CRC and high-grade intraepithelial neoplasia (HIN) were significantly higher than that in the control group, suggesting that abnormally high piR-54,265 may be an early event in CRC development [236]. piR-24,000 is highly expressed in CRC, positively correlated with tumor stage and poor differentiation. In addition, distant metastases of tumors had a significant correlation with high piR-24,000 expression [237]. Compared with the healthy control group, piR-823 in tumor tissues correlated with tumor stage and differentiation, especially in poorly differentiated tumor tissues and stages III and IV. In serum, piR-823 content correlated with the advanced clinical stages (III and IV), poor differentiation, and lymph node metastasis of CRC [90, 115, 238]. Overexpression of PIWIL1 inhibits the apoptosis of CRC cells, promotes angiogenesis, and increases tumor volume and weight in nude mice, thus promoting the malignant progression of CRC [239, 240].
Pancreatic ductal adenocarcinoma (PDAC) is the most common form of pancreatic cancer, with a doubled global burden in the past 25 years [281,282,283]. About 50% of new diagnoses are metastatic, with a mean survival of less than one year [284, 285]. piR-017061 was reported to be inversely associated with long-term survival in PDAC [91]. High expression of PIWIL2 protein in pancreatic cancer patients is statistically significantly correlated with PFS and overall survival than those with low expression. Low PIWIL2 expression is associated with higher T stage, significantly correlating with other pathological features, such as vascular and perineural invasion, tumor stage, or lymph node involvement, leading to tumor aggressiveness [243]. Li et al. demonstrated how high PIWIL1 expression enhances the invasiveness of pancreatic cancer cells, promoting cancer progression [109]. Patients with low PIWIL3 or PIWIL4 expression showed shorter PFS and overall survival than those with high expression of both proteins. In addition, low PIWIL3 levels were significantly associated with neuronal invasion. Low PIWIL4 expression correlates with a higher risk of postoperative recurrence. They were significantly related to patients with T3-stage [244].
Pancreatic fibrosis is a prominent pathological feature of pancreatic cancer caused by pancreatic stellate cells (PSCs) [286,287,288]. PIWIL1 group significantly reduced the expression levels of type I and III collagen and α-smooth muscle actin, and then inhibited PSCs’ invasion and migration ability. This observation indicates PIWIL1 to be a potential therapeutic target for pancreatic fibrosis to prevent pancreatic cancer indirectly [289].
Intrahepatic cholangiocarcinoma (ICC) is a malignant hepatobiliary tumor from the bile duct with difficult diagnoses, poor prognosis, and a high mortality rate [290,291,292]. PIWIL4 upregulation was identified as a critical biomarker in this cancer, proportional to patient survival [241]. In addition, the levels of piR-10,506,469 in plasma exosomes of CCA and GBC patients were significantly increased. In contrast, the levels of piR-10,506,469 and piR-20,548,188 in postoperative patients were significantly decreased, as detected by comparing the differential expression of extracellular piRNA in patient plasma before and after surgery. This suggests that the expression levels of piR-10,506,469 and piR-20,548,188 negatively correlated with the patient prognosis [242].
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related death worldwide [129, 293]. The expression of PIWIL1 was significantly upregulated in HCC tissues, accelerating the proliferation of HCC cells and inducing colony formation in vitro, increasing the volume of HCC grafts in vivo [118].
Respiratory cancers
Lung cancer cases and deaths are on the rise globally, causing the most cancer deaths in the population [294, 295]. Of all types of lung cancer, non-small cell lung cancer (NSCLC) is the most common subtype [296,297,298].
piR-651 is highly expressed in NSCLC, positively correlated with distant metastasis and disease recurrence, and negatively associated with overall survival [97, 98]. As a tumor suppressor, patients with high expression of piR-55,490 have more prolonged survival and better prognoses [103]. piR-hsa-211,106 inhibits proliferation, promotes apoptosis, and attenuates the invasion ability of Lung adenocarcinoma (LUAD) cells. In addition, it can also increase the sensitivity of cells to Cisplatin to improve the prognosis of patients with drug resistance [116]. piR-57,125 is mainly related to the invasiveness of lung adenocarcinomas (LAC). piR-57,125 expression is significantly down-regulated in metastatic LACs compared with non-metastatic LACs [299]. Moreover, the tumor promoter RASSF1C upregulates the oncogenic factors piR-34,871 and piR-52,200 via the RASSF1C-PIWIL1-PIRNA axis to promote LC stem cell proliferation, colony formation, and EMT [254]. However, the clinical impact of piR-34,871 and piR-52,200 on lung cancer has not been reported.
Urinary system cancers
Clear cell renal carcinoma (ccRCC) is the sixth most common cancer in the world, often with a poor prognosis [253, 300,301,302]. Moreover, ccRCC is highly resistant to chemotherapy and radiotherapy [303, 304]. piR-57,125 was reported to be down-regulated in ccRCC, especially in metastatic tumors, implying a plausible role of piR-57,125 as a tumor suppressor by inhibiting tumor metastasis [104]. In contrast, piR-30,924 and piR-38,756 are highly expressed in primary metastatic tumors and have decreased expression in non-metastatic tumors compared to normal tissue. Their high expression is also significantly associated with tumor recurrence and poor overall survival [305].
Prostate cancer (PCa) is the second most common cancer and the fifth leading cause of cancer-related death in men [245, 306]. PIWIL2 has a high expression in PCa, which positively correlates with Gleason score, lymph node metastasis, and advanced TNM stage (T3-T4) [108]. piR-19,166 is down-regulated in PCa and closely related to organ and lymph node metastasis [105]. Moreover, upregulation of piR-001773/piR-017184 leads to shorter overall survival [101].
Bladder cancer (BLCA) is a common malignancy with high morbidity and mortality [307,308,309]. Sabo et al. used next-generation sequencing to piR-hsa-5936 upregulation in plasma EVs to be a risk factor for bladder cancer [310]. As a newly discovered tumor suppressor in bladder cancer, piR-60,152 has not been proven to have clinical significance. However, in vivo experiments have shown that its overexpression inhibits the proliferation and colony formation of bladder cancer cells and promotes apoptosis [246].
Reproductive system cancers
Breast cancer was the most common cancer and the leading cause of cancer-related death among women worldwide [247, 311,312,313]. piR-36,712 is significantly down-regulated in breast cancer. A slightly higher expression of piR-36,712 significantly affects progression-free survival, while its low expression is closely related to axillary lymph node metastasis [92]. In BC patients receiving chemotherapy, the exosome piR-17,560 enhances resistance of BC cells, developing drug resistance and leading to poor prognosis [112].
More than half a million women are diagnosed with cervical cancer each year, resulting in 300,000 deaths, with metastatic and recurrent patients having the worst overall prognosis [314,315,316]. Xie et al. reported that piRNA-14,633 is highly expressed in cervical cancer and is closely related to the advanced TNM stage and increased tumor size [141]. In recent years, PIWIL2 has been proved to promote the reprogramming of normal cervical stem cells to become cervical cancer cells. PIWIL2 was significantly increased in CIN2/3 and weakly expressed in CIN1, indicating the tissue specificity of PIWIL2 [248, 317].
Ovarian cancer (OCa) is a fatal gynecological cancer, the leading cause of death in women over the past few years [249, 318]. The most common type is epithelial ovarian cancer (EOCa), with endometrioid ovarian cancer (ENOCa) and serous ovarian cancer (SOCa) being the most lethal subtypes [319,320,321]. piR-52,207 upregulates ENOCa and promotes cell proliferation, migration, and tumorigenesis. At the same time, piR-52,207 and piR-33,733 are upregulated in SOCa, leading to apoptosis inhibition in the cells [322].
Endometrial cancer (EC) is one of the most common reproductive system malignancies [250, 323]. Clinically, most endometrial cancers are type I, also known as estrogen-dependent endometrioid adenocarcinoma [324, 325]. PIWIL1 acts as an oncogene in endometrial cancers, inducing EMT and conferring stemness characteristics on EC cells, promoting cancer growth [113]. In addition, PIWIL1 inhibits its downstream target PTEN and can act as a downstream target of ERα and participate in E2-stimulated endometrial carcinogenesis [137, 138].
Hematologic cancers
Hematologic malignancies have a highly variable prognosis and continuous post-treatment relapse [326,327,328]. Among them, leukemias is the most common pediatric cancer [329]. Hsa_piR_011186 significantly pushes the cell cycle into the S phase and inhibits apoptosis, thereby promoting the occurrence and development of acute leukemia [133].
Diffuse large B-cell lymphoma (DLBCL) is the most common subtype of lymphoid malignancies [330]. And current treatment methods for advanced patients still keep 40% of patients uncured [331, 332]. It is reported that high expression of piR-30,473 is associated with poor long-term prognosis of DLBCL patients, with the overall survival being inversely proportional to the expression level [140].
Multiple myeloma (MM) is an incurable malignant tumor derived from B cells [333]. The fundamental difficulty of its treatment is bolstered by the unlimited self-renewal ability of a few cancer stem cells (CSCs) [334]. piR-004800 is highly expressed in the bone marrow supernatant exosomes of MM patients and significantly increases in ISS III patients, suggesting its upregulation be correlated with MM progression [93]. High expression of piR-823 was associated with a higher ISS stage and shorter survival time. Granulocytic MDSCs (G-MDSCs) trigger piRNA-823 expression, increasing the tumorigenic potential of MM cells [111, 129]. PIWIL1 expression was significantly high in ISSII and III stage MM patients than in stage I, suggesting it to be associated with an advanced stage of the disease. In addition, higher PIWIL1 levels in refractory MM patients than in newly diagnosed suggested a gradual increase in PIWIL1 expression from freshly diagnosed to relapsed/refractory MM patients [26].
Classical Hodgkin lymphoma (cHL) accounts for a small percentage of all lymphomas [251, 335]. The low expression of piR-651, a tumor suppressor, is associated with shorter disease-free and overall survival [336].
Nervous system cancers
Glioblastoma (GBM), the most lethal form of glioma in adults, is the most common primary brain tumor [252, 337, 338]. Overexpression of piR-8041 inhibited long-term colony formation and glioma cell proliferation. In addition, tumors induced in mouse brains showed that overexpression of piR-8041 significantly inhibited tumor volume. The effectiveness of piR-8041 requires maintained periodic therapy, which in clinical practice largely depends on the efficacy of the vector in crossing the blood-brain barrier (BBB) [94]. Bartos et al. found that low expression of piR-23,231 was significantly associated with decreased survival. In addition, piR-hsa-9491 and piR-has-12,488 significantly reduced the colony formation ability of GBM cells in vitro [339]. The high PIWIL1 expression in GBM correlates with larger tumor diameter and higher tumor grade, promoting the malignant proliferation of glioma stem cells (GSCs). It also reduces the effect of drug therapy by reducing the blood-tumor barrier (BTB) [95, 255, 340]. The tumor-suppressor PIWIL3 has a low expression in glioma and a negative correlation with the pathological grade of the disease. In contrast, overexpression of PIWIL3 significantly inhibited the progression of glioma [96].
Neuroblastoma, cancer originating in the neural crest, is the most common extracranial solid tumor in children [256, 257]. Although the lesion may spontaneously regress in some cases, it can sometimes lead to severe attacks [341, 342]. As a cancer-promoting factor, piR-39,980 overexpression significantly improves the survival of NB cells, promoting invasion and metastasis. Moreover, the higher expression of piR-39,980 reduced the sensitivity of NB cells to doxorubicin. It may contribute to the development of chemotherapy resistance, which is closely related to NB’s current clinical drug resistance [343].
Other kinds of cancers
Fibrosarcoma represents only a tiny percentage of all soft tissue sarcomas [344]. Currently, the treatment of metastatic fibrosarcoma mainly relies on chemotherapy [258, 345]. piR-39,980 is downregulated in fibrosarcoma cells, causing doxorubicin resistance. Conversely, high expression of piR-39,980 attenuates doxorubicin resistance [119]. Only 5% of fibrosarcoma patients show high PIWIL4 expression, which positively correlates with survival, disease-free survival, disease-specific survival, and progression-free survival [120].
Osteosarcoma (OS) is the most common malignancy of the skeletal system with high invasion and metastasis ability [346, 347]. The prognosis of metastatic or recurrent osteosarcoma is still unsatisfactory [348, 349]. The high expression of piR-39,980 correlates to osteosarcoma’s high invasion and metastasis. Silencing the tumor suppressor SERPINB1 and MMP-2 induction induces the invasion and metastasis ability of osteosarcoma cells [106]. Chang et al. showed the high expression of piR-13,643 and piR-21,238 in thyroid cancer, associated with clinical stages [350].
Association of piRNAs/PIWI proteins with clinical characterization in diverse non-cancer diseases
Nervous system diseases
Alzheimer’s disease (AD), the most common form of dementia, is characterized by progressive memory loss and other cognitive abilities [259, 351]. Cytochrome C (CYCS), released from the mitochondrial membrane, induces the piR-38,240- and piR-34,393-mediated regulation of apoptosis by triggering the activation of caspase cascades. In addition, piR-38,240 reduces the expression of another protective factor, KPNA6, which regulates NRF2-dependent antioxidant responses. Together, these two mechanisms contribute to nerve cell damage in AD [352].
Amyotrophic lateral sclerosis (ALS) is a fatal neuromuscular disease and results in muscle dysfunction [260, 353, 354]. In Sporadic Amyotrophic Lateral Sclerosis (sALS), the expressions of the protective factors PIWIL1 and PIWIL4 are significantly downregulated. In the absence of this neurodegenerative disease, PIWIL1 is highly expressed in anterior horn cells. In contrast, the lumbar spine of sALS patients shows a drastic reduction in the number of anterior horn cells, which are PIWIL1-negative. Previous research has implicated the crucial role of PIWIL1 in neuronal development and protection in the migration of microtubules [355]. This observation suggested that PIWIL1 expression change can act as a compensatory mechanism for axonal dysfunction [356].
Cardiovascular and circulatory systems diseases
T.cruzi, which can penetrate the interstitial vascular system and eventually invade cardiomyocytes, causes heart disease and results in myocardial damage and conduction abnormalities [261, 357]. T. cruzi has a highly differential expression of the fibrosis gene, such as metalloproteinase families MMP2, MMP9 and MMP12, as well as abnormal expression of TGF-β, which ultimately causes cardiac fibroblasts to differentiate into myofibroblasts [358,359,360]. The expression of some novel piRNAs is significantly induced at the early stage of the infection. Studies have identified a novel piRNA (npiR46) that binds to NFATC2 and FOS, and npiR573, and npiR587 bind to TGFβ1, promoting fibrosis. These piRNAs are important biomarkers for the early stage of T.cruzi infection [361].
Chronic hypertrophy and its associated myocardial remodeling significantly contribute to cardiac dysfunction, leading to severe heart failure and death [266, 362]. Overexpression of piR-141,981 enhanced Ang II-induced cardiomyocyte hypertrophy and increased Ang II-induced ANP expression, encoding the atrial natriuretic peptide. In addition, it inhibits the activation of RNA epigenetic modules of anti-hypertrophic molecules such as GSK3β and upregulates PARP10 as a pro-hypertrophic factor [142]. Rajan et al. found piR_2106027 to be significantly upregulated in patients with myocardial infarction and correlated with troponin I levels, indicating piR_2106027 expression to be associated with the degree of myocardial injury [363]. This has the potential to become a new clinical biomarker for patients with myocardial infarction.
Abnormal proliferation of pulmonary artery smooth muscle cells (PASMCs) and hypoxia-induced abnormal secretion of extracellular matrix proteins increase pulmonary vascular resistance and pulmonary hypertension (PH) [265, 364, 365]. A higher expression of piR-63,076 was seen in the medial region of hypoxic pulmonary arteries but not in normal lung tissues. piR-63,076 knockdown attenuated the development of pulmonary hypertension by inhibiting the proliferation of PASMCs, implicating its potential role as a biomarker and a therapeutic target for pulmonary hypertension [136].
Radiotherapy is considered to be an effective treatment for thoracic cancers such as that of the lung and breast [297, 366,367,368]. However, a significant complication of radiotherapy, radiation-induced pulmonary fibrosis (RILF), not only limits radiotherapy efficacy but also seriously affects patients’ quality of life [369, 370]. Upregulation of Nrf2 abolishes RILF in vitro and in vivo. Nrf2 activates its downstream target PIWIL2 to increase its expression. More importantly, the knockdown of PIWIL2 significantly reverses the protective effect of Nrf2. In conclusion, upregulation of Nrf2 and PIWIL2 might potentially have a protective role in RILF reversal [117].
Stroke is the second leading cause of death globally, with ischemic strokes accounting for at least 80% of all cases [371]. In this condition, there is an irreversible loss of brain cells due to ischemia and hypoxia [372,373,374]. Downregulated PIWIL2 contributes to neuroprotection against transient global cerebral ischemia (tGCI) in CA1 neurons induced by hypoxic postconditioning (HPC). HPC entirely prevents the tGCI-induced increase in PIWIL2 mRNA and enhanced hippocampal dendritic spine plasticity, improving memory function after tGCI [139].
Reproductive system diseases
Preeclampsia is an idiopathic pregnancy disorder that occurs after 20 weeks of gestation, severely affecting the health of the pregnant woman and the fetus [375,376,377]. PIWIL1 is highly expressed in trophoblasts, promoting their proliferation and invasion of trophoblasts and inhibiting apoptosis. PIWIL1 is reduced in preeclampsia, inhibits the proliferation of trophoblast cells, and promotes apoptosis of placental trophoblast cells, leading to shallow placenta accreta and the development of preeclampsia [378].
Adenomyosis is a benign reproductive disease characterized by the invasion of endometrial tissue into the myometrium [379]. PIWIL2 is highly expressed in adenomyosis and associated with disease progression. It is hypothesized that PIWIL2 may be involved in endometrial cell proliferation, critically mediating the disease pathogenesis; however, the specific underlying mechanisms need further study [262]. PIWIL1 is a critical E3 ligase, chelating histone ubiquitination in the cytoplasm of early sperm cells. The level of its mutants is related to the degree of developmental defects in sperms. Degradation of PIWIL1 by APC/C in advanced sperm cells releases RNF8 into the nucleus to mediate subsequent histone-to-protamine conversion. In contrast, PIWIL1 mutations block the RNF8 nuclear translocation and RNF8-mediated histone to induce protamine conversion in advanced sperm cells, resulting in impaired sperm count and quality [380].
Other kinds of diseases
PIWIL4 has been implicated in playing a significant role in suppressing HIV-1 transcription by recruiting various repressors. It promotes the latent state of HIV-1-infected cells. Endogenous PIWIL4 inhibits HIV-1 pro-viral DNA expression and maintains HIV-1 latency in CD4 + T lymphocytes. PIWIL4 knockdown enhances HIV-1 transcription and reverses HIV-1 latency in Jurkat T cells and primary CD4 + T lymphocytes. Mechanically, PIWIL4 depletion reactivates HIV-1 replication in latently infected primary CD4 + T lymphocytes [122]. This may provide new ideas for the treatment of AIDS, by promoting the expression of PIWIL4 to enforce the maintenance of HIV virus in the latent state.
Clinical application of piRNAs
Early detection and rapid evaluation of the progress of an acute cardiovascular disease or highly fatal cancer provide a valuable reference for future treatment [263, 264, 381, 382]. The expression of piRNA, a newly discovered small ncRNA, shows a statistically significant correlation between serum or body fluids and disease [383,384,385]. In this section, we summarize the potential applications of piRNA in clinical applications (Tables 4 and 5).
piRNAs as diagnostic and prognostic biomarkers in cancer
piRNAs/PIWI proteins are great potential clinical markers possessing the following characteristics: sensitivity, specificity, expression stability, and broad applicability [386]. According to their various sources, they can be roughly divided into tissue-derived, serum-derived, and body-fluid-derived ones (Fig. 4).
Potential value of piRNAs/PIWI proteins as clinical markers. piRNAs/PIWI proteins are mainly detected in tissues, serum, gastric juice and urine as biomarkers. As a biomarker, it also needs to meet the characteristics of high sensitivity, specificity, and convenient detection. Among them, the red targets are tumor suppressor factors, and their expression is positively proportional to the prognosis of patients. The black target is the pro-cancer factor, and the higher its expression, the worse the prognosis of the patient
Tissue-derived biomarkers are often stably expressed with high specificity [387]. Su et al. further tested the diagnostic value of piR-823 in ESCC using ROC plots, which showed piR-823 to be a valuable biomarker with a sensitivity of 62.96% and a specificity of 76.93%, accurately distinguishing ESCC patients from normal individuals [130]. piR-24,000 in CRC tissues could significantly distinguish patients from normal subjects. In the late group (stages III and IV), the AUC value is 0.8405, while in the early group (stages I and II), the AUC value was slightly lower at 0.7796 [237]. piR-57,125, piR-30,924, and piR-38,756 are potential prognostic biomarkers of ccRCC because the high expression of piR-30,924 and piR-38,756 and low expression of piR-57,125 significantly correlates with the metastatic tumors and bone metastasis. The prognostic significance of piR-38,756 was particularly indicative in nonmetastatic patients [305]. piR-9491 showed the best specificity and sensitivity for identifying GBM samples from non-tumor samples. Its low expression significantly correlated with shorter survival [339]. The piRNA sequencing data of papillary thyroid cancer revealed two upregulated piRNAs, piR-13,643 and piR-21,238, showing better specificity than the current biomarker HBME1 in distinguishing benign and malignant nodules and were closely related to prognosis [350].
Serum-derived biomarkers often show high sensitivity and prognostic associations, reflecting the status of patients on time [388,389,390]. In addition, they can also reflect the status of patients in a timely manner. Most people with early-stage colorectal cancer have few or no symptoms [391, 392]. The traditional serum tumor markers CEA and CA19-9 are still unsuitable for colorectal cancer screening due to their low sensitivity [393, 394]. The AUC of piR-020619 and piR-020450 were 0.883 and 0.913, respectively. The sensitivity and specificity of these piRNAs were higher than the currently used biomarkers CEA and CA199. And the serum levels of piR-020619 and piR-020450 of colorectal cancer patients after surgery also decreased. In addition, for early-stage and small CRC tumors less than 3 cm, the sensitivity of the piRNA group was much higher than that of CEA and CA19-9, and the specificity of the piRNA group was 90.94%, similar to that of CEA and CA19-9. This phenomenon is best demonstrated by the piRNA panel’s ability to distinguish CEA and CA19-9-negative CRC patients from normal controls [235]. Stability is a critical characteristic of biomarkers, playing a crucial role in clinical convenience [395, 396]. Circulating piR-54,265 is stable in CRC serum to be readily and reliably measured. In addition, Mai et al. also verified the sensitivity and specificity of piR-54,265 to be significantly higher in the serum of CRC patients than in all other control groups [107]. Mai et al. further investigated the specificity of piR-54,265 in CRC precancerous lesions, which revealed that the expression level of piR-54,265 in patients with HIN or stage I CRC was significantly higher than that in controls, implying that abnormal overexpression of piR-54,265 may be an early event in CRC development. The serum piR-54,265 level in postoperative CRC patients showed a dynamic decline. In contrast, serum indicators also showed a synchronous rise in recurrent patients, and its dynamic changes were more accurate than the current CEA, CA19-9, and CA125, indicating that serum piR-54,265 are potential new effective indicators of CRC recurrence [236]. Compared with the healthy control group, the expression of piR-651 in the serum of the CHL group was low. After the complete response, the expression of piR-651 was upregulated again and reached almost the same standard as that of the healthy control group, indicating that piR-651 has the potential to be a prognostic marker [336]. Specifically, piR-823 was differentially expressed in tissues and serum compared with normal tissues. Tissue expression of piR-823 was significantly associated with poorly differentiated tumor tissue and stage III and IV. And serum piR-823 content was associated with the advanced clinical stage (III and IV), poor differentiation, and lymph node metastasis. Correlation analysis demonstrated that the increase of piR-823 in tissues correlated with its increase in serum, promising to replace invasive colonoscopy with noninvasive blood sampling [238].
Finally, body fluid-derived biomarkers, such as piRNAs from gastric juice and urine, hold the potential to improve the convenience of detection. They can significantly avoid the invasive examination caused by routine biomarker collection. piR-1245 is a highly expressed oncogenic factor in gastric cancer, and its high expression positively correlates with overall poor survival, suggesting poor prognosis. Statistical results show that the sensitivity and specificity of piR-1245 were higher than those of CEA and CA724, commonly used gastric cancer biomarkers [233]. Therefore, it can potentially replace the currently known gastric cancer markers and help early-stage detection of gastric cancer in patients. Cystoscopy and urine cytology are the gold standards for diagnosing BLCA, but with limited convenience. Therefore, it is necessary to find highly sensitive and specific body fluid markers to facilitate high-risk screening groups. Compared with the control group, the expression of piR-hsa-5936 in BLCA cases showed an upward trend, resulting in an increased risk of BLCA and a shorter survival period. However, only plasma EV showed its differential expression but not urine [310].
piRNAs and cancer therapy resistance
Despite significant advances in tumor treatments, tumor therapy resistance remains a major challenge. Although most tumors regress rapidly and significantly after initial treatment, clinical drug resistance sets in at a later stage of the treatment, leading to treatment failure and, ultimately, patient death (Fig. 5).
The green targets in the figure are piRNAs/PIWI proteins that increase chemotherapy resistance. PIWIL1 inhibits the pro-apoptotic effect of chemotherapeutic drugs on MM cells by inhibiting mitochondrial apoptosis, thereby promoting proliferation and inhibiting apoptosis. PIWIL1/piR-DQ593109 can reduce the blood drug concentration in glioma by reducing the permeability of blood-tumor barrier, thus playing a role in chemotherapy resistance. piR-39,980, PIWIL2 and piR-1037 attenuated the efficacy of chemotherapy by reducing the sensitivity of tumor cells to chemotherapeutic drugs. piR-39,980 and piR-hsa-211,106 enhanced the chemotherapeutic effect by increasing the accumulation of chemotherapeutic drugs in tumor cells, thereby achieving anti-proliferation and pro-apoptosis effects
Overexpression of PIWIL1 leads to resistance of MM cells after treatment with chemotherapeutic drugs, whereas its downregulation leads to re-sensitization of MM cells to chemotherapy. Mechanistically, PIWIL1 overexpression reduced the phosphorylation of autophagy-related proteins (mTOR and AKT-Ser473) and increased the expression of PINK1/Parkin pathway protein Parkin, mitophagy-related protein (optineurin) and p-TBK-1, promoting MM cell drug resistance by regulating mitochondrial anti-apoptosis [26]. MM drug resistance is also caused by the blood-tumor barrier (BTB) that restricts the effective delivery of anti-glioma drugs to the affected brain tissues. The knockdown of PIWILI and piR-DQ593109 increased the permeability of BTB by reducing the expression level of tight junction-associated proteins, such as ZO-1, Occludin, and Claudin-5 [255]. High level of piR-39,980 reduced the sensitivity of NB cells to doxorubicin. It increased NB deterioration under doxorubicin treatment by inducing cell growth and enhancing metastasis by targeting JAK3 [343]. The low expression of piR-39,980 leads to increased resistance to chemotherapeutic drugs in DOX-resistance fibrosarcoma cell lines. In terms of mechanism, piR-3998 reduced the resistance of fibrosarcoma cells to DOX and increased DOX sensitivity and intracellular concentrations by targeting and down-regulating RRM2 and CYP1A2. Overexpression of piR-39,980 suppressed CYP1A2, promoting DOX accumulation and increased sensitivity [119]. piR-hsa-211,106 overexpression LUAD cells are sensitive, while piR-has-211,106 knockdown increases LUAD resistance to PDD. piR-hsa 211,106 showed synergistic pro-apoptotic effects with PDD in vitro and in vivo [116]. High levels of piR-1037 promote the resistance of OSCC to CDDP chemotherapy. piR-1037 inhibits CDDP-induced apoptosis by inhibiting Caspase-8 and Caspase-1-dependent death signaling pathways [114]. In HPV-infected cervical epithelium, the oncoproteins E6 and E7 activate PIWIL2, which then reprogram the cells that initiate tumor-initiating cells (TICs) and tumorigenesis of the cervix. In cervical neoplasia, PIWIL2 activates Wnt3a signaling to enhance reprogramming through CBP/ β-catenin interactions and further maintain the stemness of TICs. In addition, blocking the CBP/β-catenin interaction can induce early cell differentiation and increase the sensitivity of cisplatin to a certain extent [317].
The name and annotation of piRNAs: database exploration and correction
piRNA is one of three major classes of small regulatory RNAs that are encoded in discrete genomic clusters, many of which are present in the genomic regions of humans [32]. Surprisingly, previous studies have found that a significant number of sequences in different human piRNA databases show a high degree of identity with other ncRNAs, which means that the actual expression of piRNAs may be greatly overestimated [397]. Therefore, constructing a more accurate database plays an extremely important role in the study of piRNAs. In fact, the database has been changed and corrected for generations, and a large number of the above fake piRNAs have been excluded, making the results of the database more realistic. The most commonly used database is piRBase release v3.0. (http://bigdata.ibp.ac.cn/piRBase) [398]. In addition, this database further enriched piRNA targets and realized the visualization of the regulatory network of the targets, which made it more convenient for researchers to explore piRNA functions [399].
Conclusion and perspective
To date, multiple piRNAs are being continuously discovered and can be accurately identified as abnormal expressions in human diseases. Generally, piRNA and PIWI proteins maintain physiological balance through germ- and somatic cell-mediated synthesis and degradation. However, the disease conditions may occur when the expression of piRNA or PIWI protein is disturbed. We elaborate on recent studies of the abnormal regulatory mechanisms of piRNA/PIWI proteins in various diseases. The preliminary stage of the current research on piRNA/PIWI protein fails to cover its great clinical prospect. Recently, some studies have shown that many piRNAs are highly differentially expressed in fluid, serum, and tissue samples. The specificity and sensitivity of some piRNAs have exceeded that of currently commonly used clinical tumor markers, such as piR-020619 and piR-020450, which are highly expressed in CRC serum, and their sensitivity and specificity have been proved to be better than those of traditional biomarkers CEA and CA-199.
This observation crucially underlines the significant potential of piRNAs as alternatives to current tumor biomarkers. In addition, piRNA/PIWI protein also enhances chemotherapy tolerance by regulating the drug resistance of tumor cells, significantly contributing toward solving the problem of the current widespread clinical chemotherapy resistance and further improving the prognosis of patients. Although the current research on piRNA and PIWI proteins is in the preliminary stage, this cannot mask their great potential in future clinical applications. With the continuous progress of research, we believe that more piRNAs/PIWI protein complexes will be discovered and studied, and eventually be used as specific targets for clinical treatment.
Availability of data and materials
Not applicable.
Abbreviations
- piRNAs:
-
Piwi-interacting RNAs
- RISC:
-
RNA-induced silencing complex
- Pol II:
-
RNA polymerase II
- H3K9:
-
Histone H3 Lys9
- HP1:
-
Heterochromatin protein 1
- RDC:
-
Rhino-Deadlock-Cutoff
- Trf2:
-
TATA box-binding protein-associated Factor 2
- flam:
-
Flamenco
- Aub:
-
Aubergine
- Ago3:
-
Argonaute 3
- zuc:
-
Zucchini
- Armi:
-
Armitage
- TE:
-
Transposable element
- CpG:
-
Cytosine phospho-guanine
- DNMTs:
-
DNA methyltransferases
- MDSCs:
-
Myeloid-derived suppressor cells
- MMSCs:
-
Multiple myeloma stem cells
- TSGs:
-
Tumor-suppressor genes
- APC:
-
Adenomatous polyposis coli
- PTEN:
-
Phosphatase and tensin homolog
- GSTP1:
-
Glutathione S-transferase pi 1
- tGCI:
-
Transient global cerebral ischemia
- HPC:
-
Hypoxic postconditioning
- WTAP:
-
Wilms’ tumor 1-associating protein
- DLBCL:
-
Diffuse large B-cell lymphoma
- FTO:
-
Fat mass and obesity-associated protein
- CHAPIR:
-
Cardiac-hypertrophy-associated piRNA
- HSF1:
-
Heat shock factor 1
- MM:
-
Multiple myeloma
- S1PR:
-
Sphingosine-1-phosphate receptor
- IKK:
-
IκB kinase
- ESCC:
-
Esophageal cell squamous carcinoma
- CCL3:
-
Chemokine (C-C motif) ligand 3
- ccRCC:
-
Clear cell renal cell carcinoma
- PCa:
-
Prostate carcinoma
- MMP:
-
Matrix metalloproteinases
- PDAC:
-
Pancreatic ductal adenocarcinoma
- EMT:
-
Epithelial-mesenchymal transition
- TME:
-
Tumor microenvironment
- EVs:
-
Extracellular vesicles
- DOX:
-
Doxorubicin
- RR:
-
Ribonucleotide reductase
- ROS:
-
Reactive oxygen species
- PC:
-
Pyruvate carboxylase
- GSH:
-
Glutathione
- MMF:
-
Mycophenolate mofetil
- LTR:
-
Long terminal repeat
- FAO:
-
Fatty acid β-oxidation
- TAM:
-
Tumor-associated macrophages
- CDDP:
-
Cisplatin
- GC:
-
Gastric cancer
- CRC:
-
Colorectal cancer
- HIN:
-
High-grade intraepithelial neoplasia
- PSCs:
-
pancreatic stellate cells
- ICC:
-
Intrahepatic cholangiocarcinoma
- HCC:
-
Hepatocellular carcinoma
- NSCLC:
-
non-small cell lung cancer
- LUAD:
-
Lung adenocarcinoma
- LAC:
-
lung adenocarcinomas
- BLCA:
-
Bladder cancer
- OCa:
-
Ovarian cancer
- EOCa:
-
Epithelial ovarian cancer
- ENOCa:
-
Endometrioid ovarian cancer
- EC:
-
Endometrial cancer
- CSCs:
-
Cancer stem cells
- cHL:
-
Classical Hodgkin lymphoma
- GBM:
-
Glioblastoma
- BBB:
-
Blood-brain barrier
- BTB:
-
Blood-tumor barrier
- OS:
-
Osteosarcoma
- AD:
-
Alzheimer’s disease
- CYCS:
-
Cytochrome C
- ALS:
-
Amyotrophic lateral sclerosis
- SALS:
-
Sporadic Amyotrophic Lateral Sclerosis
- PASMCs:
-
Pulmonary artery smooth muscle cells
- PH:
-
Pulmonary hypertension
- RILF:
-
Radiation-induced pulmonary fibrosis
- tGCI:
-
Transient global cerebral ischemia
- HPC:
-
Hypoxic postconditioning
- TICs:
-
Tumor-initiating cells
References
Lynch SV, Pedersen O. The human intestinal microbiome in Health and Disease. N Engl J Med. 2016;375:2369–79.
Zhang L, Lu Q, Chang C. Epigenetics in Health and Disease. Adv Exp Med Biol. 2020;1253:3–55.
Nie P, Li Z, Wang Y, Zhang Y, Zhao M, Luo J, Du S, Deng Z, Chen J, Wang Y, et al. Gut microbiome interventions in human health and Diseases. Med Res Rev. 2019;39:2286–313.
Liu C, Du L, Wang S, Kong L, Zhang S, Li S, Zhang W, Du G. Differences in the prevention and control of cardiovascular and cerebrovascular Diseases. Pharmacol Res. 2021;170:105737.
Forte M, Schirone L, Ameri P, Basso C, Catalucci D, Modica J, Chimenti C, Crotti L, Frati G, Rubattu S, et al. The role of mitochondrial dynamics in Cardiovascular Diseases. Br J Pharmacol. 2021;178:2060–76.
Liberale L, Ministrini S, Carbone F, Camici GG, Montecucco F. Cytokines as therapeutic targets for cardio- and cerebrovascular Diseases. Basic Res Cardiol. 2021;116:23.
Vogel B, Acevedo M, Appelman Y, Bairey Merz CN, Chieffo A, Figtree GA, Guerrero M, Kunadian V, Lam CSP, Maas A, et al. The Lancet women and Cardiovascular Disease commission: reducing the global burden by 2030. Lancet. 2021;397:2385–438.
Sun L, Zhang H, Gao P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell. 2022;13:877–919.
Prasher D, Greenway SC, Singh RB. The impact of epigenetics on Cardiovascular Disease. Biochem Cell Biol. 2020;98:12–22.
Anzelon TA, Chowdhury S, Hughes SM, Xiao Y, Lander GC, MacRae IJ. Structural basis for piRNA targeting. Nature. 2021;597:285–9.
Huang X, Fejes Tóth K, Aravin AA. piRNA Biogenesis in Drosophila melanogaster. Trends Genet. 2017;33:882–94.
Pippadpally S, Venkatesh T. Deciphering piRNA biogenesis through cytoplasmic granules, mitochondria and exosomes. Arch Biochem Biophys. 2020;695:108597.
Genzor P, Konstantinidou P, Stoyko D, Manzourolajdad A, Marlin Andrews C, Elchert AR, Stathopoulos C, Haase AD. Cellular abundance shapes function in piRNA-guided genome defense. Genome Res. 2021;31:2058–68.
Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, Zamore PD. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet. 2019;20:89–108.
Parhad SS, Theurkauf WE. Rapid evolution and conserved function of the piRNA pathway. Open Biol. 2019;9:180181.
Iwasaki YW, Siomi MC, Siomi H. PIWI-Interacting RNA: its Biogenesis and functions. Annu Rev Biochem. 2015;84:405–33.
Zeng Q, Wan H, Zhao S, Xu H, Tang T, Oware KA, Qu S. Role of PIWI-interacting RNAs on cell survival: proliferation, apoptosis, and cycle. IUBMB Life. 2020;72:1870–8.
Tóth KF, Pezic D, Stuwe E, Webster A. The piRNA Pathway guards the germline genome against transposable elements. Adv Exp Med Biol. 2016;886:51–77.
Wang K, Wang T, Gao XQ, Chen XZ, Wang F, Zhou LY. Emerging functions of piwi-interacting RNAs in Diseases. J Cell Mol Med. 2021;25:4893–901.
Huang X, Wong G. An old weapon with a new function: PIWI-interacting RNAs in neurodegenerative Diseases. Transl Neurodegener. 2021;10:9.
Wang K, Zhou LY, Liu F, Lin L, Ju J, Tian PC, Liu CY, Li XM, Chen XZ, Wang T, et al. PIWI-Interacting RNA HAAPIR regulates Cardiomyocyte Death after Myocardial Infarction by promoting NAT10-Mediated ac(4) C acetylation of Tfec mRNA. Adv Sci (Weinh). 2022;9:e2106058.
Liu Y, Dou M, Song X, Dong Y, Liu S, Liu H, Tao J, Li W, Yin X, Xu W. The emerging role of the piRNA/piwi complex in cancer. Mol Cancer. 2019;18:123.
Ramat A, Simonelig M. Functions of PIWI proteins in Gene Regulation: new arrows added to the piRNA Quiver. Trends Genet. 2021;37:188–200.
Lin Y, Zheng J, Lin D. PIWI-interacting RNAs in human cancer. Semin Cancer Biol. 2021;75:15–28.
Mokarram P, Niknam M, Sadeghdoust M, Aligolighasemabadi F, Siri M, Dastghaib S, Brim H, Ashktorab H. PIWI interacting RNAs perspectives: a new avenues in future cancer investigations. Bioengineered. 2021;12:10401–19.
Chen S, Ben S, Xin J, Li S, Zheng R, Wang H, Fan L, Du M, Zhang Z, Wang M. The biogenesis and biological function of PIWI-interacting RNA in cancer. J Hematol Oncol. 2021;14:93.
Zhang D, Tu S, Stubna M, Wu WS, Huang WC, Weng Z, Lee HC. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science. 2018;359:587–92.
Khan C, Muliyil S, Rao BJ. Genome damage sensing leads to tissue homeostasis in Drosophila. Int Rev Cell Mol Biol. 2019;345:173–224.
Lupski JR, Liu P, Stankiewicz P, Carvalho CMB, Posey JE. Clinical genomics and contextualizing genome variation in the diagnostic laboratory. Expert Rev Mol Diagn. 2020;20:995–1002.
Goga A, Stoffel M. Therapeutic RNA-silencing oligonucleotides in metabolic Diseases. Nat Rev Drug Discov. 2022;21:417–39.
Henney NC, Banach M, Penson PE. RNA silencing in the management of Dyslipidemias. Curr Atheroscler Rep. 2021;23:69.
Czech B, Munafò M, Ciabrelli F, Eastwood EL, Fabry MH, Kneuss E, Hannon GJ. piRNA-Guided Genome Defense: from Biogenesis to silencing. Annu Rev Genet. 2018;52:131–57.
Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–39.
Gebert D, Neubert LK, Lloyd C, Gui J, Lehmann R, Teixeira FK. Large Drosophila germline piRNA clusters are evolutionarily labile and dispensable for transposon regulation. Mol Cell. 2021;81:3965–3978e3965.
Kofler R. Dynamics of transposable element invasions with piRNA clusters. Mol Biol Evol. 2019;36:1457–72.
Yu T, Fan K, Özata DM, Zhang G, Fu Y, Theurkauf WE, Zamore PD, Weng Z. Long first exons and epigenetic marks distinguish conserved pachytene piRNA clusters from other mammalian genes. Nat Commun. 2021;12:73.
Wierzbicki F, Kofler R, Signor S. Evolutionary dynamics of piRNA clusters in Drosophila. Mol Ecol. 2023;32:1306–22.
Adashev VE, Kotov AA, Bazylev SS, Shatskikh AS, Aravin AA, Olenina LV. Stellate genes and the piRNA pathway in Speciation and Reproductive isolation of Drosophila melanogaster. Front Genet. 2020;11:610665.
Penke TJ, McKay DJ, Strahl BD, Matera AG, Duronio RJ. Direct interrogation of the role of H3K9 in metazoan heterochromatin function. Genes Dev. 2016;30:1866–80.
Yu B, Lin YA, Parhad SS, Jin Z, Ma J, Theurkauf WE, et al. Structural insights into Rhino-Deadlock complex for germline piRNA cluster specification. EMBO Rep. 2018;19(7):e45418.
Tsai SY, Huang F. Acetyltransferase enok regulates transposon silencing and piRNA cluster transcription. PLoS Genet. 2021;17:e1009349.
Mohn F, Sienski G, Handler D, Brennecke J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 2014;157:1364–79.
Parhad SS, Yu T, Zhang G, Rice NP, Weng Z, Theurkauf WE. Adaptive evolution targets a piRNA precursor transcription network. Cell Rep. 2020;30:2672–2685e2675.
Pane A, Jiang P, Zhao DY, Singh M, Schüpbach T. The cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. Embo j. 2011;30:4601–15.
Chen YA, Stuwe E, Luo Y, Ninova M, Le Thomas A, Rozhavskaya E, Li S, Vempati S, Laver JD, Patel DJ, et al. Cutoff suppresses RNA polymerase II termination to ensure expression of piRNA precursors. Mol Cell. 2016;63:97–109.
Zhang Z, Wang J, Schultz N, Zhang F, Parhad SS, Tu S, Vreven T, Zamore PD, Weng Z, Theurkauf WE. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell. 2014;157:1353–63.
ElMaghraby MF, Andersen PR, Pühringer F, Hohmann U, Meixner K, Lendl T, Tirian L, Brennecke J. A heterochromatin-specific RNA export pathway facilitates piRNA production. Cell. 2019;178:964–979e920.
Ge DT, Wang W, Tipping C, Gainetdinov I, Weng Z, Zamore PD. The RNA-Binding ATPase, Armitage, couples piRNA amplification in Nuage to phased piRNA production on Mitochondria. Mol Cell. 2019;74:982–995e986.
Wang X, Lv C, Guo Y, Yuan S. Mitochondria associated germinal structures in Spermatogenesis: piRNA pathway regulation and beyond. Cells. 2020;9(2):399.
Sato K, Siomi MC. The piRNA pathway in Drosophila ovarian germ and somatic cells. Proc Jpn Acad Ser B Phys Biol Sci. 2020;96:32–42.
Sokolova OA, Ilyin AA, Poltavets AS, Nenasheva VV, Mikhaleva EA, Shevelyov YY, Klenov MS. Yb body assembly on the flamenco piRNA precursor transcripts reduces genic piRNA production. Mol Biol Cell. 2019;30:1544–54.
Goriaux C, Desset S, Renaud Y, Vaury C, Brasset E. Transcriptional properties and splicing of the flamenco piRNA cluster. EMBO Rep. 2014;15:411–8.
Teixeira FK, Okuniewska M, Malone CD, Coux RX, Rio DC, Lehmann R. piRNA-mediated regulation of transposon alternative splicing in the soma and germ line. Nature. 2017;552:268–72.
Munafò M, Lawless VR, Passera A, MacMillan S, Bornelöv S, Haussmann IU, et al. Channel nuclear pore complex subunits are required for transposon silencing in Drosophila. Elife. 2021;10:e66321.
Hirakata S, Ishizu H, Fujita A, Tomoe Y, Siomi MC. Requirements for multivalent Yb body assembly in transposon silencing in Drosophila. EMBO Rep. 2019;20:e47708.
Batki J, Schnabl J, Wang J, Handler D, Andreev VI, Stieger CE, Novatchkova M, Lampersberger L, Kauneckaite K, Xie W, et al. The nascent RNA binding complex SFiNX licenses piRNA-guided heterochromatin formation. Nat Struct Mol Biol. 2019;26:720–31.
Wang X, Zeng C, Liao S, Zhu Z, Zhang J, Tu X, Yao X, Feng X, Guang S, Xu C. Molecular basis for PICS-mediated piRNA biogenesis and cell division. Nat Commun. 2021;12:5595.
Sun YH, Wang RH, Du K, Zhu J, Zheng J, Xie LH, Pereira AA, Zhang C, Ricci EP, Li XZ. Coupled protein synthesis and ribosome-guided piRNA processing on mRNAs. Nat Commun. 2021;12:5970.
Venkei ZG, Choi CP, Feng S, Chen C, Jacobsen SE, Kim JK, Yamashita YM. A kinesin Klp10A mediates cell cycle-dependent shuttling of Piwi between nucleus and nuage. PLoS Genet. 2020;16:e1008648.
Ramat A, Garcia-Silva MR, Jahan C, Naït-Saïdi R, Dufourt J, Garret C, Chartier A, Cremaschi J, Patel V, Decourcelle M, et al. The PIWI protein aubergine recruits eIF3 to activate translation in the germ plasm. Cell Res. 2020;30:421–35.
Blom-Dahl D, Azpiazu N. The Pax protein eyegone (eyg) interacts with the pi-RNA component aubergine (aub) and controls egg chamber development in Drosophila. Dev Biol. 2018;434:267–77.
Murakami R, Sumiyoshi T, Negishi L, Siomi MC. DEAD-box polypeptide 43 facilitates piRNA amplification by actively liberating RNA from Ago3-piRISC. EMBO Rep. 2021;22:e51313.
Vrettos N, Maragkakis M, Alexiou P, Sgourdou P, Ibrahim F, Palmieri D, et al. Modulation of Aub-TDRD interactions elucidates piRNA amplification and germplasm formation. Life Sci Alliance. 2020;4(3):e202000912.
Huang X, Hu H, Webster A, Zou F, Du J, Patel DJ, Sachidanandam R, Toth KF, Aravin AA, Li S. Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex. Nat Commun. 2021;12:4061.
Xie W, Sowemimo I, Hayashi R, Wang J, Burkard TR, Brennecke J, Ameres SL, Patel DJ. Structure-function analysis of microRNA 3’-end trimming by Nibbler. Proc Natl Acad Sci U S A. 2020;117:30370–9.
Nishida KM, Sakakibara K, Iwasaki YW, Yamada H, Murakami R, Murota Y, Kawamura T, Kodama T, Siomi H, Siomi MC. Hierarchical roles of mitochondrial papi and Zucchini in Bombyx germline piRNA biogenesis. Nature. 2018;555:260–4.
Modepalli V, Fridrich A, Agron M, Moran Y. The methyltransferase HEN1 is required in Nematostella vectensis for microRNA and piRNA stability as well as larval metamorphosis. PLoS Genet. 2018;14:e1007590.
Mickute M, Nainyte M, Vasiliauskaite L, Plotnikova A, Masevicius V, Klimašauskas S, Vilkaitis G. Animal Hen1 2’-O-methyltransferases as tools for 3’-terminal functionalization and labelling of single-stranded RNAs. Nucleic Acids Res. 2018;46:e104.
Munafò M, Manelli V, Falconio FA, Sawle A, Kneuss E, Eastwood EL, Seah JWE, Czech B, Hannon GJ. Daedalus and Gasz recruit Armitage to mitochondria, bringing piRNA precursors to the biogenesis machinery. Genes Dev. 2019;33:844–56.
Ishizu H, Kinoshita T, Hirakata S, Komatsuzaki C, Siomi MC. Distinct and collaborative functions of Yb and Armitage in Transposon-Targeting piRNA Biogenesis. Cell Rep. 2019;27:1822–1835e1828.
Chen P, Kotov AA, Godneeva BK, Bazylev SS, Olenina LV, Aravin AA. piRNA-mediated gene regulation and adaptation to sex-specific transposon expression in D. Melanogaster male germline. Genes Dev. 2021;35:914–35.
Rogers AK, Situ K, Perkins EM, Toth KF. Zucchini-dependent piRNA processing is triggered by recruitment to the cytoplasmic processing machinery. Genes Dev. 2017;31:1858–69.
Chavda V, Madhwani K, Chaurasia B. PiWi RNA in Neurodevelopment and neurodegenerative disorders. Curr Mol Pharmacol. 2022;15:517–31.
Cosby RL, Chang NC, Feschotte C. Host-transposon interactions: conflict, cooperation, and cooption. Genes Dev. 2019;33:1098–116.
Mérel V, Gibert P, Buch I, Rodriguez Rada V, Estoup A, Gautier M, Fablet M, Boulesteix M, Vieira C. The Worldwide Invasion of Drosophila suzukii is accompanied by a large increase of transposable element load and a small number of Putatively adaptive insertions. Mol Biol Evol. 2021;38:4252–67.
Teefy BB, Siebert S, Cazet JF, Lin H, Juliano CE. PIWI-piRNA pathway-mediated transposable element repression in Hydra somatic stem cells. RNA. 2020;26:550–63.
Vandewege MW, Platt RN 2nd, Ray DA, Hoffmann FG. Transposable element targeting by piRNAs in Laurasiatherians with distinct transposable element histories. Genome Biol Evol. 2016;8:1327–37.
Watanabe T, Cui X, Yuan Z, Qi H, Lin H. MIWI2 targets RNAs transcribed from piRNA-dependent regions to drive DNA methylation in mouse prospermatogonia. Embo J. 2018;37(18):e95329.
Pandey RR, Homolka D, Olotu O, Sachidanandam R, Kotaja N, Pillai RS. Exonuclease Domain-containing 1 enhances MIWI2 piRNA Biogenesis via its Interaction with TDRD12. Cell Rep. 2018;24:3423–3432e3424.
Wang X, Ramat A, Simonelig M, Liu MF. Emerging roles and functional mechanisms of PIWI-interacting RNAs. Nat Rev Mol Cell Biol. 2023;24:123–41.
Weng W, Li H, Goel A. Piwi-interacting RNAs (piRNAs) and cancer: emerging biological concepts and potential clinical implications. Biochim Biophys Acta Rev Cancer. 2019;1871:160–9.
Onishi R, Yamanaka S, Siomi MC. piRNA- and siRNA-mediated transcriptional repression in Drosophila, mice, and yeast: new insights and biodiversity. EMBO Rep. 2021;22:e53062.
Xu J, Yang X, Zhou Q, Zhuang J, Han S. Biological significance of piRNA in Liver cancer: a review. Biomarkers. 2020;25:436–40.
Zhou Y, Fang Y, Dai C, Wang Y. PiRNA pathway in the cardiovascular system: a novel regulator of cardiac differentiation, repair and regeneration. J Mol Med (Berl). 2021;99:1681–90.
Fu K, Tian S, Tan H, Wang C, Wang H, Wang M, Wang Y, Chen Z, Wang Y, Yue Q, et al. Biological and RNA regulatory function of MOV10 in mammalian germ cells. BMC Biol. 2019;17:39.
Dong P, Xiong Y, Konno Y, Ihira K, Xu D, Kobayashi N, Yue J, Watari H. Critical roles of PIWIL1 in human tumors: expression, functions, mechanisms, and potential clinical implications. Front Cell Dev Biol. 2021;9:656993.
Barucci G, Cornes E, Singh M, Li B, Ugolini M, Samolygo A, Didier C, Dingli F, Loew D, Quarato P, Cecere G. Small-RNA-mediated transgenerational silencing of histone genes impairs fertility in piRNA mutants. Nat Cell Biol. 2020;22:235–45.
Cui P, Xin D, Li F, Deng L, Gao Y. Butorphanol suppresses the Proliferation and Migration of Osteosarcoma by promoting the expression of piRNA hsa_piR_006613. Front Oncol. 2022;12:775132.
Doke M, Kashanchi F, Khan MA, Samikkannu T. HIV-1 Tat and cocaine coexposure impacts piRNAs to affect astrocyte energy metabolism. Epigenomics. 2022;14:261–78.
Yin J, Jiang XY, Qi W, Ji CG, Xie XL, Zhang DX, Cui ZJ, Wang CK, Bai Y, Wang J, Jiang HQ. piR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1. Cancer Sci. 2017;108:1746–56.
Xie J, Xing S, Shen BY, Chen HT, Sun B, Wang ZT, Wang JW, Lu XX. PIWIL1 interacting RNA piR-017061 inhibits Pancreatic cancer growth via regulating EFNA5. Hum Cell. 2021;34:550–63.
Tan L, Mai D, Zhang B, Jiang X, Zhang J, Bai R, Ye Y, Li M, Pan L, Su J, et al. PIWI-interacting RNA-36712 restrains Breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol Cancer. 2019;18:9.
Ma H, Wang H, Tian F, Zhong Y, Liu Z, Liao A. PIWI-Interacting RNA-004800 is regulated by S1P receptor signaling pathway to keep Myeloma Cell Survival. Front Oncol. 2020;10:438.
Jacobs DI, Qin Q, Fu A, Chen Z, Zhou J, Zhu Y. piRNA-8041 is downregulated in human glioblastoma and suppresses Tumor growth in vitro and in vivo. Oncotarget. 2018;9:37616–26.
Huang H, Yu X, Han X, Hao J, Zhao J, Bebek G, Bao S, Prayson RA, Khalil AM, Jankowsky E, Yu JS. Piwil1 regulates glioma stem cell maintenance and Glioblastoma Progression. Cell Rep. 2021;34:108522.
Liu X, Zheng J, Xue Y, Yu H, Gong W, Wang P, Li Z, Liu Y. PIWIL3/OIP5-AS1/miR-367-3p/CEBPA feedback loop regulates the biological behavior of glioma cells. Theranostics. 2018;8:1084–105.
Li D, Luo Y, Gao Y, Yang Y, Wang Y, Xu Y, Tan S, Zhang Y, Duan J, Yang Y. piR-651 promotes Tumor formation in non-small cell lung carcinoma through the upregulation of cyclin D1 and CDK4. Int J Mol Med. 2016;38:927–36.
Zhang SJ, Yao J, Shen BZ, Li GB, Kong SS, Bi DD, Pan SH, Cheng BL. Role of piwi-interacting RNA-651 in the carcinogenesis of non-small cell Lung cancer. Oncol Lett. 2018;15:940–6.
Zhao X, Huang L, Lu Y, Jiang W, Song Y, Qiu B, Tao D, Liu Y, Ma Y. PIWIL2 interacting with IKK to regulate autophagy and apoptosis in esophageal squamous cell carcinoma. Cell Death Differ. 2021;28:1941–54.
Du X, Li H, Xie X, Shi L, Wu F, Li G, et al. piRNA-31115 promotes cell proliferation and invasion via PI3K/AKT pathway in clear cell renal carcinoma. Dis Markers. 2021;2021:6915329.
Zhang L, Meng X, Li D, Han X. piR-001773 and piR-017184 promote Prostate cancer progression by interacting with PCDH9. Cell Signal. 2020;76:109780.
Guo Z, Wang G, Wu B, Chou WC, Cheng L, Zhou C, Lou J, Wu D, Su L, Zheng J, et al. DCAF1 regulates Treg senescence via the ROS axis during immunological aging. J Clin Invest. 2020;130:5893–908.
Peng L, Song L, Liu C, Lv X, Li X, Jie J, Zhao D, Li D. piR-55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling. Tumour Biol. 2016;37:2749–56.
Ding L, Wang R, Xu W, Shen D, Cheng S, Wang H, Lu Z, Zheng Q, Wang L, Xia L, Li G. PIWI-interacting RNA 57125 restrains clear cell renal cell carcinoma Metastasis by downregulating CCL3 expression. Cell Death Discov. 2021;7:333.
Qi T, Cao H, Sun H, Feng H, Li N, Wang C, Wang L. piR-19166 inhibits migration and Metastasis through CTTN/MMPs pathway in prostate carcinoma. Aging. 2020;12:18209–20.
Das B, Jain N, Mallick B. piR-39980 promotes cell proliferation, migration and invasion, and inhibits apoptosis via repression of SERPINB1 in human osteosarcoma. Biol Cell. 2020;112:73–91.
Mai D, Ding P, Tan L, Zhang J, Pan Z, Bai R, Li C, Li M, Zhou Y, Tan W, et al. PIWI-interacting RNA-54265 is oncogenic and a potential therapeutic target in colorectal adenocarcinoma. Theranostics. 2018;8:5213–30.
Yang Y, Zhang X, Song D, Wei J. Piwil2 modulates the invasion and Metastasis of Prostate cancer by regulating the expression of matrix metalloproteinase-9 and epithelial-mesenchymal transitions. Oncol Lett. 2015;10:1735–40.
Li F, Yuan P, Rao M, Jin CH, Tang W, Rong YF, Hu YP, Zhang F, Wei T, Yin Q, et al. piRNA-independent function of PIWIL1 as a co-activator for anaphase promoting complex/cyclosome to drive Pancreatic cancer Metastasis. Nat Cell Biol. 2020;22:425–38.
Li B, Hong J, Hong M, Wang Y, Yu T, Zang S, Wu Q. piRNA-823 delivered by multiple myeloma-derived extracellular vesicles promoted tumorigenesis through re-educating endothelial cells in the Tumor environment. Oncogene. 2019;38:5227–38.
Yan H, Wu QL, Sun CY, Ai LS, Deng J, Zhang L, Chen L, Chu ZB, Tang B, Wang K, et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in Multiple Myeloma. Leukemia. 2015;29:196–206.
Ou B, Liu Y, Gao Z, Xu J, Yan Y, Li Y, Zhang J. Senescent neutrophils-derived exosomal piRNA-17560 promotes chemoresistance and EMT of Breast cancer via FTO-mediated m6A demethylation. Cell Death Dis. 2022;13:905.
Chen Z, Che Q, He X, Wang F, Wang H, Zhu M, Sun J, Wan X. Stem cell protein Piwil1 endowed endometrial cancer cells with stem-like properties via inducing epithelial-mesenchymal transition. BMC Cancer. 2015;15:811.
Li G, Wang X, Li C, Hu S, Niu Z, Sun Q, Sun M. Piwi-Interacting RNA1037 enhances chemoresistance and motility in human oral squamous cell carcinoma cells. Onco Targets Ther. 2019;12:10615–27.
Wang S, Jiang X, Xie X, Yin J, Zhang J, Liu T, Chen S, Wang Y, Zhou X, Wang Y, et al. piR-823 inhibits cell apoptosis via modulating mitophagy by binding to PINK1 in Colorectal cancer. Cell Death Dis. 2022;13:465.
Liu Y, Dong Y, He X, Gong A, Gao J, Hao X, Wang S, Fan Y, Wang Z, Li M, Xu W. Pir-hsa-211106 inhibits the progression of lung adenocarcinoma through pyruvate carboxylase and enhances chemotherapy sensitivity. Front Oncol. 2021;11:651915.
Zou GL, Zhang XR, Ma YL, Lu Q, Zhao R, Zhu YZ, Wang YY. The role of Nrf2/PIWIL2/purine metabolism axis in controlling radiation-induced lung fibrosis. Am J Cancer Res. 2020;10:2752–67.
Wang N, Tan HY, Lu Y, Chan YT, Wang D, Guo W, Xu Y, Zhang C, Chen F, Tang G, Feng Y. PIWIL1 governs the crosstalk of cancer cell metabolism and immunosuppressive microenvironment in hepatocellular carcinoma. Signal Transduct Target Ther. 2021;6:86.
Das B, Jain N, Mallick B. piR-39980 mediates doxorubicin resistance in fibrosarcoma by regulating drug accumulation and DNA repair. Commun Biol. 2021;4:1312.
Das B, Sahoo S, Mallick B. HIWI2 induces G2/M cell cycle arrest and apoptosis in human fibrosarcoma via the ROS/DNA damage/p53 axis. Life Sci. 2022;293:120353.
Liu J, Chen M, Ma L, Dang X, Du G. piRNA-36741 regulates BMP2-mediated osteoblast differentiation via METTL3 controlled m6A modification. Aging. 2021;13:23361–75.
He Z, Jing S, Yang T, Chen J, Huang F, Zhang W, et al. PIWIL4 maintains HIV-1 latency by enforcing epigenetically suppressive modifications on the 5’ long terminal repeat. J Virol. 2020;94(10):e01923–19.
Ma D, Zhou X, Wang Y, Dai L, Yuan J, Peng J, Zhang X, Wang C. Changes in the small noncoding RNAome during M1 and M2 macrophage polarization. Front Immunol. 2022;13:799733.
Jia DD, Jiang H, Zhang YF, Zhang Y, Qian LL, Zhang YF. The regulatory function of piRNA/PIWI complex in cancer and other human Diseases: the role of DNA methylation. Int J Biol Sci. 2022;18:3358–73.
Su JF, Concilla A, Zhang DZ, Zhao F, Shen FF, Zhang H, Zhou FY. PIWI-interacting RNAs: Mitochondria-based biogenesis and functions in cancer. Genes Dis. 2021;8:603–22.
Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392:777–86.
Bell CG, Lowe R, Adams PD, Baccarelli AA, Beck S, Bell JT, Christensen BC, Gladyshev VN, Heijmans BT, Horvath S, et al. DNA methylation aging clocks: challenges and recommendations. Genome Biol. 2019;20:249.
Wong KK. DNMT1: a key drug target in triple-negative Breast cancer. Semin Cancer Biol. 2021;72:198–213.
Couri T, Pillai A. Goals and targets for personalized therapy for HCC. Hepatol Int. 2019;13:125–37.
Su JF, Zhao F, Gao ZW, Hou YJ, Li YY, Duan LJ, Lun SM, Yang HJ, Li JK, Dai NT, et al. piR-823 demonstrates Tumor oncogenic activity in esophageal squamous cell carcinoma through DNA methylation induction via DNA methyltransferase 3B. Pathol Res Pract. 2020;216:152848.
Ding X, Li Y, Lü J, Zhao Q, Guo Y, Lu Z, Ma W, Liu P, Pestell RG, Liang C, Yu Z. piRNA-823 is involved in Cancer Stem Cell Regulation through altering DNA methylation in Association with luminal Breast Cancer. Front Cell Dev Biol. 2021;9:641052.
Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo Tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315:12–7.
Wu D, Fu H, Zhou H, Su J, Zhang F, Shen J. Effects of Novel ncRNA molecules, p15-piRNAs, on the methylation of DNA and histone H3 of the CDKN2B promoter region in U937 cells. J Cell Biochem. 2015;116:2744–54.
Zhang L, Meng X, Pan C, Qu F, Gan W, Xiang Z, Han X, Li D. piR-31470 epigenetically suppresses the expression of glutathione S-transferase pi 1 in Prostate cancer via DNA methylation. Cell Signal. 2020;67:109501.
Liu T, Wang J, Sun L, Li M, He X, Jiang J, Zhou Q. Piwi-interacting RNA-651 promotes cell proliferation and migration and inhibits apoptosis in Breast cancer by facilitating DNMT1-mediated PTEN promoter methylation. Cell Cycle. 2021;20:1603–16.
Ma C, Zhang L, Wang X, He S, Bai J, Li Q, Zhang M, Zhang C, Yu X, Zhang J, et al. piRNA-63076 contributes to pulmonary arterial smooth muscle cell proliferation through acyl-CoA dehydrogenase. J Cell Mol Med. 2020;24:5260–73.
Chen Z, Yang HJ, Lin Q, Zhu MJ, Yu YY, He XY, Wan XP. Estrogen-ERα signaling and DNA hypomethylation co-regulate expression of stem cell protein PIWIL1 in ERα-positive endometrial cancer cells. Cell Commun Signal. 2020;18:84.
Chen Z, Che Q, Jiang FZ, Wang HH, Wang FY, Liao Y, Wan XP. Piwil1 causes epigenetic alteration of PTEN gene via upregulation of DNA methyltransferase in type I endometrial cancer. Biochem Biophys Res Commun. 2015;463:876–80.
Zhan L, Chen M, Pang T, Li X, Long L, Liang D, et al. Attenuation of Piwil2 induced by hypoxic postconditioning prevents cerebral ischemic injury by inhibiting CREB2 promoter methylation. Brain Pathol. 2023;33(1):e13109.
Han H, Fan G, Song S, Jiang Y, Qian C, Zhang W, Su Q, Xue X, Zhuang W, Li B. piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood. 2021;137:1603–14.
Xie Q, Li Z, Luo X, Wang D, Zhou Y, Zhao J, Gao S, Yang Y, Fu W, Kong L, Sun T. piRNA-14633 promotes Cervical cancer cell malignancy in a METTL14-dependent m6A RNA methylation manner. J Transl Med. 2022;20:51.
Gao XQ, Zhang YH, Liu F, Ponnusamy M, Zhao XM, Zhou LY, Zhai M, Liu CY, Li XM, Wang M, et al. The piRNA CHAPIR regulates cardiac hypertrophy by controlling METTL3-dependent N(6)-methyladenosine methylation of Parp10 mRNA. Nat Cell Biol. 2020;22:1319–31.
Lee WP, Chiang MH, Chang LY, Lee JY, Tsai YL, Chiu TH, Chiang HC, Fu TF, Wu T, Wu CL. Mushroom body subsets encode CREB2-dependent water-reward long-term memory in Drosophila. PLoS Genet. 2020;16:e1008963.
Liu SC, Sheu ML, Tsai YC, Lin YC, Chang CW, Lai DW. Attenuation of in vitro and in vivo melanin synthesis using a Chinese herbal medicine through the inhibition of tyrosinase activity. Phytomedicine. 2022;95:153876.
Oerum S, Meynier V, Catala M, Tisné C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021;49:7239–55.
Chen M, Wong CM. The emerging roles of N6-methyladenosine (m6A) deregulation in liver carcinogenesis. Mol Cancer. 2020;19:44.
Tang Y, Chen K, Song B, Ma J, Wu X, Xu Q, Wei Z, Su J, Liu G, Rong R, et al. m6A-Atlas: a comprehensive knowledgebase for unraveling the N6-methyladenosine (m6A) epitranscriptome. Nucleic Acids Res. 2021;49:D134–d143.
Liu ZX, Li LM, Sun HL, Liu SM. Link between m6A modification and cancers. Front Bioeng Biotechnol. 2018;6:89.
Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother. 2019;112:108613.
Zhang H, Shi X, Huang T, Zhao X, Chen W, Gu N, Zhang R. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 2020;48:6251–64.
Zhang B, Jiang H, Dong Z, Sun A, Ge J. The critical roles of m6A modification in metabolic abnormality and Cardiovascular Diseases. Genes Dis. 2021;8:746–58.
Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, Yang C, Chen Y. The role of m6A modification in the biological functions and Diseases. Signal Transduct Target Ther. 2021;6:74.
Cui Y, Zhang C, Ma S, Li Z, Wang W, Li Y, Ma Y, Fang J, Wang Y, Cao W, Guan F. RNA m6A demethylase FTO-mediated epigenetic up-regulation of LINC00022 promotes tumorigenesis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2021;40:294.
Jiang ZX, Wang YN, Li ZY, Dai ZH, He Y, Chu K, Gu JY, Ji YX, Sun NX, Yang F, Li W. The m6A mRNA demethylase FTO in granulosa cells retards FOS-dependent ovarian aging. Cell Death Dis. 2021;12:744.
Yu J, Chai P, Xie M, Ge S, Ruan J, Fan X, Jia R. Histone lactylation drives oncogenesis by facilitating m(6)a reader protein YTHDF2 expression in ocular Melanoma. Genome Biol. 2021;22:85.
Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, Qiu Z, Kidwell RL, Kim LJY, Xie Q, et al. The RNA m6A reader YTHDF2 maintains Oncogene expression and is a targetable dependency in Glioblastoma Stem cells. Cancer Discov. 2021;11:480–99.
Morana O, Wood W, Gregory CD. The apoptosis Paradox in cancer. Int J Mol Sci. 2022;23(3):1328.
Pan R, Ryan J, Pan D, Wucherpfennig KW, Letai A. Augmenting NK cell-based immunotherapy by targeting mitochondrial apoptosis. Cell. 2022;185:1521–1538e1518.
Patra S, Pradhan B, Nayak R, Behera C, Panda KC, Das S, Jena M, Bhutia SK. Apoptosis and autophagy modulating dietary phytochemicals in cancer therapeutics: current evidences and future perspectives. Phytother Res. 2021;35:4194–214.
Jiang M, Qi L, Li L, Wu Y, Song D, Li Y. Caspase-8: a key protein of cross-talk signal way in PANoptosis in cancer. Int J Cancer. 2021;149:1408–20.
Nardone V, Barbarino M, Angrisani A, Correale P, Pastina P, Cappabianca S, et al. CDK4, CDK6/cyclin-D1 complex inhibition and radiotherapy for cancer control: a role for autophagy. Int J Mol Sci. 2021;22(16):8391.
Xie C, Zhou X, Liang C, Li X, Ge M, Chen Y, Yin J, Zhu J, Zhong C. Apatinib triggers autophagic and apoptotic cell death via VEGFR2/STAT3/PD-L1 and ROS/Nrf2/p62 signaling in Lung cancer. J Exp Clin Cancer Res. 2021;40:266.
Schmidt DR, Patel R, Kirsch DG, Lewis CA, Vander Heiden MG, Locasale JW. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J Clin. 2021;71:333–58.
Xue C, Li G, Zheng Q, Gu X, Bao Z, Lu J, Li L. The functional roles of the circRNA/Wnt axis in cancer. Mol Cancer. 2022;21:108.
Xu Y, Zhang M, Zhang Q, Yu X, Sun Z, He Y, Guo W. Role of main RNA methylation in Hepatocellular Carcinoma: N6-Methyladenosine, 5-Methylcytosine, and N1-Methyladenosine. Front Cell Dev Biol. 2021;9:767668.
Xia T, Wu X, Cao M, Zhang P, Shi G, Zhang J, Lu Z, Wu P, Cai B, Miao Y, Jiang K. The RNA m6A methyltransferase METTL3 promotes Pancreatic cancer cell proliferation and invasion. Pathol Res Pract. 2019;215:152666.
Patergnani S, Danese A, Bouhamida E, Aguiari G, Previati M, Pinton P, et al. Various aspects of calcium signaling in the regulation of apoptosis, autophagy, cell proliferation, and cancer. Int J Mol Sci. 2020;21(21):8323.
Vaghari-Tabari M, Ferns GA, Qujeq D, Andevari AN, Sabahi Z, Moein S. Signaling, metabolism, and cancer: an important relationship for therapeutic intervention. J Cell Physiol. 2021;236:5512–32.
Wang J, Cai H, Liu Q, Xia Y, Xing L, Zuo Q, Zhang Y, Chen C, Xu K, Yin P, Chen T. Cinobufacini inhibits Colon Cancer Invasion and Metastasis via suppressing Wnt/β-Catenin signaling pathway and EMT. Am J Chin Med. 2020;48:703–18.
Sun J, Zhang Z, Chen J, Xue M, Pan X. ELTD1 promotes invasion and Metastasis by activating MMP2 in Colorectal cancer. Int J Biol Sci. 2021;17:3048–58.
Liu F, Zhang H, Xie F, Tao D, Xiao X, Huang C, Wang M, Gu C, Zhang X, Jiang G. Hsa_circ_0001361 promotes Bladder cancer invasion and Metastasis through miR-491-5p/MMP9 axis. Oncogene. 2020;39:1696–709.
Moon EJ, Mello SS, Li CG, Chi JT, Thakkar K, Kirkland JG, Lagory EL, Lee IJ, Diep AN, Miao Y, et al. The HIF target MAFF promotes Tumor invasion and Metastasis through IL11 and STAT3 signaling. Nat Commun. 2021;12:4308.
Zhao H, Yan G, Zheng L, Zhou Y, Sheng H, Wu L, Zhang Q, Lei J, Zhang J, Xin R, et al. STIM1 is a metabolic checkpoint regulating the invasion and Metastasis of hepatocellular carcinoma. Theranostics. 2020;10:6483–99.
Yousefi H, Vatanmakanian M, Mahdiannasser M, Mashouri L, Alahari NV, Monjezi MR, Ilbeigi S, Alahari SK. Understanding the role of integrins in Breast cancer invasion, Metastasis, angiogenesis, and drug resistance. Oncogene. 2021;40:1043–63.
Liu D, Kang H, Gao M, Jin L, Zhang F, Chen D, Li M, Xiao L. Exosome-transmitted circ_MMP2 promotes hepatocellular carcinoma Metastasis by upregulating MMP2. Mol Oncol. 2020;14:1365–80.
Maybee DV, Ink NL, Ali MAM. Novel roles of MT1-MMP and MMP-2: beyond the extracellular milieu. Int J Mol Sci. 2022;23(17):9513.
Ramjiawan RR, Griffioen AW, Duda DG. Anti-angiogenesis for cancer revisited: is there a role for combinations with immunotherapy? Angiogenesis. 2017;20:185–204.
Lopes-Coelho F, Martins F, Pereira SA, Serpa J. Anti-angiogenic therapy: current challenges and Future perspectives. Int J Mol Sci. 2021;22(7):3765.
Olejarz W, Kubiak-Tomaszewska G, Chrzanowska A, Lorenc T. Exosomes in angiogenesis and anti-angiogenic therapy in cancers. Int J Mol Sci. 2020;21(16):5840.
Varone E, Decio A, Chernorudskiy A, Minoli L, Brunelli L, Ioli F, Piotti A, Pastorelli R, Fratelli M, Gobbi M, et al. The ER stress response mediator ERO1 triggers cancer Metastasis by favoring the angiogenic switch in hypoxic conditions. Oncogene. 2021;40:1721–36.
Chong ZX, Yeap SK, Ho WY. Unraveling the roles of miRNAs in regulating epithelial-to-mesenchymal transition (EMT) in osteosarcoma. Pharmacol Res. 2021;172:105818.
Pan G, Liu Y, Shang L, Zhou F, Yang S. EMT-associated microRNAs and their roles in cancer stemness and drug resistance. Cancer Commun (Lond). 2021;41:199–217.
Han ML, Zhao YF, Tan CH, Xiong YJ, Wang WJ, Wu F, Fei Y, Wang L, Liang ZQ. Cathepsin L upregulation-induced EMT phenotype is associated with the acquisition of cisplatin or paclitaxel resistance in A549 cells. Acta Pharmacol Sin. 2016;37:1606–22.
Bakir B, Chiarella AM, Pitarresi JR, Rustgi AK. EMT, MET, plasticity, and Tumor Metastasis. Trends Cell Biol. 2020;30:764–76.
Babaei G, Aziz SG, Jaghi NZZ. EMT, cancer stem cells and autophagy; the three main axes of metastasis. Biomed Pharmacother. 2021;133:110909.
Aiello NM, Kang Y. Context-dependent EMT programs in cancer Metastasis. J Exp Med. 2019;216:1016–26.
Yousefzadeh M, Henpita C, Vyas R, Soto-Palma C, Robbins P, Niedernhofer L. DNA damage-how and why we age? Elife. 2021;10:e62852.
Shmulevich R, Krizhanovsky V. Cell senescence, DNA damage, and metabolism. Antioxid Redox Signal. 2021;34:324–34.
Davis CK, Vemuganti R. DNA damage and repair following traumatic brain injury. Neurobiol Dis. 2021;147:105143.
Gillman R, Lopes Floro K, Wankell M, Hebbard L. The role of DNA damage and repair in Liver cancer. Biochim Biophys Acta Rev Cancer. 2021;1875:188493.
Pessina F, Gioia U, Brandi O, Farina S, Ceccon M, Francia S. d’Adda Di Fagagna F: DNA damage triggers a New Phase in Neurodegeneration. Trends Genet. 2021;37:337–54.
Juretschke T, Beli P. Causes and consequences of DNA damage-induced autophagy. Matrix Biol. 2021;100–101:39–53.
Zhan Y, Jiang L, Jin X, Ying S, Wu Z, Wang L, Yu W, Tong J, Zhang L, Lou Y, Qiu Y. Inhibiting RRM2 to enhance the anticancer activity of chemotherapy. Biomed Pharmacother. 2021;133:110996.
Jiang X, Li Y, Zhang N, Gao Y, Han L, Li S, Li J, Liu X, Gong Y, Xie C. RRM2 silencing suppresses malignant phenotype and enhances radiosensitivity via activating cGAS/STING signaling pathway in lung adenocarcinoma. Cell Biosci. 2021;11:74.
Liu Q, Guo L, Qi H, Lou M, Wang R, Hai B, Xu K, Zhu L, Ding Y, Li C, et al. A MYBL2 complex for RRM2 transactivation and the synthetic effect of MYBL2 knockdown with WEE1 inhibition against Colorectal cancer. Cell Death Dis. 2021;12:683.
Yu J, Xia X, Dong Y, Gong Z, Li G, Chen GG, Lai PBS. CYP1A2 suppresses hepatocellular carcinoma through antagonizing HGF/MET signaling. Theranostics. 2021;11:2123–36.
Yu J, Wang N, Gong Z, Liu L, Yang S, Chen GG, Lai PBS. Cytochrome P450 1A2 overcomes nuclear factor kappa B-mediated sorafenib resistance in hepatocellular carcinoma. Oncogene. 2021;40:492–507.
Moloney JN, Cotter TG. ROS signalling in the biology of cancer. Semin Cell Dev Biol. 2018;80:50–64.
Zorov DB, Juhaszova M, Sollott SJ. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev. 2014;94:909–50.
Cheung EC, Vousden KH. The role of ROS in tumour development and progression. Nat Rev Cancer. 2022;22:280–97.
Kiran KR, Deepika VB, Swathy PS, Prasad K, Kabekkodu SP, Murali TS, Satyamoorthy K, Muthusamy A. ROS-dependent DNA damage and repair during germination of NaCl primed seeds. J Photochem Photobiol B. 2020;213:112050.
Hayman TJ, Baro M, MacNeil T, Phoomak C, Aung TN, Cui W, Leach K, Iyer R, Challa S, Sandoval-Schaefer T, et al. STING enhances cell death through regulation of reactive oxygen species and DNA damage. Nat Commun. 2021;12:2327.
Lei X, Du L, Yu W, Wang Y, Ma N, Qu B. GSTP1 as a novel target in radiation induced lung injury. J Transl Med. 2021;19:297.
Cui J, Li G, Yin J, Li L, Tan Y, Wei H, Liu B, Deng L, Tang J, Chen Y, Yi L. GSTP1 and cancer: expression, methylation, polymorphisms and signaling (review). Int J Oncol. 2020;56:867–78.
Judge A, Dodd MS, Metabolism. Essays Biochem. 2020;64:607–47.
Hood DA, Memme JM, Oliveira AN, Triolo M. Maintenance of Skeletal Muscle Mitochondria in Health, Exercise, and aging. Annu Rev Physiol. 2019;81:19–41.
Boyman L, Karbowski M, Lederer WJ. Regulation of mitochondrial ATP production: ca(2+) signaling and Quality Control. Trends Mol Med. 2020;26:21–39.
Burke PJ. Mitochondria, Bioenergetics and apoptosis in Cancer. Trends Cancer. 2017;3:857–70.
Rosenzweig R, Nillegoda NB, Mayer MP, Bukau B. The Hsp70 chaperone network. Nat Rev Mol Cell Biol. 2019;20:665–80.
Brünnert D, Langer C, Zimmermann L, Bargou RC, Burchardt M, Chatterjee M, Stope MB. The heat shock protein 70 inhibitor VER155008 suppresses the expression of HSP27, HOP and HSP90β and the androgen receptor, induces apoptosis, and attenuates Prostate cancer cell growth. J Cell Biochem. 2020;121:407–17.
Lin TY, Hua WJ, Yeh H, Tseng AJ. Functional proteomic analysis reveals that fungal immunomodulatory protein reduced expressions of heat shock proteins correlates to apoptosis in Lung cancer cells. Phytomedicine. 2021;80:153384.
Barazzuol L, Giamogante F, Brini M, Calì T. PINK1/Parkin mediated mitophagy, Ca(2+) signalling, and ER-Mitochondria contacts in Parkinson’s Disease. Int J Mol Sci. 2020;21(5):1772.
Wang SH, Zhu XL, Wang F, Chen SX, Chen ZT, Qiu Q, Liu WH, Wu MX, Deng BQ, Xie Y, et al. LncRNA H19 governs mitophagy and restores mitochondrial respiration in the heart through Pink1/Parkin signaling during obesity. Cell Death Dis. 2021;12:557.
Eiyama A, Okamoto K. PINK1/Parkin-mediated mitophagy in mammalian cells. Curr Opin Cell Biol. 2015;33:95–101.
Yi S, Zheng B, Zhu Y, Cai Y, Sun H, Zhou J. Melatonin ameliorates excessive PINK1/Parkin-mediated mitophagy by enhancing SIRT1 expression in granulosa cells of PCOS. Am J Physiol Endocrinol Metab. 2020;319:E91–e101.
Fu A, van Rooyen L, Evans L, Armstrong N, Avizonis D, Kin T, Bird GH, Reddy A, Chouchani ET, Liesa-Roig M, et al. Glucose metabolism and pyruvate carboxylase enhance glutathione synthesis and restrict oxidative stress in pancreatic islets. Cell Rep. 2021;37:110037.
Kiesel VA, Sheeley MP, Coleman MF, Cotul EK, Donkin SS, Hursting SD, Wendt MK, Teegarden D. Pyruvate carboxylase and cancer progression. Cancer Metab. 2021;9:20.
Liu C, Zhou X, Pan Y, Liu Y, Zhang Y. Pyruvate carboxylase promotes thyroid cancer aggressiveness through fatty acid synthesis. BMC Cancer. 2021;21:722.
Schwörer S, Pavlova NN, Cimino FV, King B, Cai X, Sizemore GM, Thompson CB. Fibroblast pyruvate carboxylase is required for collagen production in the tumour microenvironment. Nat Metab. 2021;3:1484–99.
Lodyga M, Hinz B. TGF-β1 - A truly transforming growth factor in fibrosis and immunity. Semin Cell Dev Biol. 2020;101:123–39.
Gifford CC, Tang J, Costello A, Khakoo NS, Nguyen TQ, Goldschmeding R, Higgins PJ, Samarakoon R. Negative regulators of TGF-β1 signaling in renal fibrosis; pathological mechanisms and novel therapeutic opportunities. Clin Sci (Lond). 2021;135:275–303.
Yanagihara T, Tsubouchi K, Gholiof M, Chong SG, Lipson KE, Zhou Q, Scallan C, Upagupta C, Tikkanen J, Keshavjee S, et al. Connective-tissue growth factor contributes to TGF-β1-induced lung fibrosis. Am J Respir Cell Mol Biol. 2022;66:260–70.
Atkins AJ, Allen AG, Dampier W, Haddad EK, Nonnemacher MR, Wigdahl B. HIV-1 cure strategies: why CRISPR? Expert Opin Biol Ther. 2021;21:781–93.
Xiao T, Cai Y, Chen B. HIV-1 entry and membrane Fusion inhibitors. Viruses. 2021;13(5):735.
Zhang J, Crumpacker C. HIV UTR, LTR, and epigenetic immunity. Viruses. 2022;14(5):1084.
Chen CJ, Chiu ML, Hung CH, Liang WM, Ho MW, Lin TH, Liu X, Tsang H, Liao CC, Huang SM, et al. Effect of Xanthium Strumarium on HIV-1 5’-LTR transcriptional activity and viral reactivation in latently infected cells. Front Pharmacol. 2021;12:720821.
Griffin GK, Wu J, Iracheta-Vellve A, Patti JC, Hsu J, Davis T, Dele-Oni D, Du PP, Halawi AG, Ishizuka JJ, et al. Epigenetic silencing by SETDB1 suppresses tumour intrinsic immunogenicity. Nature. 2021;595:309–14.
Myers KV, Amend SR, Pienta KJ. Targeting Tyro3, Axl and MerTK (TAM receptors): implications for macrophages in the Tumor microenvironment. Mol Cancer. 2019;18:94.
Aehnlich P, Powell RM, Peeters MJW, Rahbech A, Thor Straten P. TAM receptor inhibition-implications for cancer and the immune system. Cancers (Basel). 2021;13(6):1195.
Xia Y, Rao L, Yao H, Wang Z, Ning P, Chen X. Engineering macrophages for Cancer Immunotherapy and Drug Delivery. Adv Mater. 2020;32:e2002054.
Wang S, Li F, Fan H, Xu J, Hu Z. Expression of PIWIL2 in Oral cancer and leukoplakia: prognostic implications and insights from tumors. Cancer Biomark. 2019;26:11–20.
Zhou S, Yang S, Li F, Hou J, Chang H. P-element Induced WImpy protein-like RNA-mediated gene silencing 2 regulates Tumor cell progression, apoptosis, and Metastasis in oral squamous cell carcinoma. J Int Med Res. 2021;49:3000605211053158.
Fang N, Shi Y, Fan Y, Long T, Shu Y, Zhou J. Circ_0072088 Promotes Proliferation, Migration, and Invasion of Esophageal Squamous Cell Cancer by Absorbing miR-377. J Oncol 2020, 2020:8967126.
Shi S, Yang ZZ, Liu S, Yang F, Lin H. PIWIL1 promotes gastric cancer via a piRNA-independent mechanism. Proc Natl Acad Sci U S A. 2020;117(36):22390–401.
Wang Z, Yang H, Ma D, Mu Y, Tan X, Hao Q, Feng L, Liang J, Xin W, Chen Y, et al. Serum PIWI-Interacting RNAs piR-020619 and piR-020450 are promising novel biomarkers for early detection of Colorectal Cancer. Cancer Epidemiol Biomarkers Prev. 2020;29:990–8.
Mai D, Zheng Y, Guo H, Ding P, Bai R, Li M, Ye Y, Zhang J, Huang X, Liu D, et al. Serum piRNA-54265 is a New Biomarker for early detection and clinical surveillance of human Colorectal Cancer. Theranostics. 2020;10:8468–78.
Iyer DN, Wan TM, Man JH, Sin RW, Li X, Lo OS, et al. Small RNA profiling of piRNAs in Colorectal cancer identifies consistent overexpression of piR-24000 that correlates clinically with an aggressive disease phenotype. Cancers (Basel). 2020;12(1):188.
Sabbah NA, Abdalla WM, Mawla WA, AbdAlMonem N, Gharib AF, Abdul-Saboor A, et al. piRNA-823 is a unique potential diagnostic non-invasive biomarker in Colorectal Cancer patients. Genes (Basel). 2021;12(4):598.
Jiang H, Liu H, Jiang B. Long non-coding RNA FALEC promotes Colorectal cancer progression via regulating miR-2116-3p-targeted PIWIL1. Cancer Biol Ther. 2020;21:1025–32.
Sellitto A, Geles K, D’Agostino Y, Conte M, Alexandrova E, Rocco D, et al. Molecular and functional characterization of the somatic PIWIL1/piRNA pathway in Colorectal cancer cells. Cells. 2019;8(11):1390.
Zou W, Wang Z, Zhang X, Xu S, Wang F, Li L, Deng Z, Wang J, Pan K, Ge X, et al. PIWIL4 and SUPT5H combine to predict prognosis and immune landscape in intrahepatic cholangiocarcinoma. Cancer Cell Int. 2021;21:657.
Gu X, Wang C, Deng H, Qing C, Liu R, Liu S, Xue X. Exosomal piRNA profiling revealed unique circulating piRNA signatures of cholangiocarcinoma and gallbladder carcinoma. Acta Biochim Biophys Sin (Shanghai). 2020;52:475–84.
Li W, Martinez-Useros J, Garcia-Carbonero N, Fernandez-Aceñero MJ, Ortega-Medina L, Garcia-Botella S, et al. The prognosis value of PIWIL1 and PIWIL2 expression in Pancreatic cancer. J Clin Med. 2019;8(9):1275.
Li W, Martinez-Useros J, Garcia-Carbonero N, Fernandez-Aceñero MJ, Orta A, Ortega-Medina L, et al. The clinical significance of PIWIL3 and PIWIL4 expression in Pancreatic cancer. J Clin Med. 2020;9(5):1252.
Haffner MC, Zwart W, Roudier MP, True LD, Nelson WG, Epstein JI, De Marzo AM, Nelson PS, Yegnasubramanian S. Genomic and phenotypic heterogeneity in Prostate cancer. Nat Rev Urol. 2021;18:79–92.
Chu H, Hui G, Yuan L, Shi D, Wang Y, Du M, Zhong D, Ma L, Tong N, Qin C, et al. Identification of novel piRNAs in Bladder cancer. Cancer Lett. 2015;356:561–7.
Thorat MA, Balasubramanian R. Breast cancer prevention in high-risk women. Best Pract Res Clin Obstet Gynaecol. 2020;65:18–31.
Feng D, Yan K, Zhou Y, Liang H, Liang J, Zhao W, Dong Z, Ling B. Piwil2 is reactivated by HPV oncoproteins and initiates cell reprogramming via epigenetic regulation during Cervical cancer tumorigenesis. Oncotarget. 2016;7:64575–88.
Kuroki L, Guntupalli SR. Treatment of epithelial Ovarian cancer. BMJ. 2020;371:m3773.
Makker V, MacKay H, Ray-Coquard I, Levine DA, Westin SN, Aoki D, Oaknin A. Endometrial cancer. Nat Rev Dis Primers. 2021;7:88.
Wang HW, Balakrishna JP, Pittaluga S, Jaffe ES. Diagnosis of Hodgkin Lymphoma in the modern era. Br J Haematol. 2019;184:45–59.
McKinnon C, Nandhabalan M, Murray SA, Plaha P. Glioblastoma: clinical presentation, diagnosis, and management. BMJ. 2021;374:n1560.
Jonasch E, Walker CL, Rathmell WK. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nat Rev Nephrol. 2021;17:245–61.
Reeves ME, Firek M, Jliedi A, Amaar YG. Identification and characterization of RASSF1C piRNA target genes in Lung cancer cells. Oncotarget. 2017;8:34268–82.
Shen S, Yu H, Liu X, Liu Y, Zheng J, Wang P, Gong W, Chen J, Zhao L, Xue Y. PIWIL1/piRNA-DQ593109 regulates the permeability of the blood-tumor barrier via the MEG3/miR-330-5p/RUNX3 Axis. Mol Ther Nucleic Acids. 2018;10:412–25.
Zafar A, Wang W, Liu G, Wang X, Xian W, McKeon F, Foster J, Zhou J, Zhang R. Molecular targeting therapies for neuroblastoma: Progress and challenges. Med Res Rev. 2021;41:961–1021.
Chung C, Boterberg T, Lucas J, Panoff J, Valteau-Couanet D, Hero B, Bagatell R. Hill-Kayser CE: Neuroblastoma. Pediatr Blood Cancer. 2021;68(Suppl 2):e28473.
Nourmohammadi E, Khoshdel-Sarkarizi H, Nedaeinia R, Darroudi M, Kazemi Oskuee R. Cerium oxide nanoparticles: a promising tool for the treatment of fibrosarcoma in-vivo. Mater Sci Eng C Mater Biol Appl. 2020;109:110533.
Khan S, Barve KH, Kumar MS. Recent advancements in Pathogenesis, Diagnostics and Treatment of Alzheimer’s Disease. Curr Neuropharmacol. 2020;18:1106–25.
Norris SP, Likanje MN, Andrews JA. Amyotrophic Lateral Sclerosis: update on clinical management. Curr Opin Neurol. 2020;33:641–8.
Bonney KM, Luthringer DJ, Kim SA, Garg NJ, Engman DM. Pathology and Pathogenesis of Chagas Heart Disease. Annu Rev Pathol. 2019;14:421–47.
Mattia MMC, Fernandes ACP, Genro VK, de Souza CAB, da Rocha Olsen P, Cunha-Filho JS. PIWIL2 is overexpressed in adenomyotic lesions of women with diffuse adenomyosis. Arch Gynecol Obstet. 2020;302:925–33.
Gong FF, Vaitenas I, Malaisrie SC, Maganti K. Mechanical Complications of Acute Myocardial Infarction: a review. JAMA Cardiol. 2021;6:341–9.
Wander GS, Bansal M, Kasliwal RR. Prediction and early detection of Cardiovascular Disease in South asians with Diabetes Mellitus. Diabetes Metab Syndr. 2020;14:385–93.
Vonk MC, Vandecasteele E, van Dijk AP. Pulmonary Hypertension in connective tissue Diseases, new evidence and challenges. Eur J Clin Invest. 2021;51:e13453.
McLellan MA, Skelly DA, Dona MSI, Squiers GT, Farrugia GE, Gaynor TL, Cohen CD, Pandey R, Diep H, Vinh A, et al. High-resolution transcriptomic profiling of the heart during chronic stress reveals Cellular drivers of Cardiac Fibrosis and Hypertrophy. Circulation. 2020;142:1448–63.
Stasiewicz M, Karpiński TM. The oral microbiota and its role in carcinogenesis. Semin Cancer Biol. 2022;86:633–42.
Gangwar SK, Kumar A, Jose S, Alqahtani MS, Abbas M, Sethi G, Kunnumakkara AB. Nuclear receptors in oral cancer-emerging players in tumorigenesis. Cancer Lett. 2022;536:215666.
Rogers JE, Sewastjanow-Silva M, Waters RE, Ajani JA. Esophageal cancer: emerging therapeutics. Expert Opin Ther Targets. 2022;26:107–17.
Yamamoto S, Kato K. Pembrolizumab for the treatment of Esophageal cancer. Expert Opin Biol Ther. 2020;20:1143–50.
Yang T, Hui R, Nouws J, Sauler M, Zeng T, Wu Q. Untargeted metabolomics analysis of esophageal squamous cell cancer progression. J Transl Med. 2022;20:127.
Han H, Yang C, Ma J, Zhang S, Zheng S, Ling R, Sun K, Guo S, Huang B, Liang Y, et al. N(7)-methylguanosine tRNA modification promotes esophageal squamous cell carcinoma tumorigenesis via the RPTOR/ULK1/autophagy axis. Nat Commun. 2022;13:1478.
Thrift AP, El-Serag HB. Burden of gastric Cancer. Clin Gastroenterol Hepatol. 2020;18:534–42.
Sexton RE, Al Hallak MN, Diab M, Azmi AS. Gastric cancer: a comprehensive review of current and future treatment strategies. Cancer Metastasis Rev. 2020;39:1179–203.
Shaukat A, Kahi CJ, Burke CA, Rabeneck L, Sauer BG, Rex DK. ACG clinical guidelines: Colorectal Cancer Screening 2021. Am J Gastroenterol. 2021;116:458–79.
Basak D, Uddin MN, Hancock J. The role of oxidative stress and its counteractive utility in Colorectal Cancer (CRC). Cancers (Basel). 2020;12(11):3336.
Gausman V, Dornblaser D, Anand S, Hayes RB, O’Connell K, Du M, Liang PS. Risk factors Associated with Early-Onset Colorectal Cancer. Clin Gastroenterol Hepatol. 2020;18:2752–2759e2752.
Si H, Yang Q, Hu H, Ding C, Wang H, Lin X. Colorectal cancer occurrence and treatment based on changes in intestinal flora. Semin Cancer Biol. 2021;70:3–10.
Lu C, Schardey J, Zhang T, Crispin A, Wirth U, Karcz KW, Bazhin AV, Andrassy J, Werner J, Kühn F. Survival outcomes and clinicopathological features in inflammatory bowel disease-associated Colorectal Cancer: a systematic review and Meta-analysis. Ann Surg. 2022;276:e319–30.
Xu S, Yin W, Zhang Y, Lv Q, Yang Y, He J. Foes or friends? Bacteria enriched in the tumor microenvironment of Colorectal cancer. Cancers (Basel). 2020;12(2):372.
Chandana S, Babiker HM, Mahadevan D. Therapeutic trends in pancreatic ductal adenocarcinoma (PDAC). Expert Opin Investig Drugs. 2019;28:161–77.
Malinova A, Veghini L, Real FX, Corbo V. Cell lineage infidelity in PDAC progression and Therapy Resistance. Front Cell Dev Biol. 2021;9:795251.
Panchal K, Sahoo RK, Gupta U, Chaurasiya A. Role of targeted immunotherapy for pancreatic ductal adenocarcinoma (PDAC) treatment: an overview. Int Immunopharmacol. 2021;95:107508.
Koikawa K, Kibe S, Suizu F, Sekino N, Kim N, Manz TD, Pinch BJ, Akshinthala D, Verma A, Gaglia G, et al. Targeting Pin1 renders Pancreatic cancer eradicable by synergizing with immunochemotherapy. Cell. 2021;184:4753–4771e4727.
Freed-Pastor WA, Lambert LJ, Ely ZA, Pattada NB, Bhutkar A, Eng G, Mercer KL, Garcia AP, Lin L, Rideout WM, et al. The CD155/TIGIT axis promotes and maintains immune evasion in neoantigen-expressing Pancreatic cancer. Cancer Cell. 2021;39:1342–1360e1314. 3rd.
Huang C, Iovanna J, Santofimia-Castaño P. Targeting fibrosis: the bridge that connects Pancreatitis and Pancreatic cancer. Int J Mol Sci. 2021;22(9):4970.
Yang X, Chen J, Wang J, Ma S, Feng W, Wu Z, Guo Y, Zhou H, Mi W, Chen W, et al. Very-low-density lipoprotein receptor-enhanced lipid metabolism in pancreatic stellate cells promotes pancreatic fibrosis. Immunity. 2022;55:1185–1199e1188.
Singh VK, Yadav D, Garg PK. Diagnosis and management of chronic Pancreatitis: a review. JAMA. 2019;322:2422–34.
Xue R, Zhou J, Wu J, Meng Q, Gong J, Shen L. P-element-Induced Wimpy-Testis-Like Protein 1 regulates the activation of Pancreatic stellate cells through the PI3K/AKT/mTOR signaling pathway. Dig Dis Sci. 2023;68:1339–50.
Kelley RK, Bridgewater J, Gores GJ, Zhu AX. Systemic therapies for intrahepatic cholangiocarcinoma. J Hepatol. 2020;72:353–63.
Mejia JC, Pasko J. Primary liver cancers: Intrahepatic Cholangiocarcinoma and Hepatocellular Carcinoma. Surg Clin North Am. 2020;100:535–49.
Chen Q, Wang H, Li Z, Li F, Liang L, Zou Y, Shen H, Li J, Xia Y, Cheng Z, et al. Circular RNA ACTN4 promotes intrahepatic cholangiocarcinoma progression by recruiting YBX1 to initiate FZD7 transcription. J Hepatol. 2022;76:135–47.
Chalasani NP, Ramasubramanian TS, Bhattacharya A, Olson MC, Edwards VD, Roberts LR, Kisiel JB, Reddy KR, Lidgard GP, Johnson SC, Bruinsma JJ. A novel blood-based panel of methylated DNA and Protein Markers for Detection of Early-Stage Hepatocellular Carcinoma. Clin Gastroenterol Hepatol. 2021;19:2597–2605e2594.
Nooreldeen R, Bach H. Current and future development in lung cancer diagnosis. Int J Mol Sci. 2021;22(16):8661.
Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398:535–54.
Bade BC, Dela Cruz CS. Lung Cancer 2020: epidemiology, etiology, and Prevention. Clin Chest Med. 2020;41:1–24.
Vinod SK, Hau E. Radiotherapy treatment for Lung cancer: current status and future directions. Respirology. 2020;25(Suppl 2):61–71.
Thakur SK, Singh DP, Choudhary J. Lung cancer identification: a review on detection and classification. Cancer Metastasis Rev. 2020;39:989–98.
Daugaard I, Venø MT, Yan Y, Kjeldsen TE, Lamy P, Hager H, Kjems J, Hansen LL. Small RNA sequencing reveals metastasis-related microRNAs in lung adenocarcinoma. Oncotarget. 2017;8:27047–61.
Wettersten HI, Aboud OA, Lara PN Jr., Weiss RH. Metabolic reprogramming in clear cell renal cell carcinoma. Nat Rev Nephrol. 2017;13:410–9.
Wolf MM, Kimryn Rathmell W, Beckermann KE. Modeling clear cell renal cell carcinoma and therapeutic implications. Oncogene. 2020;39:3413–26.
Rysz J, Franczyk B, Ławiński J, Gluba-Brzózka A. Characteristics of Clear Cell Papillary Renal Cell Carcinoma (ccpRCC). Int J Mol Sci. 2021;23(1):151.
Makhov P, Joshi S, Ghatalia P, Kutikov A, Uzzo RG, Kolenko VM. Resistance to systemic therapies in Clear Cell Renal Cell Carcinoma: mechanisms and management strategies. Mol Cancer Ther. 2018;17:1355–64.
Sharma R, Kadife E, Myers M, Kannourakis G, Prithviraj P, Ahmed N. Determinants of resistance to VEGF-TKI and immune checkpoint inhibitors in metastatic renal cell carcinoma. J Exp Clin Cancer Res. 2021;40:186.
Busch J, Ralla B, Jung M, Wotschofsky Z, Trujillo-Arribas E, Schwabe P, Kilic E, Fendler A, Jung K. Piwi-interacting RNAs as novel prognostic markers in clear cell renal cell carcinomas. J Exp Clin Cancer Res. 2015;34:61.
Desai K, McManus JM, Sharifi N. Hormonal therapy for Prostate Cancer. Endocr Rev. 2021;42:354–73.
Lenis AT, Lec PM, Chamie K, Mshs MD. Bladder Cancer: a review. JAMA. 2020;324:1980–91.
Patel VG, Oh WK, Galsky MD. Treatment of muscle-invasive and advanced Bladder cancer in 2020. CA Cancer J Clin. 2020;70:404–23.
Richters A, Aben KKH, Kiemeney L. The global burden of urinary Bladder cancer: an update. World J Urol. 2020;38:1895–904.
Sabo AA, Birolo G, Naccarati A, Dragomir MP, Aneli S, Allione A, et al. Small non-coding RNA profiling in plasma Extracellular vesicles of Bladder Cancer patients by Next-Generation sequencing: expression levels of mir-126-3p and piR-5936 increase with higher histologic grades. Cancers (Basel). 2020;12(6):1507.
Barzaman K, Karami J, Zarei Z, Hosseinzadeh A, Kazemi MH, Moradi-Kalbolandi S, Safari E, Farahmand L. Breast cancer: Biology, biomarkers, and treatments. Int Immunopharmacol. 2020;84:106535.
Tray N, Taff J, Adams S. Therapeutic landscape of metaplastic Breast cancer. Cancer Treat Rev. 2019;79:101888.
Gradishar WJ, Anderson BO, Abraham J, Aft R, Agnese D, Allison KH, Blair SL, Burstein HJ, Dang C, Elias AD, et al. Breast Cancer, Version 3.2020, NCCN Clinical Practice guidelines in Oncology. J Natl Compr Canc Netw. 2020;18:452–78.
Johnson CA, James D, Marzan A, Armaos M. Cervical Cancer: an overview of pathophysiology and management. Semin Oncol Nurs. 2019;35:166–74.
Bedell SL, Goldstein LS, Goldstein AR, Goldstein AT. Cervical Cancer screening: past, Present, and Future. Sex Med Rev. 2020;8:28–37.
Feng CH, Mell LK, Sharabi AB, McHale M, Mayadev JS. Immunotherapy with Radiotherapy and Chemoradiotherapy for Cervical Cancer. Semin Radiat Oncol. 2020;30:273–80.
Feng D, Yan K, Liang H, Liang J, Wang W, Yu H, Zhou Y, Zhao W, Dong Z, Ling B. CBP-mediated Wnt3a/β-catenin signaling promotes cervical oncogenesis initiated by Piwil2. Neoplasia. 2021;23:1–11.
Barani M, Bilal M, Sabir F, Rahdar A, Kyzas GZ. Nanotechnology in Ovarian cancer: diagnosis and treatment. Life Sci. 2021;266:118914.
Sipos A, Ujlaki G, Mikó E, Maka E, Szabó J, Uray K, Krasznai Z, Bai P. The role of the microbiome in Ovarian cancer: mechanistic insights into oncobiosis and to bacterial metabolite signaling. Mol Med. 2021;27:33.
Gong G, Lin T, Yuan Y. Integrated analysis of gene expression and DNA methylation profiles in Ovarian cancer. J Ovarian Res. 2020;13:30.
Kim SI, Kim JW. Role of Surgery and hyperthermic intraperitoneal chemotherapy in Ovarian cancer. ESMO Open. 2021;6:100149.
Singh G, Roy J, Rout P, Mallick B. Genome-wide profiling of the PIWI-interacting RNA-mRNA regulatory networks in epithelial ovarian cancers. PLoS ONE. 2018;13:e0190485.
Brooks RA, Fleming GF, Lastra RR, Lee NK, Moroney JW, Son CH, Tatebe K, Veneris JL. Current recommendations and recent progress in endometrial cancer. CA Cancer J Clin. 2019;69:258–79.
Lu KH, Broaddus RR. Endometrial Cancer. N Engl J Med. 2020;383:2053–64.
Passarello K, Kurian S, Villanueva V. Endometrial Cancer: an overview of Pathophysiology, Management, and Care. Semin Oncol Nurs. 2019;35:157–65.
Holstein SA, Lunning MA. CAR T-Cell therapy in hematologic malignancies: a voyage in Progress. Clin Pharmacol Ther. 2020;107:112–22.
El-Jawahri A, Nelson AM, Gray TF, Lee SJ, LeBlanc TW. Palliative and End-of-life care for patients with hematologic malignancies. J Clin Oncol. 2020;38:944–53.
Wei AH, Roberts AW, Spencer A, Rosenberg AS, Siegel D, Walter RB, Caenepeel S, Hughes P, McIver Z, Mezzi K, et al. Targeting MCL-1 in hematologic malignancies: Rationale and progress. Blood Rev. 2020;44:100672.
Seth R, Singh A. Leukemias in Children. Indian J Pediatr. 2015;82:817–24.
Miao Y, Medeiros LJ, Li Y, Li J, Young KH. Genetic alterations and their clinical implications in DLBCL. Nat Rev Clin Oncol. 2019;16:634–52.
Li S, Young KH, Medeiros LJ. Diffuse large B-cell Lymphoma. Pathology. 2018;50:74–87.
Liu Y, Barta SK. Diffuse large B-cell Lymphoma: 2019 update on diagnosis, risk stratification, and treatment. Am J Hematol. 2019;94:604–16.
van de Donk N, Pawlyn C, Yong KL. Multiple Myeloma. Lancet. 2021;397:410–27.
Gu W, Qu R, Meng F, Cornelissen J, Zhong Z. Polymeric nanomedicines targeting hematological malignancies. J Control Release. 2021;337:571–88.
Brice P, de Kerviler E, Friedberg JW. Classical Hodgkin Lymphoma. Lancet. 2021;398:1518–27.
Cordeiro A, Navarro A, Gaya A, Díaz-Beyá M, Gonzalez-Farré B, Castellano JJ, Fuster D, Martínez C, Martínez A, Monzó M. PiwiRNA-651 as marker of treatment response and survival in classical Hodgkin Lymphoma. Oncotarget. 2016;7:46002–13.
Le Rhun E, Preusser M, Roth P, Reardon DA, van den Bent M, Wen P, Reifenberger G, Weller M. Molecular targeted therapy of glioblastoma. Cancer Treat Rev. 2019;80:101896.
Venkataramani V, Yang Y, Schubert MC, Reyhan E, Tetzlaff SK, Wißmann N, Botz M, Soyka SJ, Beretta CA, Pramatarov RL, et al. Glioblastoma hijacks neuronal mechanisms for brain invasion. Cell. 2022;185:2899–2917e2831.
Bartos M, Siegl F, Kopkova A, Radova L, Oppelt J, Vecera M, Kazda T, Jancalek R, Hendrych M, Hermanova M, et al. Small RNA sequencing identifies PIWI-Interacting RNAs deregulated in glioblastoma-piR-9491 and piR-12488 reduce Tumor Cell colonies in Vitro. Front Oncol. 2021;11:707017.
Zhou H, Zhang Y, Lai Y, Xu C, Cheng Y. Circ_101064 regulates the proliferation, invasion and migration of glioma cells through miR-154-5p/ PIWIL1 axis. Biochem Biophys Res Commun. 2020;523:608–14.
Irwin MS, Naranjo A, Zhang FF, Cohn SL, London WB, Gastier-Foster JM, Ramirez NC, Pfau R, Reshmi S, Wagner E, et al. Revised neuroblastoma risk classification system: a Report from the children’s Oncology Group. J Clin Oncol. 2021;39:3229–41.
Ponzoni M, Bachetti T, Corrias MV, Brignole C, Pastorino F, Calarco E, Bensa V, Giusto E, Ceccherini I, Perri P. Recent advances in the developmental origin of neuroblastoma: an overview. J Exp Clin Cancer Res. 2022;41:92.
Roy J, Das B, Jain N, Mallick B. PIWI-interacting RNA 39980 promotes Tumor progression and reduces drug sensitivity in neuroblastoma cells. J Cell Physiol. 2020;235:2286–99.
Bernardo B, Joaquim S, Garren J, Boucher M, Houle C, LaCarubba B, Qiao S, Wu Z, Esquejo RM, Peloquin M, et al. Characterization of cachexia in the human fibrosarcoma HT-1080 mouse tumour model. J Cachexia Sarcopenia Muscle. 2020;11:1813–29.
Khan MA, Aljarbou AN, Aldebasi YH, Alorainy MS, Khan A. Combination of glycosphingosomes and liposomal doxorubicin shows increased activity against dimethyl-α-benzanthracene-induced fibrosarcoma in mice. Int J Nanomedicine. 2015;10:6331–8.
Gill J, Gorlick R. Advancing therapy for osteosarcoma. Nat Rev Clin Oncol. 2021;18:609–24.
Eaton BR, Schwarz R, Vatner R, Yeh B, Claude L, Indelicato DJ, Laack N. Osteosarcoma. Pediatr Blood Cancer. 2021;68(Suppl 2):e28352.
Yang C, Tian Y, Zhao F, Chen Z, Su P, Li Y, et al. Bone microenvironment and osteosarcoma metastasis. Int J Mol Sci. 2020;21(19):6985.
Lilienthal I, Herold N. Targeting molecular mechanisms underlying treatment efficacy and resistance in osteosarcoma: a review of current and future strategies. Int J Mol Sci. 2020;21(18):6885.
Chang Z, Ji G, Huang R, Chen H, Gao Y, Wang W, Sun X, Zhang J, Zheng J, Wei Q. PIWI-interacting RNAs piR-13643 and piR-21238 are promising diagnostic biomarkers of papillary thyroid carcinoma. Aging. 2020;12:9292–310.
Breijyeh Z, Karaman R. Comprehensive review on Alzheimer’s Disease: causes and treatment. Molecules. 2020;25(24):5789.
Roy J, Sarkar A, Parida S, Ghosh Z, Mallick B. Small RNA sequencing revealed dysregulated piRNAs in Alzheimer’s Disease and their probable role in pathogenesis. Mol Biosyst. 2017;13:565–76.
Feldman EL, Goutman SA, Petri S, Mazzini L, Savelieff MG, Shaw PJ, Sobue G. Amyotrophic Lateral Sclerosis. Lancet. 2022;400:1363–80.
Goutman SA, Hardiman O, Al-Chalabi A, Chió A, Savelieff MG, Kiernan MC, Feldman EL. Recent advances in the diagnosis and prognosis of Amyotrophic Lateral Sclerosis. Lancet Neurol. 2022;21:480–93.
Sohn EJ, Jo YR, Park HT. Downregulation MIWI-piRNA regulates the migration of Schwann cells in peripheral nerve injury. Biochem Biophys Res Commun. 2019;519:605–12.
Abdelhamid RF, Ogawa K, Beck G, Ikenaka K, Takeuchi E, Yasumizu Y, Jinno J, Kimura Y, Baba K, Nagai Y, et al. piRNA/PIWI Protein Complex as a potential biomarker in sporadic Amyotrophic Lateral Sclerosis. Mol Neurobiol. 2022;59:1693–705.
Echavarría NG, Echeverría LE, Stewart M, Gallego C, Saldarriaga C. Chagas Disease: chronic Chagas Cardiomyopathy. Curr Probl Cardiol. 2021;46:100507.
Choudhuri S, Garg NJ. Trypanosoma cruzi Induces the PARP1/AP-1 pathway for upregulation of metalloproteinases and transforming growth factor β in macrophages: role in cardiac fibroblast differentiation and fibrosis in Chagas Disease. mBio. 2020;11(6):e01853–20.
P MF, L MN, O CM MGL, Pereira MC, de Mendonça-Lima L, T CA-J MCW. Inhibition of TGF-β pathway reverts extracellular matrix remodeling in T. cruzi-infected cardiac spheroids. Exp Cell Res. 2018;362:260–7.
Ferreira RR, Waghabi MC, Bailly S, Feige JJ, Hasslocher-Moreno AM, Saraiva RM, Araujo-Jorge TC. The search for biomarkers and treatments in Chagas Disease: insights from TGF-Beta studies and immunogenetics. Front Cell Infect Microbiol. 2021;11:767576.
Rayford KJ, Cooley A, Arun A, Rachakonda G, Kleschenko Y, Villalta F, et al. Trypanosoma Cruzi modulates PIWI-Interacting RNA expression in primary human cardiac myocytes during the early phase of Infection. Int J Mol Sci. 2020;21(24):9439.
Cluntun AA, Badolia R, Lettlova S, Parnell KM, Shankar TS, Diakos NA, Olson KA, Taleb I, Tatum SM, Berg JA, et al. The pyruvate-lactate axis modulates cardiac hypertrophy and Heart Failure. Cell Metab. 2021;33:629–648e610.
Rajan KS, Velmurugan G, Gopal P, Ramprasath T, Babu DD, Krithika S, Jenifer YC, Freddy A, William GJ, Kalpana K, Ramasamy S. Abundant and altered expression of PIWI-Interacting RNAs during Cardiac Hypertrophy. Heart Lung Circ. 2016;25:1013–20.
Poch D, Mandel J. Pulmonary Hypertension. Ann Intern Med. 2021;174:Itc49–itc64.
Haque A, Kiely DG, Kovacs G, Thompson AAR, Condliffe R. Pulmonary hypertension phenotypes in patients with systemic sclerosis. Eur Respir Rev. 2021;30(161):210053.
Brown S, Banfill K, Aznar MC, Whitehurst P, Faivre Finn C. The evolving role of radiotherapy in non-small cell Lung cancer. Br J Radiol. 2019;92:20190524.
Yang WC, Hsu FM, Yang PC. Precision radiotherapy for non-small cell Lung cancer. J Biomed Sci. 2020;27:82.
Corradini S, Krug D, Meattini I, Matuschek C, Bölke E, Francolini G, Baumann R, Figlia V, Pazos M, Tonetto F, et al. Preoperative radiotherapy: a paradigm shift in the treatment of Breast cancer? A review of literature. Crit Rev Oncol Hematol. 2019;141:102–11.
Hanania AN, Mainwaring W, Ghebre YT, Hanania NA, Ludwig M. Radiation-Induced Lung Injury: Assessment and Management. Chest. 2019;156:150–62.
Türkkan G, Willems Y, Hendriks LEL, Mostard R, Conemans L, Gietema HA, Mitea C, Peeters S, De Ruysscher D. Idiopathic Pulmonary Fibrosis: current knowledge, future perspectives and its importance in radiation oncology. Radiother Oncol. 2021;155:269–77.
Saini V, Guada L, Yavagal DR. Global Epidemiology of Stroke and Access to acute ischemic Stroke interventions. Neurology. 2021;97:6–s16.
Simonsen CZ, Leslie-Mazwi TM, Thomalla G. Which Imaging Approach should be used for Stroke of unknown time of Onset? Stroke. 2021;52:373–80.
Lioutas VA, Ivan CS, Himali JJ, Aparicio HJ, Leveille T, Romero JR, Beiser AS, Seshadri S. Incidence of transient ischemic Attack and Association with Long-Term risk of Stroke. JAMA. 2021;325:373–81.
Roaldsen MB, Lindekleiv H, Mathiesen EB. Intravenous thrombolytic treatment and endovascular thrombectomy for ischaemic wake-up Stroke. Cochrane Database Syst Rev. 2021;12:Cd010995.
Jung E, Romero R, Yeo L, Gomez-Lopez N, Chaemsaithong P, Jaovisidha A, Gotsch F, Erez O. The etiology of preeclampsia. Am J Obstet Gynecol. 2022;226:844–s866.
Opichka MA, Rappelt MW, Gutterman DD, Grobe JL, McIntosh JJ. Vascular dysfunction in Preeclampsia. Cells. 2021;10(11):3055.
Matsubara K, Matsubara Y, Uchikura Y, Sugiyama T. Pathophysiology of Preeclampsia: the role of exosomes. Int J Mol Sci. 2021;22(5):2572.
Lin J, Zhou Y, Gu W. Novel piRNA regulates PIWIL1 to modulate the behavior of placental trophoblast cells and participates in preeclampsia. Oxid Med Cell Longev. 2022;2022:7856290.
Szubert M, Koziróg E, Olszak O, Krygier-Kurz K, Kazmierczak J, Wilczynski J. Adenomyosis and infertility-review of medical and surgical approaches. Int J Environ Res Public Health. 2021;18(3):1235.
Gou LT, Kang JY, Dai P, Wang X, Li F, Zhao S, Zhang M, Hua MM, Lu Y, Zhu Y, et al. Ubiquitination-deficient mutations in human piwi cause male infertility by impairing histone-to-Protamine Exchange during Spermiogenesis. Cell. 2017;169:1090–1104e1013.
Celermajer DS, Chow CK, Marijon E, Anstey NM, Woo KS. Cardiovascular Disease in the developing world: prevalences, patterns, and the potential of early Disease detection. J Am Coll Cardiol. 2012;60:1207–16.
Campos-Carrillo A, Weitzel JN, Sahoo P, Rockne R, Mokhnatkin JV, Murtaza M, Gray SW, Goetz L, Goel A, Schork N, Slavin TP. Circulating Tumor DNA as an early cancer detection tool. Pharmacol Ther. 2020;207:107458.
Ameli Mojarad M, Ameli Mojarad M, Shojaee B, Nazemalhosseini-Mojarad E. piRNA: a promising biomarker in early detection of gastrointestinal cancer. Pathol Res Pract. 2022;230:153757.
Kuo MC, Liu SC, Hsu YF, Wu RM. The role of noncoding RNAs in Parkinson’s Disease: biomarkers and associations with pathogenic pathways. J Biomed Sci. 2021;28:78.
Fonseca Cabral G, Azevedo Dos Santos Pinheiro J, Vidal AF, Santos S. Ribeiro-Dos-Santos Â: piRNAs in gastric Cancer: a New Approach towards Translational Research. Int J Mol Sci 2020, 21.
Weng W, Liu N, Toiyama Y, Kusunoki M, Nagasaka T, Fujiwara T, Wei Q, Qin H, Lin H, Ma Y, Goel A. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of Disease progression, and a potential prognostic biomarker in Colorectal cancer. Mol Cancer. 2018;17:16.
James JP, Riis LB, Malham M, Høgdall E, Langholz E, Nielsen BS. MicroRNA biomarkers in IBD-Differential diagnosis and prediction of colitis-Associated Cancer. Int J Mol Sci 2020, 21.
Zhao L, Shi J, Chang L, Wang Y, Liu S, Li Y, Zhang T, Zuo T, Fu B, Wang G, et al. Serum-derived exosomal proteins as potential candidate biomarkers for Hepatocellular Carcinoma. ACS Omega. 2021;6:827–35.
Li C, Li C, Zhi C, Liang W, Wang X, Chen X, Lv T, Shen Q, Song Y, Lin D, Liu H. Clinical significance of PD-L1 expression in serum-derived exosomes in NSCLC patients. J Transl Med. 2019;17:355.
Kerr N, García-Contreras M, Abbassi S, Mejias NH, Desousa BR, Ricordi C, Dietrich WD, Keane RW, de Rivero Vaccari JP. Inflammasome Proteins in serum and serum-derived extracellular vesicles as biomarkers of Stroke. Front Mol Neurosci. 2018;11:309.
Tauriello DV, Calon A, Lonardo E, Batlle E. Determinants of metastatic competency in Colorectal cancer. Mol Oncol. 2017;11:97–119.
Skrede OJ, De Raedt S, Kleppe A, Hveem TS, Liestøl K, Maddison J, Askautrud HA, Pradhan M, Nesheim JA, Albregtsen F, et al. Deep learning for prediction of Colorectal cancer outcome: a discovery and validation study. Lancet. 2020;395:350–60.
Baassiri A, Nassar F, Mukherji D, Shamseddine A, Nasr R, Temraz S. Exosomal non coding RNA in LIQUID biopsies as a promising biomarker for Colorectal cancer. Int J Mol Sci. 2020;21(4):1398.
Liu H, Ye D, Chen A, Tan D, Zhang W, Jiang W, Wang M, Zhang X. A pilot study of new promising non-coding RNA diagnostic biomarkers for early-stage colorectal cancers. Clin Chem Lab Med. 2019;57:1073–83.
Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–74.
Hanin A, Lambrecq V, Denis JA, Imbert-Bismut F, Rucheton B, Lamari F, Bonnefont-Rousselot D, Demeret S, Navarro V. Cerebrospinal fluid and blood biomarkers of status epilepticus. Epilepsia. 2020;61:6–18.
Geles K, Palumbo D, Sellitto A, Giurato G, Cianflone E, Marino F, Torella D, Mirici Cappa V, Nassa G, Tarallo R et al. WIND (Workflow for pIRNAs aNd beyonD): a strategy for in-depth analysis of small RNA-seq data. F1000Res 2021, 10:1.
Wang J, Zhang P, Lu Y, Li Y, Zheng Y, Kan Y, Chen R, He S. piRBase: a comprehensive database of piRNA sequences. Nucleic Acids Res. 2019;47:D175–d180.
Wang J, Shi Y, Zhou H, Zhang P, Song T, Ying Z, Yu H, Li Y, Zhao Y, Zeng X, et al. piRBase: integrating piRNA annotation in all aspects. Nucleic Acids Res. 2022;50:D265–d272.
Acknowledgements
None.
Funding
This work was funded by Leading Talents of Zhongyuan Science and Technology Innovation (214200510027), Science and technology department of Henan Province (222102310365), Henan Medical Science and Technology Joint Building Program (LHGJ20210324), Gandan Xiangzhao Research Fund (GDXZ2023001), Henan Provincial Medical Science and Technology Research Plan (SBGJ2018002 and SBGJ202102117), and Outstanding Foreign Scientist Studio in Henan Province (GZS2020004).
Author information
Authors and Affiliations
Contributions
WG, SZ and YH designed and guided the review. ZW and YH wrote and edited the manuscript. ZW helped with reference collection and draw the figures. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wu, Z., Yu, X., Zhang, S. et al. Novel roles of PIWI proteins and PIWI-interacting RNAs in human health and diseases. Cell Commun Signal 21, 343 (2023). https://doi.org/10.1186/s12964-023-01368-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12964-023-01368-x