Abstract
Background
Despite the global prevalence of Klebsiella pneumoniae Carbapenemase (KPC)-type class A β-lactamases, occurrences of KPC-3-producing isolates in China remain infrequent. This study aims to explore the emergence, antibiotic resistance profiles, and plasmid characteristics of blaKPC-3-carrying Pseudomonas aeruginosa.
Methods
Species identification was performed by MALDI-TOF-MS, and antimicrobial resistance genes (ARGs) were identified by polymerase chain reaction (PCR). The characteristics of the target strain were detected by whole-genome sequencing (WGS) and antimicrobial susceptibility testing (AST). Plasmids were analyzed by S1-nuclease pulsed-field gel electrophoresis(S1-PFGE), Southern blotting and transconjugation experiment.
Results
Five P. aeruginosa strains carrying blaKPC-3 were isolated from two Chinese patients without a history of travelling to endemic areas. All strains belonged to the novel sequence type ST1076. The blaKPC-3 was carried on a 395-kb IncP-2 megaplasmid with a conserved structure (IS6100-ISKpn27-blaKPC-3-ISKpn6-korC-klcA), and this genetic sequence was identical to many plasmid-encoded KPC of Pseudomonas species. By further analyzing the genetic context, it was supposed that the original of blaKPC-3 in our work was a series of mutation of blaKPC-2.
Conclusions
The emergence of a multidrug resistance IncP-2 megaplasmid and clonal transmission of blaKPC-3-producing P. aeruginosa in China underlined the crucial need for continuous monitoring of blaKPC-3 for prevention and control of its further dissemination in China.
Similar content being viewed by others
Background
Due to the irrational use of antibiotics, the quantity of multidrug-resistant (MDR) bacteria [1] is on rise, and the largest proportion of them is Enterobacteriaceae. For infections caused by MDR bacteria, carbapenems are often the preferred treatment. Consequently, the appearance of carbapenem-resistant organisms (CRO) invalidates many commonly used antibiotics and leads to clinical treatment failure, which causes a high mortality rate in several regions, especially developing countries [2, 3]. The leading cause of CRO is carbapenem-hydrolyzing enzymes, which account for about 90%. The presence of the high-yield AmpC enzyme combined with the deletion of membrane pore proteins or the high expression of the excretion system accounts for a minor portion of CRO [4]. There are many types of carbapenemases, including KPC, NDM, IMP, VIM, OXA-48 and so on [5].
KPC, one of the carbapenemases family representatives, was first identified in Klebsiella pneumoniae from a hospital in North Carolina in 2001 [6]. Since then, different variants of blaKPC genes have been identified globally [7,8,9]. KPC-3, a common type of KPC, which differs from KPC-2 with a His (272) -Tyr substitution [10, 11]. KPC-3 was initially reported in a New York Medical Center in 2004 and had been detected in Spain, Brazil, and Africa [12,13,14]. However, there have been few reports about KPC-3 in China until now [15, 16].
As an important pathogen causing nosocomial infections worldwide, Pseudomonas aeruginosa is known to become carbapenem-resistant by the overexpression of resistance-nodulation-division (RND) efflux systems, the absence of porin (OprD), AmpC-lactamase encoding and carbapenemase production [17]. This is one of the reasons for the high mortality of infections caused by P. aeruginosa. Of which, the acquisition of antimicrobial in P. aeruginosa mainly through the horizontal transfer of plasmids [18]. Currently, IncP incompatibility groups have been reported in P. aeruginosa, include IncP-1 to IncP-7 and IncP-9 to IncP-14 [19].
Herein, we identified P. aeruginosa isolates carrying blaKPC-3, and performed whole-genome sequencing (WGS). Meanwhile, this study reported the antimicrobial resistance profiles and genomics characterization of blaKPC-3-harboring P. aeruginosa with an IncP-2 megaplasmid.
Materials and methods
Clinical isolates and antimicrobial susceptibility testing
From March 2021 to December 2021, a total of 37 carbapenem-non-susceptible P. aeruginosa isolates were obtained from a teaching hospital of Zhejiang University. All isolates were inoculated on the agar plate and cultured at 37 °C for 18 h. Species were identified using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) (Bruker, Bremen, Germany). Carbapenemases encoding genes including blaKPC, blaNDM, blaOXA-48, blaVIM, and blaIMP were identified using polymerase chain reaction (PCR) and genomic sequencing, as described previously [20]. Susceptibility testing of polymyxin was performed by broth dilution method, and other antibiotics such as aztreonam, imipenem, meropenem, ceftazidime, levofloxacin, ciprofloxacin, gentamicin, piperacillin/tazobactam, cefepime, amikacin, ceftazidime/avibactam were performed by agar dilution method. Interpretation of susceptibility results was according to Clinical and Laboratory Standards Institute (CLSI) (https://clsi.org/) guidelines, except polymyxin which the susceptibility breakpoints were proposed by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org) [21]. P. aeruginosa ATCC 27853 were used as a quality control standard.
Location of bla KPC-3 gene and transferability of plasmids carrying bla KPC-3 gene
In order to analyze the homology of strains, we performed the PFGE experiments. The size and number of plasmids were identified by the S1-nuclease digestion and pulsed-field gel electrophoresis (S1-PFGE). After electrophoresis, the location of blaKPC-3 gene was determined by Southern blotting and hybridization with digoxigenin-labeled blaKPC-3 specific probe. Salmonella strain H9812 was used as a control strain and size marker [20].
The transferability of the plasmid carrying blaKPC-3 was verified by conjugation experiments using rifampin-resistant P. aeruginosa PAO1Ri and E. coli 600 as the recipient strain. Mueller–Hinton medium containing 300 mg/L rifampicin and 2 mg/L meropenem was used as the screening medium for transconjugants. The transconjugants were identified by MALDI-TOF-MS, and then the blaKPC-3 was screened by PCR to confirm whether the plasmids were successfully transferred [22].
Whole-genome sequencing and plasmid analysis
Genomic DNA was extracted by using a Bacterial DNA Kit (QIAGEN, Hilden, Germany). Then the DNA was sequenced both on the Illumina NovaSeq 6000 (Illumina, San Diego, CA, United States) and Oxford Nanopore platforms (Oxford Nanopore Technologies, Oxford, United Kingdom) to obtain short-read data and long-read data, respectively. The complete genome sequence was assembled with Unicycler v0.4.7. The bacterial genome was annotated by the RAST server (https://rast.nmpdr.org/). Genotyping based on the analysis of internal fragments of seven housekeeping genes (acsA, aroE, guaA, mutL, nuoD, ppsA and trpE) was performed via the online service MLST (https://cge.cbs.dtu.dk/services/MLST/) [23]. Additionally, the acquired ARGs were detected by ResFinder 4.1 (https://cge.food.dtu.dk/services/ResFinder/), and the plasmid replicon type was identified by PCR [24]. The analysis of virulence factors was performed on VFDB website (http://www.mgc.ac.cn/cgi-bin/VFs/v5/main.cgi). The origin of transfers in DNA sequences of bacterial mobile genetic elements was identified by oriTfinder tool (https://tool-mml.sjtu.edu.cn/oriTfinder/oriTfinder.html). The transposon and insertion sequence were detected using the ISFinder database (http://www-is.biotoul.fr/). Finally, the comparison figures of the genetic context surrounding blaKPC-3 were generated by Easyfig2.0 software, and BLAST Ring Image Generator (BRIG) was used to generate the circular image of multiple plasmids comparisons.
Nucleotide sequence accession numbers
The complete sequence of pLHL1-KPC-3 has been submitted to GenBank under accession no. CP099961.
Biofilm formation assays
Biofilm formation was measured in accordance with the assays outlined in previous researches [25]. In a word, the bacterial overnight culture was diluted in LB and the 200 μL of the mixture was dispensed into the each well of 96-well plate. After static culture at 37℃ for 24 h, PBS was utilized to clean the microwells three times in order to eliminate all non-adherent bacteria. Methanol was used for fixation, and 0.1% crystal violet solution were added to each well for subsequent stain. After washing the plate with PBS three times and discarding the washing solution, 100 μL of DMSO were added to dissolve the crystal violet attached to the biofilm, and the plate should be incubated for 5 min. Finally, the OD (optical density) was measured at 590 nm. LB broth was used for the negative control. ODc is the mean OD of the negative control, ODs is the mean OD of samples. The experimental results were interpreted using the following criteria: ODc ≥ ODs: non biofilm formation, ODc < ODs ≤ 2*ODc: weak biofilm formation, 2*ODc < ODs ≤ 4*ODc: moderate biofilm formation, 4*ODc < ODs: strong biofilm formation.
Results
Isolation and identification of KPC-3-producing P. aeruginosa
Five KPC-3-producing P. aeruginosa strains were obtained from two different patients. The clinical characteristics of the strains are shown in Additional file 6: Table S1. The first strain of blaKPC-3-carrying P. aeruginosa LHL-1 was isolated from the sputum sample of patient 1 in the neurological ward on May 11, 2021. Two weeks later, we obtained another strain of P. aeruginosa LHL-11 from the sputum culture of patient 1 again. Three months later, while patient 1 was admitted to ICU, two drug-resistant strains LHL-20, LHL-28 were isolated. In contrast, LHL-37 strain was isolated from patient 2 in the neurological ward on October 9, 2021.
Antimicrobial susceptibility profiles
The minimum inhibitory concentration (MIC) values of antibiotics for five isolates are shown in Table 1. All five isolates had similar resistance profiles, resistant to aztreonam, imipenem, meropenem, ceftazidime, levofloxacin, ciprofloxacin, gentamicin, piperacillin/tazobactam, cefepime, but sensitive to amikacin, ceftazidime/avibactam, polymyxin.
Biofilm formation assays
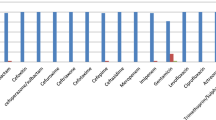
The LHL-1 from patient 1 and LHL-37 from patient 2 were selected for biofilm formation assay. The biofilm assay shown in Fig. 1 revealed that LHL-1 and LHL-37 had a moderate capacity for biofilm formation, with all mean OD values of trial groups greater than 2*ODc.
Genomic characteristics of LHL-1
The results of PFGE showed the five strains were highly homologous (Additional file 5: Fig. S5). By comparing the whole-genome data, we found that the sequences of the five strains were highly consistent and all belonged to a novel sequence type ST1076, so the strain LHL-1 was selected for further genomics analysis. It was found that the genome consists of a 7,232,504 bp circular chromosome with an average G + C content of 65.5% and a 394,987 kb megaplasmid with 491 predicted coding regions. By further analyzing the genome sequence of LHL-1, there were many ARGs such as coding for aminoglycoside (aph(3')-IIb), ciprofloxacin (crpP), chloramphenicol (catB7), beta-lactam (blaOXA-395, blaPAO) and fosfomycin (fosA) encoding on the chromosome. Besides, only one carbapenem resistance gene, blaKPC-3, was encoded on the megaplasmid. Virulence profiles of LHL-1 mainly including pilD (Type IV pili biosynthesis), chpA, pilG (Type IV pili twitching motility related proteins) and csrA (Carbon storage regulator A), as in LHL-37.
Characterization of megaplasmids harboring bla KPC-3
According to the S1-PFGE, it can be determined that the five isolates all harbor two plasmids with different sizes, and the results of Southern blotting revealed that blaKPC-3 gene was located on the megaplasmid with 394,987 bp in length (Additional files 1, 2, 3, 4 and Fig. 2). The blaKPC-3-harboring megaplasmids in isolates LHL-1 and LHL-37 are identical, containing 353 predicted open reading frames and a G + C content of 56.57%. Therefore, we chose the pLHL1-KPC-3 for further experiment.
We tried to transfer plasmid pLHL1-KPC-3 to the recipient strain P. aeruginosa PAO1Ri and E. coli 600. Nevertheless, no blaKPC-3-positive transformant was obtained in our study. Many functional genes were distributed on the skeleton megaplasmid pLHL-1-KPC-3, relating to replication and partitioning. By searching the core plasmid region against those in GenBank, the pLHL1-KPC-3 backbone showed 91% query coverage and 99% nucleotide identity to plasmid pRBL16 from P. citronellolis strain SJTE-3 (GenBank accession number CP015879), 94% query coverage and 100% nucleotide identity to pOZ176 which belonged to the IncP-2 group with IMP-9-mediated carbapenem resistance from P. aeruginosa strain PA96 (GenBank accession number KC543497), 92% query coverage and 98.27% nucleotide identity to plasmid unnamed-2 from P. aeruginosa strain AR439 (GenBank accession number CP029096) (Fig. 3). The information about the 20 related megaplasmids is shown in Table 2.
Genetic environment analysis showed that ISKpn6 (upstream), ISKpn27 (downstream) and several mobile elements including IS6100 and TnAs1 are densely distributed around blaKPC-3. Notably, a conserved structure sequence (IS6100-ISKpn27-blaKPC-3-ISKpn6-korC-klcA) was observed in the plasmid structure (Fig. 4). The genetic environment surrounding the blaKPC-3 in pLHL1-KPC-3 was related to the region of plasmid unnamed-1 from P. aeruginosa strain P9W (> 99% identities and > 99% query coverage; GenBank accession number CP081203) and plasmid pNK546-KPC from P. aeruginosa strain PAB546 (> 99% identities and > 99% query coverage; GenBank accession number MN433457), isolated from China.
The genetic context of blaKPC-3 gene on plasmid pLHL1-KPC-3, plasmid pNK546-KPC (MN433457), and plasmid unnamed-1 (CP081203). Open reading frames (ORFs) are shown as arrows and indicated according to their putative functions. Black and yellow indicates antimicrobial resistance genes and blue indicates genes related to mobile elements. The pink represents other functional genes. Regions with a high degree of homology are indicated by light green shading
Discussion
On the list for research and development of new antibiotics reported by the World Health Organization in 2017, carbapenem-resistant P. aeruginosa (CRPA) was ranked as one of the top antibiotic-resistant bacteria [26]. In contrast to K. pneumoniae, the incidence of plasmid-mediated blaKPC-3 was comparatively less frequent in Pseudomonas spp. To date, there have been no reports of KPC-3-producing P. aeruginosa infection in China. Using the keywords “pseudomonas”, “KPC-3”, and “China” to search the PubMed database (last accessed on 5 Dec. 2022), it retrieved zero results. Here, we collected the blaKPC-3 producing P. aeruginosa isolates, investigated the molecular characterization and genetic background of the blaKPC-3-harboring megaplasmid.
Over the period of five months, five identical strains of drug-resistant bacteria were isolated from two distinct patients over five months. Simultaneously, neither patient had a history of foreign residency. Considering the similarity of the genome and plasmid characteristics of the five bacterial strains, we inferred that clonal transmission occurred between the two patients. All five strains of P. aeruginosa carrying blaKPC-3 with a novel sequence type ST1076. Up to now, the blaKPC-3 has been detected in various ST-type strains, such as ST11 [27], ST147 [28], and ST258 [29]. To our knowledge, the study about ST1076 is limited, except one research has reported that ST1076 is associated with epidemiologically well-described outbreaks in burn units [30]. There is no obvious relationship between the new clone and the spread of blaKPC-3. However, the new ST type reminded us that mutations occurred in P. aeruginosa which deserve our attention. Biofilm forming ability partly reflects the virulence ability of bacteria. In the study, LHL-1 and LHL-37 both exhibited a moderate ability to form biofilms and virulence profiles were also identical. The contained virulence factors were primarily contributed to the biofilm formation and colonization in infection [31]. Infection caused by P. aeruginosa with certain biofilm capacity poses a threat to clinical treatment.
To the best of knowledge, the blaKPC gene can be either plasmid-mediated or on the chromosome of the host. Many types of plasmid replicons which was relative to Enterobacteriaceae carried the blaKPC, including IncX3, IncFrepB, IncQ and IncFIIK2-FIB [32, 33]. Unlike the common plasmid replicon types, blaKPC-harboring plasmid in P. aeruginosa often could be recognized as other different incompatibility groups, or could not be classified. A previous study reported a non-conjugative but mobilizable IncP-6-type resistance plasmid p10265-KPC in China [19]. Hu et al. described a novel plasmid carrying carbapenem-resistant gene blaKPC-2 in P. aeruginosa [34]. Moreover, there were many researches represented megaplasmids family carrying different ARGs in Pseudomonas spp. [35, 36]. Our study firstly identified a blaKPC-3-carrying IncP-2 megaplasmid. There have been several reports on ARGs-associated IncP-2 megaplasmids which were commonly found in the isolates of P. aeruginosa [37, 38]. Currently, most of the reported ARGs carried by IncP-2 megaplasmid encoded metallo-β-lactamases (MBLs),such as IMP and VIM. [39]. The discovery of blaKPC-3-carrying IncP-2 megaplasmid complements a novel example of IncP-2 megaplasmid family and its evolution, which deserves further investigation.
The oriTfinder result showed there no Origin site of DNA transfer (oriT), Relaxase, Type IV coupling protein (T4CP) and Type IV secretion system (T4SS) were found in the pLHL-KPC-3. The lack of conjugal transfer gene regions in pLHL-1 backbone was consistent with the fact that the vitro conjugation experiments were unsuccessful [40, 41]. By investigating the backbone structure of pLHL-1-KPC-3, the repA gene encodes the plasmid replication initiator protein and the parB encodes the protein that binds to the centromere-like sites, respectively. The ter gene was associated with the tellurite resistance as part of the megaplasmid family core genome. The pliT gene encodes PilT domain-containing protein. The merP was responsible for mercuric transport protein periplasmic component while the merT gene was related with mercuric transport protein [42].
Unlike the previous literature that blaKPC gene was embedded on Tn4401 generally, a member of the Tn3 family transposon involved in gene acquisition and dissemination, blaKPC-3 gene with the linear structure IS6100-ISKpn27-blaKPC-3-ISKpn6-korC-klcA in this work was found adjacent to another Tn3 family transposase TnAs1 [43, 44]. IS6100, belonging to the IS6 family members, is also one of the insertion sequences closely related to ARGs and can mediate sequence transfer and recombination between chromosomes or plasmids in different bacteria [45]. BLAST results showed that this region IS6100-ISKpn27-blaKPC-3-ISKpn6-korC-klcA was highly similar to those in the blaKPC-2-harboring element which played a key role in its transmission [46]. It was reasonable to presume that the sequence may also be involved in the transmission of blaKPC-3. By comparing the closely resemble plasmid backbone of pLHL1-KPC-3 and the other two plasmids, the major difference was the different ARGs variants they carried (Fig. 3). These results suggested the megaplasmid can capture diverse segment of ARGs based on the formation of backbone and then transfer to different host strains.
Although the isolates of KPC-3-producing P. aeruginosa in our work were not susceptible to multiple antibiotics, they remained susceptible to ceftazidime-avibactam (CAZ-AVI). In 2019, CAZ/AVI was used for clinical treatment in China. However, the resistance to CAZ-AVI has emerged and is more likely to occur in blaKPC-3-producing strains. Shields has reported the emergence of CAZ-AVI resistance in KPC-3-carbapenem-resistant K. pneumoniae during the treatment [47]. Thus, CAZ/AVI should be applied cautiously in the clinical treatment of infections caused by KPC-3-producing P. aeruginosa.
Conclusion
To our knowledge, this is the first report about KPC-3 producing P. aeruginosa in China. And our work revealed the characterization of an IncP-2 megaplasmid carrying blaKPC-3. This study reminded us that blaKPC-3 has gradually appeared in China with related to various bacterial species. Furthermore, it contributes to study the significance of megaplasmids in the evolution and resistance acquisition by P. aeruginosa. At the same time, we should also strengthen surveillance of CRPA spread in hospitals to prevent outbreaks of nosocomial infections.
Availability of data and materials
The complete sequence of pLHL1-KPC-3 has been submitted to GenBank under accession no. CP099961.
Abbreviations
- KPC:
-
Klebsiella pneumoniae Carbapenemase
- ARGs:
-
Antimicrobial resistance genes
- AST:
-
Antimicrobial susceptibility testing
- S1-PFGE:
-
S1-nuclease pulsed-field gel electrophoresis
- MDR:
-
Multidrug-resistant
- CRO:
-
Carbapenem-resistant organisms
- RND:
-
Resistance-nodulation-division
- WGS:
-
Whole-genome sequencing
- PCR:
-
Polymerase chain reaction
- MALDI-TOF-MS:
-
Matrix-assisted laser desorption ionization time-of-flight mass spectrometry
- CLSI:
-
Clinical and Laboratory Standards Institute
- EUCAST:
-
European Committee on Antimicrobial Susceptibility Testing
- MIC:
-
Minimum inhibitory concentration
- CRPA:
-
Carbapenem-resistant P. aeruginosa
- CAZ-AVI:
-
Ceftazidime-avibactam
References
Neut C. Carriage of multidrug-resistant bacteria in healthy people: recognition of several risk groups. Antibiotics. 2021;10(10):1163.
Ara-Montojo MF, Escosa-García L, Alguacil-Guillén M, Seara N, Zozaya C, Plaza D, et al. Predictors of mortality and clinical characteristics among carbapenem-resistant or carbapenemase-producing Enterobacteriaceae bloodstream infections in Spanish children. J Antimicrob Chemother. 2021;76(1):220–5.
Aguilera-Alonso D, Escosa-García L, Saavedra-Lozano J, Cercenado E, Baquero-Artigao F. Carbapenem-resistant gram-negative bacterial infections in children. Antimicrob Agents Chemother. 2020;64(3):e02183.
Suay-García B, Pérez-Gracia MT. Present and future of carbapenem-resistant Enterobacteriaceae (CRE) infections. Antibiotics. 2019;8(3):122.
Lutgring JD. Carbapenem-resistant Enterobacteriaceae: an emerging bacterial threat. Semin Diagn Pathol. 2019;36(3):182–6.
Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(4):1151–61.
Álvarez VE, Campos J, Galiana A, Borthagaray G, Centrón D, Márquez VC. Genomic analysis of the first isolate of KPC-2-producing Klebsiella pneumoniae from Uruguay. J Glob Antimicrob Resist. 2018;15:109–10.
Jousset AB, Oueslati S, Emeraud C, Bonnin RA, Dortet L, Iorga BI, et al. KPC-39-mediated resistance to ceftazidime-avibactam in a Klebsiella pneumoniae ST307 clinical isolate. Antimicrob Agents Chemother. 2021;65(12): e0116021.
Gartzonika K, Bozidis P, Priavali E, Sakkas H. Rapid detection of bla(KPC-9) allele from clinical isolates. Pathogens. 2021;10(4):487.
Woodford N, Tierno PM Jr, Young K, Tysall L, Palepou MF, Ward E, et al. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A beta-lactamase, KPC-3, in a New York medical center. Antimicrob Agents Chemother. 2004;48(12):4793–9.
Sotgiu G, Are BM, Pesapane L, Palmieri A, Muresu N, Cossu A, et al. Nosocomial transmission of carbapenem-resistant Klebsiella pneumoniae in an Italian university hospital: a molecular epidemiological study. J Hosp Infect. 2018;99(4):413–8.
Pérez-Vazquez M, Oteo-Iglesias J, Sola-Campoy PJ, Carrizo-Manzoni H, Bautista V, Lara N, et al. Characterization of carbapenemase-producing Klebsiella oxytoca in Spain, 2016–2017. Antimicrob Agents Chemother. 2019;63(6):e02529.
Migliorini LB, de Sales RO, Koga PCM, Doi AM, Poehlein A, Toniolo AR, et al. Prevalence of bla(KPC-2), bla(KPC-3) and bla(KPC-30)-carrying plasmids in Klebsiella pneumoniae isolated in a Brazilian hospital. Pathogens. 2021;10(3):332.
Ben Yahia H, Chairat S, Gharsa H, Alonso CA, Ben Sallem R, Porres-Osante N, et al. First report of KPC-2 and KPC-3-producing Enterobacteriaceae in wild birds in Africa. Microb Ecol. 2020;79(1):30–7.
Du H, Chen L, Chavda KD, Pandey R, Zhang H, Xie X, et al. Genomic characterization of Enterobacter cloacae isolates from China that coproduce KPC-3 and NDM-1 carbapenemases. Antimicrob Agents Chemother. 2016;60(4):2519–23.
Chen L, Ai W, Zhou Y, Wu C, Guo Y, Wu X, et al. Outbreak of IncX8 plasmid-mediated KPC-3-producing Enterobacterales infection. China Emerg Infect Dis. 2022;28(7):1421–30.
Breidenstein EB, de la Fuente-Núñez C, Hancock RE. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 2011;19(8):419–26.
Li M, Guan C, Song G, Gao X, Yang W, Wang T, et al. Characterization of a conjugative multidrug resistance IncP-2 Megaplasmid, pPAG5, from a clinical Pseudomonas aeruginosa isolate. Microbiol Spectr. 2022;10(1): e0199221.
Dai X, Zhou D, Xiong W, Feng J, Luo W, Luo G, et al. The IncP-6 plasmid p10265-KPC from Pseudomonas aeruginosa carries a novel ΔISEc33-associated bla KPC-2 gene cluster. Front Microbiol. 2016;7:310.
Guo X, Wang Q, Xu H, He X, Guo L, Liu S, et al. Emergence of IMP-8-producing Comamonas thiooxydans causing urinary tract infection in China. Front Microbiol. 2021;12: 585716.
Liu R, Xu H, Guo X, Liu S, Qiao J, Ge H, et al. Genomic characterization of two Escherichia fergusonii isolates harboring mcr-1 gene from farm environment. Front Cell Infect Microbiol. 2022;12: 774494.
Zheng B, Feng C, Xu H, Yu X, Guo L, Jiang X, et al. Detection and characterization of ESBL-producing Escherichia coli expressing mcr-1 from dairy cows in China. J Antimicrob Chemother. 2019;74(2):321–5.
Liang G, Rao Y, Wang S, Chi X, Xu H, Shen Y. Co-occurrence of NDM-9 and MCR-1 in a human gut colonized Escherichia coli ST1011. Infect Drug Resist. 2021;14:3011–7.
Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–28.
Zhao J, Zheng B, Xu H, Li J, Sun T, Jiang X, et al. Emergence of a NDM-1-producing ST25 Klebsiella pneumoniae strain causing neonatal sepsis in China. Front Microbiol. 2022;13: 980191.
Mancuso G, Midiri A, Gerace E, Biondo C. Bacterial antibiotic resistance: the most critical pathogens. Pathogens. 2021;10(10):1310.
Garcia-Fulgueiras V, Zapata Y, Papa-Ezdra R, Ávila P, Caiata L, Seija V, et al. First characterization of K. pneumoniae ST11 clinical isolates harboring bla(KPC-3) in Latin America. Rev Argent Microbiol. 2020;52(3):211–6.
Guerra AM, Lira A, Lameirão A, Selaru A, Abreu G, Lopes P, et al. Multiplicity of carbapenemase-producers three years after a KPC-3-producing K. pneumoniae ST147-K64 hospital outbreak. Antibiotics. 2020;9(11):806.
Gomez-Simmonds A, Annavajhala MK, McConville TH, Dietz DE, Shoucri SM, Laracy JC, et al. Carbapenemase-producing Enterobacterales causing secondary infections during the COVID-19 crisis at a New York City hospital. J Antimicrob Chemother. 2021;76(2):380–4.
Tissot F, Blanc DS, Basset P, Zanetti G, Berger MM, Que YA, et al. New genotyping method discovers sustained nosocomial Pseudomonas aeruginosa outbreak in an intensive care burn unit. J Hosp Infect. 2016;94(1):2–7.
McDaniel MS, Lindgren NR, Billiot CE, Valladares KN, Sumpter NA, Swords WE. Pseudomonas aeruginosa promotes persistence of Stenotrophomonas maltophilia via increased adherence to depolarized respiratory epithelium. Microbiol Spectr. 2023;11(1): e0384622.
Venditti C, Fortini D, Villa L, Vulcano A, D’Arezzo S, Capone A, et al. Circulation of bla(KPC-3)-carrying IncX3 plasmids among Citrobacter freundii isolates in an Italian hospital. Antimicrob Agents Chemother. 2017. https://doi.org/10.1128/AAC.00505-17.
Seiffert SN, Wüthrich D, Gerth Y, Egli A, Kohler P, Nolte O. First clinical case of KPC-3-producing Klebsiella michiganensis in Europe. New Microbes New Infect. 2019;29: 100516.
Hu YY, Wang Q, Sun QL, Chen GX, Zhang R. A novel plasmid carrying carbapenem-resistant gene bla (KPC-2) in Pseudomonas aeruginosa. Infect Drug Resist. 2019;12:1285–8.
Zou H, Berglund B, Xu H, Chi X, Zhao Q, Zhou Z, et al. Genetic characterization and virulence of a carbapenem-resistant Raoultella ornithinolytica isolated from well water carrying a novel megaplasmid containing bla(NDM-1). Environ Pollut. 2020;260: 114041.
Li R, Peng K, Xiao X, Liu Y, Peng D, Wang Z. Emergence of a multidrug resistance efflux pump with carbapenem resistance gene blaVIM-2 in a Pseudomonas putida megaplasmid of migratory bird origin. J Antimicrob Chemother. 2021;76(6):1455–8.
Urbanowicz P, Bitar I, Izdebski R, Baraniak A, Literacka E, Hrabák J, et al. Epidemic territorial spread of IncP-2-Type VIM-2 carbapenemase-encoding megaplasmids in nosocomial Pseudomonas aeruginosa populations. Antimicrob Agents Chemother. 2021. https://doi.org/10.1128/AAC.02122-20.
Zhang X, Wang L, Li D, Li P, Yuan L, Yang F, et al. An IncP-2 plasmid sublineage associated with dissemination of bla(IMP-45) among carbapenem-resistant Pseudomonas aeruginosa. Emerg Microbes Infect. 2021;10(1):442–9.
Urbanowicz P, Izdebski R, Biedrzycka M, Literacka E, Hryniewicz W, Gniadkowski M. Genomic epidemiology of MBL-producing Pseudomonas putida group isolates in Poland. Infect Dis Ther. 2022;11(4):1725–40.
Giakkoupi P, Pappa O, Polemis M, Vatopoulos AC, Miriagou V, Zioga A, et al. Emerging Klebsiella pneumoniae isolates coproducing KPC-2 and VIM-1 carbapenemases. Antimicrob Agents Chemother. 2009;53(9):4048–50.
Zheng B, Zhang J, Ji J, Fang Y, Shen P, Ying C, et al. Emergence of Raoultella ornithinolytica coproducing IMP-4 and KPC-2 carbapenemases in China. Antimicrob Agents Chemother. 2015;59(11):7086–9.
Cazares A, Moore MP, Hall JPJ, Wright LL, Grimes M, Emond-Rhéault JG, et al. A megaplasmid family driving dissemination of multidrug resistance in Pseudomonas. Nat Commun. 2020;11(1):1370.
Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. Genetic structures at the origin of acquisition of the beta-lactamase bla KPC gene. Antimicrob Agents Chemother. 2008;52(4):1257–63.
Hu X, Xu H, Shang Y, Guo L, Song L, Zhang H, et al. First genome sequence of a bla(KPC-2)-carrying Citrobacter koseri isolate collected from a patient with diarrhoea. J Glob Antimicrob Resist. 2018;15:166–8.
Güneş G, Smith B, Dyson P. Genetic instability associated with insertion of IS6100 into one end of the Streptomyces lividans chromosome. Microbiology. 1999;145(Pt 9):2203–8.
Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, et al. Nationwide surveillance of clinical Carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106.
Shields RK, Chen L, Cheng S, Chavda KD, Press EG, Snyder A, et al. Emergence of ceftazidime-avibactam resistance due to plasmid-borne bla(KPC-3) mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother. 2017. https://doi.org/10.1128/AAC.02097-16.
Acknowledgements
Not applicable.
Funding
This study was supported by the Henan Province Medical Science and Technology Research Project Joint Construction Project (No. LHGJ20190232), National Natural Science Foundation of China (82072314), the Research Project of Jinan Microecological Biomedicine Shandong Laboratory (JNL-2022011B), the Fundamental Research Funds for the Central Universities (2022ZFJH003), and CAMS Innovation Fund for Medical Sciences (2019-I2M-5–045).
Author information
Authors and Affiliations
Contributions
BZ and XG conceived and designed the experiments. HG JQ and JZ wrote the main manuscript text. RC and CL collected samples and performed the experiments. HX, RL and JZ analyzed the data, prepared the Figs. 1, 2, 3. BZ and XG reviewed and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
All other authors report no potential conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
The original gel picture of P. aeruginosa LHL1, LHL11, LHL20 and LHL28.
Additional file 2.
The original gel picture of P. aeruginosa LHL37.
Additional file 3.
The original blot image of P. aeruginosa LHL1, LHL11, LHL20 and LHL28.
Additional file 4.
The original blot image of P. aeruginosa LHL37.
Additional file 5.
The PFGE result of five KPC-3-producing P. aeruginosa strains.
Additional file 6: Table S1.
The clinical characteristics of the clinical strains.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ge, H., Qiao, J., Zheng, J. et al. Emergence and clonal dissemination of KPC-3-producing Pseudomonas aeruginosa in China with an IncP-2 megaplasmid. Ann Clin Microbiol Antimicrob 22, 31 (2023). https://doi.org/10.1186/s12941-023-00577-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12941-023-00577-z