Abstract
Background
Klebsiella variicola is considered a newly emerging human pathogen. Clinical isolates of carbapenemase and broad-spectrum β-lactamase-producing K. variicola remain relatively uncommon. A strain of K. variicola 4253 was isolated from a clinical sample, and was identified to carry the blaIMP−4 and blaSFO−1 genes. This study aims to discern its antibiotic resistance phenotype and genomic characteristics.
Methods
Species identification was conducted using MALDI-TOF/MS. PCR identification confirmed the presence of the blaIMP−4 and blaSFO−1 genes. Antibiotic resistance phenotype and genomic characteristics were detected by antimicrobial susceptibility testing and whole-genome sequencing. Plasmid characterization was carried out through S1-PFGE, conjugation experiments, Southern blot, and comparative genomic analysis.
Results
K. variicola 4253 belonged to ST347, and demonstrated resistance to broad-spectrum β-lactamase drugs and tigecycline while being insensitive to imipenem and meropenem. The blaIMP−4 and blaSFO−1 genes harbored on the plasmid p4253-imp. The replicon type of p4253-imp was identified as IncHI5B, representing a multidrug-resistant plasmid capable of horizontal transfer and mediating the dissemination of drug resistance. The blaIMP−4 gene was located on the In809-like integrative element (Intl1-blaIMP−4-aacA4-catB3), which circulates in Acinetobacter and Enterobacteriaceae.
Conclusions
This study reports the presence of a strain of K. variicola, which is insensitive to tigecycline, carrying a plasmid harboring blaIMP−4 and blaSFO−1. It is highly likely that the strain acquired this plasmid through horizontal transfer. The blaIMP−4 array (Intl1-blaIMP−4-aacA4-catB3) is also mobile in Acinetobacter and Enterobacteriaceae. So it is essential to enhance clinical awareness and conduct epidemiological surveillance on multidrug-resistant K. variicola, conjugative plasmids carrying blaIMP−4, and the In809 integrative element.
Similar content being viewed by others
Introduction
The Klebsiella pneumoniae complex is a member of the Klebsiella genus within the family Enterobacteriaceae, including Klebsiella pneumoniae, Klebsiella quasipneumoniae, and Klebsiella variicola. All members of this complex exhibit similar biochemical and phenotypic characteristics, making them indistinguishable using conventional microbiological methods [1, 2]. Furthermore, due to the inability of current microbiological automated detection systems to effectively differentiate Klebsiella species, there is a possibility of misidentifying certain isolates of K. variicola as K. pneumoniae in clinical settings [3,4,5], leading to an underestimation of the clinical prevalence of K. variicola. K. variicola was first described in 2004 and is considered a newly emerging pathogen in humans [6]. It is a gram-negative, facultative anaerobic, non-spore-forming, and non-motile rod-shaped bacterium that forms circular, convex, and smooth colonies [7]. In recent years, with the updating of the MALDI-TOF database and the widespread use of genomic sequencing, there has been an increasing number of reports on isolates of K. variicola, including the emergence of highly virulent K. variicola strains resistant to colistin [8], the presence of a hypervirulent conjugative plasmid in K. variicola [9], and the coexistence of blaNDM−1 and blaIMP−4 genes on one plasmid in K. variicola [10, 11]. Compared to K. pneumoniae, typically, K. variicola isolates exhibit lower antibiotic resistance rates [12], but studies have shown a high mortality rate associated with bloodstream infections caused by multidrug-resistant and extended-spectrum β-lactamase-producing K. variicola [13, 14]. Therefore, it is crucial to pay close attention to and monitor multidrug-resistant and extended-spectrum β-lactamase-producing K. variicola. In this study, the K. variicola 4253 strain was isolated from a stool sample of a female leukemia patient in a teaching hospital. The strain was identified to carry the blaIMP−4 and blaSFO−1 genes through PCR, and its antibiotic resistance phenotype and genomic characteristics were extensively characterized.
Materials and methods
Species identification and antimicrobial susceptibility testing
During routine monitoring targeting at carbapenem-resistant Enterobacteriaceae (CRE) in Hangzhou, Zhejiang Province, China, K. variicola 4253 was collected from a stool sample of a female leukemia patient at the First Affiliated Hospital of Zhejiang University. Species identification was conducted using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF/MS) (Bruker Daltonik GmbH, Bremen, Germany), while PCR was employed to identify the presence of the carbapenemase gene blaIMP−4 and the broad-spectrum β-lactamase gene blaSFO−1 (The primer sequences were showed in Table S1).
Agar dilution method and microbroth dilution method were utilized for antimicrobial susceptibility testing (AST), with Escherichia coli ATCC25922 and Pseudomonas aeruginosa ATCC27853 serving as the quality control strain. AST results were interpreted according to the guidelines provided by the Clinical and Laboratory Standards Institute (CLSI 2020) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST 2020).
Plasmid characterization and conjugation experiment
The number and size of plasmids in K. variicola 4253 were assessed using S1-PFGE. The location of blaIMP−4 and blaSFO−1 on the plasmid was determined through Southern blotting and gene hybridization with a digoxigenin-labeled specific probe (The probe sequences were showed in Table S1). To investigate the transferability of the plasmid, E. coli J53 was employed as the recipient strain in conjugation experiments. Transconjugants were selected using Mueller-Hinton agar (OXOID, Hampshire, United Kingdom) supplemented with 200 mg/L NaN3 and 8 mg/L cefepime. Their identification as E. coli and the presence of the blaIMP−4 and blaSFO−1 genes were confirmed through PCR and further validated using MALDI-TOF/MS.
Whole genome sequencing and analysis
Genomic DNA was extracted using the Genomic DNA Isolation Kit (QIAGEN, Hilden, Germany), and sequencing was performed using the Illumina Novaseq6000 platform (Illumina, San Diego, CA, USA) and the Oxford Nanopore platforms (Oxford Nanopore Technologies, Oxford, United Kingdom). Following sequencing, the short and long reads were subjected to hybrid assembly using Unicycler v0.4.7 to obtain the complete genome sequence. Annotation was carried out using the RAST 2.0 and Prokka tools. Kleborate v2.0.1 and Kaptive v0.7.3 were utilized for subspecies, MLST, wzi allele, capsule (K), and O antigen (LPS) serotype analysis. Resistance genes were identified using Resfinder. Virulence genes were obtained from the VFDB website, while plasmid replicon types were determined using PlasmidFinder 2.1 and KpVR. Integrons and insertion sequence elements were disclosed using INTEGRALL and ISfinder, respectively. Comparative analysis of different plasmid genome sequences was performed using the BLAST Ring Image Generator (BRIG). Then, the genetic context encompassing the blaIMP−4 and blaSFO−1 genes was visualized using Easyfig 2.3.
Results
Isolation of K. Variicola 4253 strain and antimicrobial susceptibility testing
The K. variicola 4253 was recovered from a fecal sample of a 22-year-old female patient with acute lymphocytic leukemia on November 8th, 2021. During hospitalization, the patient experienced high fever and received intravenous administration of cefoperazone sodium, sulbactam sodium, and vancomycin for treatment. After improvement of symptoms, the patient was discharged. The strain was identified as K. variicola using MALDI-TOF/MS, and further characterization through third-generation sequencing, using the Kleborate tool, confirmed it as K. variicola subsp. variicola. PCR and sequencing revealed the presence of the blaIMP−4 and blaSFO−1 genes.
As shown in Table 1, the antimicrobial susceptibility results demonstrated that both the strain and its transmissible conjugate exhibited resistance to broad-spectrum β-lactam drugs, including ceftriaxone, cefotaxime, ceftazidime, cefepime, aztreonam, piperacillin/tazobactam, and amoxicillin-clavulanic acid. However, they were susceptible to meropenem and imipenem. They also showed sensitivity to quinolones (levofloxacin, ciprofloxacin), sulfonamides (trimethoprim/sulfamethoxazole), and aminoglycosides (except for gentamicin, which showed resistance but amikacin showed sensitivity). Both the strain and its conjugate exhibited sensitivity to colistin. K. variicola 4253 demonstrated resistance to fosfomycin and tigecycline, while its conjugate remained susceptible to them.
Genomic features of the strain
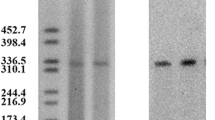
Genomic characteristics of K. variicola 4253 were explored through whole-genome sequencing. The genomic sequence analysis revealed that the isolated strain belongs to ST347 (gapA-infB-mdh-pgi-phoE-rpoB-tonB, allele no. 16-24-21-27-47-22-67). The capsule serotype was identified as wzi269/KL25/K25, and the O antigen serotype was classified as OL5/O5. The strain possessed a circular chromosome with a size of 5,533,843 bp and three plasmids, namely p4253-imp, p4253-2, and p4253-3. The sizes of the plasmids were 334,271 bp, 175,117 bp, and 4,655 bp, respectively (p4253-3 is not visible in S1-PFGE due to its small size) (Fig. 1). The average G + C content of the chromosome and plasmids were 57.3%, 48.7%, 51.5%, and 42.8%, respectively. Notably, the blaIMP−4 and blaSFO−1 genes were located on the same plasmid (p4253-imp). There were 2,651 protein-coding genes, 171 tRNA genes, and 61 rRNA genes on the chromosome. (Table S2).
A total of nine different antimicrobial resistance genes were identified in the strain, including beta-lactamase genes (blaOXA−1, blaIMP−4, blaSFO−1, blaTEM−214, blaLEN−25), aminoglycoside resistance genes (aac [3]-IId, aac(6’)-Ib-cr, aac(6’)-Ib4, aph [6]-Id, and aph(3’’)-Ib), macrolide resistance genes (mphA, mphE, and msrE), phenicol resistance gene (catB3), rifampicin resistance gene (arr-3), sulfonamide resistance gene (sul1), tetracycline resistance gene (tet(D)), fosfomycin resistance gene (fosA), and efflux pump-associated genes (oqxA and oqxB), of which seven were encoded on plasmid p4253-imp (Table 2), indicating that p4253-imp is a multidrug-resistant plasmid.
Furthermore, a total of 56 virulence genes were identified in the strain (Table S3). The chromosome harbored genes such as type 3 fimbriae (mrkABCDFHIJ), type 1 fimbriae (fimABCDEFGHIK), fimbrial adherence determinants (stcB, stcC), efflux pump (acrA, acrB), aerobactin (iutA), enterobactin siderophore (entABCDEFS, fepABCDG, fes), salmochelin (iroE), magnesium uptake (mgtB), regulation (rcsA, rcsB), T6SS-I (tssJGFM), T6SS-II (clpV), T6SS-III (dotU, icmF, impJGHFA, ompA, sciN, vgrG). The plasmid p4253-2 carried the T6SS-I gene (tssH), while no virulence genes were found on other plasmids.
Characterization of the p4253-imp plasmid
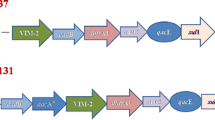
The size of the p4253-imp plasmid was 334,271 bp. It encoded 318 protein-coding genes and had a G + C content of 48.7%. No hits were found in the Plasmidfinder database for plasmid typing; however, analysis using the KpVR online tool revealed that the p4253-imp replicon belonged to the IncHI5B subtype. Based on this information, it can be inferred that the plasmid is a multidrug-resistant plasmid, carrying carbapenemase gene blaIMP−4 and broad-spectrum β-lactamase gene blaSFO−1. Consistent with the genomic features, S1-PFGE and Southern blot confirmed the size of the plasmid and the presence of blaIMP−4 and blaSFO−1 on the p4253-imp plasmid (Fig. 1). BLASTN search showed a high similarity between p4253-imp and pNDM-IMP-1 (CP050681.1), pKOX7525_1 (CP065475.1), pWH11 (ON882017.1), pIMP4-KP294 (CP083446.1), pKP1814-1 (KX839207.1), pA (CP068445.1), p2019SCSN059_tmexCD (ON169978.1), and pFK2020ZBJ35_tmexCD (ON169979.1) (More details about the 8 plasmids are showed in Table S4), with identity and coverage both exceeding 90%. Among these, p4253-imp exhibited higher similarity to pNDM-IMP-1, with 99% identity and 100% query coverage. pNDM-IMP-1 is a plasmid carrying blaIMP−4 from a urine catheter isolate of K. variicola SHET-01, obtained from a 2-year-old girl in the pediatric intensive care unit of a teaching hospital in Shanghai, China [11]. Gene comparison analysis (Fig. 2) revealed that p4253-imp and pNDM-IMP-1 share a similar plasmid backbone, including resistance determinants, insertion elements, conjugative transfer gene clusters, and related functional genes. Similar to the pNDM-IMP-1 plasmid, blaIMP−4 was carried by a Tn6406-like composite transposon, flanked by IS5075 elements, and containing an In809-like integron (Intl1-blaIMP−4-aacA4-catB3). The upstream sequence was identical to Tn5393-blaSFO−1-ampR-IS5075, where Tn5393 harbored aph [6]-Id and aph(3’’)-Ib resistance genes. Furthermore, conjugation experiments demonstrated the transferability of p4253-imp, and the antimicrobial susceptibility results indicated that this plasmid mediated the spread of resistance.
Discussion
The antimicrobial susceptibility test indicated that K. variicola 4253 exhibited.
insensitivity to tigecycline (MIC 8 mg/L). Through whole-genome analysis, no plasmid-mediated tet(X) gene encoding tetracycline inactivation enzyme or resistance-nodulation-division (RND) genes conferring tigecycline resistance were identified [15, 16]. Additionally, no conjugate conferring tigecycline insensitivity were obtained in the conjugation experiment. Therefore, it is postulated that the insensitivity of this strain to tigecycline is mediated by chromosomally carried genes. Previously reported mechanisms of tigecycline resistance in K. pneumoniae included mutations in the ribosomal protein S10 encoding gene rpsJ, which mediates tigecycline resistance [17], the involvement of the efflux pump OqxAB in low to moderate levels of tigecycline resistance [18], overexpression of the AcrAB efflux pump affecting sensitivity to tigecycline in K. pneumoniae [19,20,21], and mutations, insertions, deletions, and upregulation of transcriptional regulatory factors and related genes such as rarA, acrR, ramA, ramR, marA, and soxS, which can regulate the expression levels of the OqxAB and AcrAB efflux pumps and influence sensitivity to tigecycline [22,23,24]. In this study, the chromosomal genome of the strain contained the efflux pump genes acrAB-tolC and oqxAB. However, whether the tigecycline-insensitive phenotype in this strain is caused by overexpression of the efflux pump or mutations in related genes remains to be determined and requires further experimentation.
Although the isolated strain K. variicola 4253 produces carbapenemase, the antimicrobial susceptibility test revealed its sensitivity to imipenem and meropenem (MIC 1 mg/L). Previous literature reports have shown heterogeneity in the sensitivity of Enterobacteriaceae isolates carrying blaIMP−4 gene to carbapenems. For instance, Yang et al. reported that most of the 25 IMP-producing K. pneumoniae (IMPKp) isolates (including 16 IMP-4 strains) collected from a teaching hospital in China between 2009 and 2016 exhibited low levels of resistance and susceptibility to various carbapenem drugs [25]. Among them, three strains demonstrated intermediate to high-level resistance (MIC ≥ 8 mg/L) to imipenem and meropenem. Qiao et al. reported a Enterobacter hormaechei strain carrying blaIMP−4, which showed intermediate susceptibility to imipenem and sensitivity to meropenem [26]. Additionally, K. pneumoniae 1220, isolated from a three-month-old infant’s blood specimen and carrying the blaIMP−4 gene on a plasmid, was capable of conjugation into E. coli EC600. The antimicrobial susceptibility results indicated that both the strain and its conjugative form exhibited resistance to imipenem and meropenem (MIC 8 mg/L) [27]. It is speculated that IMP-4 hydrolyzing enzymes may have weak hydrolytic capabilities against carbapenems, and the production of IMP-4 is not the sole determinant for high-level carbapenem resistance. Other resistance mechanisms, including overexpression of efflux systems, porin protein deficiencies, and the presence of other broad-spectrum β-lactamases, may contribute to this process [28]. Currently, the extent to which different resistance mechanisms impact the carbapenem-resistant phenotype in Enterobacteriaceae strains carrying blaIMP−4 remains unclear.
The genetic context of blaIMP−4 in K. variicola 4253 was represented by Intl1-blaIMP−4-aacA4-catB3, which is similar to the integrative element In809 (Intl1-blaIMP−4-qacG-aacA4-catB3). It is possible that the qacG gene cassette was lost during the dissemination process. In809 was first described in Acinetobacter species isolated from Hong Kong between 1997 and 2000 [29] and has been subsequently reported in various Acinetobacter [30]and Enterobacteriaceae isolates [11, 25, 31]. This suggests that the blaIMP−4 array may be mobilized in Acinetobacter and Enterobacteriaceae through homologous recombination or plasmid-mediated horizontal transfer. Plasmid-mediated dissemination of carbapenemase-encoding genes plays a significant role in the emergence and spread of carbapenem-resistant Enterobacteriaceae (CRE). Previously reported incompatible plasmid types carrying the blaIMP gene include HI2/HI5, FI/FII, L/M, A/C, P, and N [32, 33]. However, in this study, the plasmid carrying blaIMP−4, p4253-imp, belonged to the IncHI5B type. IncHI plasmids are conjugative plasmids typically larger than 200 kb, serving as important vehicles for the dissemination of heavy metal resistance genes and antibiotic resistance genes. They are considered broad-host-range plasmids [34]. The conjugation experiments in this study also confirmed that p4253-imp was a conjugative plasmid capable of mediating horizontal transfer of resistance genes. BLAST search revealed an extremely high similarity between p4253-imp and the plasmid pNDM-IMP-1 isolated from K. variicola SHET-01 in Shanghai [11]. Given the geographic proximity between Shanghai and Hangzhou, it raises the possibility of clonal dissemination. Based on multilocus sequence typing (MLST), K. variicola SHET-01 belonged to ST3936, while K. variicola 4253 in this study belonged to ST347. Therefore, it is inferred that the strain isolated in this study is more likely to have been acquired through plasmid-mediated horizontal transfer.
Conclusion
We have reported the presence of a strain of K. variicola, which was insensitive to tigecycline, carrying a plasmid harboring blaIMP−4 and blaSFO−1. We have provided a detailed exposition of its drug resistance phenotype and genomic characteristics. This strain was classified as a multidrug-resistant strain, and its one plasmid coexisting blaIMP−4 and blaSFO−1, possessed the ability of horizontal transfer, thereby mediating the dissemination of drug resistance. Additionally, it is highly probable that K. variicola 4253 acquired this plasmid through plasmid-mediated horizontal transfer. Furthermore, the blaIMP−4 array (Intl1-blaIMP−4-aacA4-catB3) is also mobile in Acinetobacter and Enterobacteriaceae. Thus, it is imperative to enhance clinical awareness and conduct epidemiological surveillance in this regard.
Data availability
The complete sequence of K. variicola 4253 has been submitted to GenBank under accession no. CP135068-CP135071.
Abbreviations
- S1-PFGE:
-
S1-nuclease pulsed-filed gel electrophoresis
- PCR:
-
Polymerase chain reaction
- MALDI-TOF/MS:
-
Matrix-assisted laser desorption ionization time-of-flight mass
- MLST:
-
Multilocus sequence typing
- CRE:
-
Carbapenem-resistant Enterobacteriaceae
- AST:
-
Antimicrobial susceptibility testing
- WGS:
-
Whole-genome sequencing
- CLSI:
-
Clinical and Laboratory Standards Institute
- EUCAST:
-
European Committee on Antimicrobial Susceptibility Testing
- MIC:
-
Minimum inhibitory concentration
References
Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA. 2015;112(27):E3574–81.
Brisse S, Passet V, Grimont PAD. Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int J Syst Evol MicroBiol. 2014;64(Pt 9):3146–52.
Long SW, Linson SE, Ojeda Saavedra M, Cantu C, Davis JJ, Brettin T et al. Whole-genome sequencing of human clinical Klebsiella pneumoniae isolates reveals misidentification and misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mSphere. 2017;2(4).
Berry GJ, Loeffelholz MJ, Williams-Bouyer N. An investigation into Laboratory Misidentification of a bloodstream Klebsiella variicola infection. J Clin Microbiol. 2015;53(8):2793–4.
Seki M, Gotoh K, Nakamura S, Akeda Y, Yoshii T, Miyaguchi S, et al. Fatal sepsis caused by an unusual Klebsiella species that was misidentified by an automated identification system. J Med Microbiol. 2013;62(Pt 5):801–3.
Rosenblueth M, Martínez L, Silva J, Martínez-Romero E. Klebsiella variicola, a novel species with clinical and plant-associated isolates. Syst Appl Microbiol. 2004;27(1):27–35.
Lin L, Wei C, Chen M, Wang H, Li Y, Li Y et al. Complete genome sequence of endophytic nitrogen-fixing Klebsiella variicola strain DX120E. Standards in genomic sciences. 2015;10:22.
Lu Y, Feng Y, McNally A, Zong Z. Occurrence of colistin-resistant hypervirulent Klebsiella variicola. J Antimicrob Chemother. 2018;73(11):3001–4.
Yang X, Wai-Chi Chan E, Zhang R, Chen S. A conjugative plasmid that augments virulence in Klebsiella pneumoniae. Nat Microbiol. 2019;4(12):2039–43.
Xiao T, Peng K, Chen Q, Hou X, Huang W, Lv H, et al. Coexistence of tmexCD-toprJ, bla(NDM-1), and bla(IMP-4) in one plasmid carried by clinical Klebsiella spp. Microbiol Spectr. 2022;10(3):e0054922.
Wang B, Pan F, Han D, Zhao W, Shi Y, Sun Y, et al. Genetic characteristics and Microbiological Profile of Hypermucoviscous Multidrug-resistant Klebsiella variicola coproducing IMP-4 and NDM-1 Carbapenemases. Microbiol Spectr. 2022;10(1):e0158121.
Rodríguez-Medina N, Barrios-Camacho H, Duran-Bedolla J, Garza-Ramos U. Klebsiella variicola: an emerging pathogen in humans. Emerg Microbes Infections. 2019;8(1):973–88.
Maatallah M, Vading M, Kabir MH, Bakhrouf A, Kalin M, Nauclér P, et al. Klebsiella variicola is a frequent cause of bloodstream infection in the Stockholm area, and associated with higher mortality compared to K. pneumoniae. PLoS ONE. 2014;9(11):e113539.
Farzana R, Jones LS, Rahman MA, Andrey DO, Sands K, Portal E, et al. Outbreak of Hypervirulent Multidrug-resistant Klebsiella variicola causing high mortality in neonates in Bangladesh. Clin Infect Diseases: Official Publication Infect Dis Soc Am. 2019;68(7):1225–7.
Mohsin M, Hassan B, Martins W, Li R, Abdullah S, Sands K, et al. Emergence of plasmid-mediated tigecycline resistance tet(X4) gene in Escherichia coli isolated from poultry, food and the environment in South Asia. Sci Total Environ. 2021;787:147613.
Sun S, Gao H, Liu Y, Jin L, Wang R, Wang X, et al. Co-existence of a novel plasmid-mediated efflux pump with colistin resistance gene mcr in one plasmid confers transferable multidrug resistance in Klebsiella pneumoniae. Emerg Microbes Infections. 2020;9(1):1102–13.
Villa L, Feudi C, Fortini D, García-Fernández A, Carattoli A. Genomics of KPC-producing Klebsiella pneumoniae sequence type 512 clone highlights the role of RamR and ribosomal S10 protein mutations in conferring tigecycline resistance. Antimicrob Agents Chemother. 2014;58(3):1707–12.
Sheng ZK, Hu F, Wang W, Guo Q, Chen Z, Xu X, et al. Mechanisms of tigecycline resistance among Klebsiella pneumoniae clinical isolates. Antimicrob Agents Chemother. 2014;58(11):6982–5.
Ruzin A, Visalli MA, Keeney D, Bradford PA. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2005;49(3):1017–22.
Chetri S, Das BJ, Bhowmik D, Chanda DD, Chakravarty A, Bhattacharjee A. Transcriptional response of mar, sox and Rob regulon against concentration gradient carbapenem stress within Escherichia coli isolated from hospital acquired infection. BMC Res Notes. 2020;13(1):168.
Chetri S, Bhowmik D, Paul D, Pandey P, Chanda DD, Chakravarty A, et al. AcrAB-TolC efflux pump system plays a role in carbapenem non-susceptibility in Escherichia coli. BMC Microbiol. 2019;19(1):210.
Hentschke M, Wolters M, Sobottka I, Rohde H, Aepfelbacher M. ramR mutations in clinical isolates of Klebsiella pneumoniae with reduced susceptibility to tigecycline. Antimicrob Agents Chemother. 2010;54(6):2720–3.
Camp J, Schuster S, Vavra M, Schweigger T, Rossen JWA, Reuter S et al. Limited Multidrug Resistance Efflux Pump overexpression among Multidrug-Resistant Escherichia coli strains of ST131. Antimicrob Agents Chemother. 2021;65(4).
Chetri S, Singha K, Bhowmik D, Chanda DD, Chakravarty A, Bhattacharjee A. Sub-inhibitory concentration of ertapenem induces overexpression of regulator of antibiotic resistance A in Escherichia coli. Ind J Med Microbiol. 2018;36(4):569–71.
Yang L, Zhang G, Zhao Q, Guo L, Yang J. Molecular characteristics of clinical IMP-producing Klebsiella pneumoniae isolates: novel IMP-90 and integron In2147. Ann Clin Microbiol Antimicrob. 2023;22(1):38.
Qiao J, Ge H, Xu H, Guo X, Liu R, Li C, et al. Detection of IMP-4 and SFO-1 co-producing ST51 Enterobacter hormaechei clinical isolates. Front Cell Infect Microbiol. 2022;12:998578.
Feng W, Zhou D, Wang Q, Luo W, Zhang D, Sun Q, et al. Dissemination of IMP-4-encoding pIMP-HZ1-related plasmids among Klebsiella pneumoniae and Pseudomonas aeruginosa in a Chinese teaching hospital. Sci Rep. 2016;6:33419.
Zavascki AP, Carvalhaes CG, Picão RC, Gales AC. Multidrug-resistant Pseudomonas aeruginosa and Acinetobacter baumannii: resistance mechanisms and implications for therapy. Expert Rev anti-infective Therapy. 2010;8(1):71–93.
Houang ET, Chu YW, Lo WS, Chu KY, Cheng AF. Epidemiology of rifampin ADP-ribosyltransferase (arr-2) and metallo-beta-lactamase (blaIMP-4) gene cassettes in class 1 integrons in Acinetobacter strains isolated from blood cultures in 1997 to 2000. Antimicrob Agents Chemother. 2003;47(4):1382–90.
Koh TH, Sng LH, Wang GC, Hsu LY, Zhao Y. IMP-4 and OXA beta-lactamases in Acinetobacter baumannii from Singapore. J Antimicrob Chemother. 2007;59(4):627–32.
Espedido BA, Partridge SR, Iredell JR. Bla(IMP-4) in different genetic contexts in Enterobacteriaceae isolates from Australia. Antimicrob Agents Chemother. 2008;52(8):2984–7.
Zhao WH, Hu ZQ. IMP-type metallo-β-lactamases in Gram-negative bacilli: distribution, phylogeny, and association with integrons. Crit Rev Microbiol. 2011;37(3):214–26.
Matsumura Y, Peirano G, Motyl MR, Adams MD, Chen L, Kreiswirth B et al. Global molecular epidemiology of IMP-Producing Enterobacteriaceae. Antimicrob Agents Chemother. 2017;61(4).
Cain AK, Hall RM. Evolution of IncHI2 plasmids via acquisition of transposons carrying antibiotic resistance determinants. J Antimicrob Chemother. 2012;67(5):1121–7.
Acknowledgements
Not applicable.
Funding
This study was supported by the Fundamental Research Funds for the Central Universities (No. 2022ZFJH003), Shandong Provincial Laboratory Project (SYS202202), and Research Project of Jinan Microecological Biomedicine Shandong Laboratory (JNL-2022001 A).
Author information
Authors and Affiliations
Contributions
LL and BZ conceived and designed the experiments. HC wrote the main manuscript text. HC, HX, RL and JS collected samples and performed the experiments. HC analyzed the data, prepared the Figs. LL and BZ reviewed and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Ethical approval
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Chen, H., Xu, H., Liu, R. et al. Coexistence of blaIMP−4 and blaSFO−1 in an IncHI5B plasmid harbored by tigecycline-non-susceptible Klebsiella variicola strain. Ann Clin Microbiol Antimicrob 23, 24 (2024). https://doi.org/10.1186/s12941-024-00680-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12941-024-00680-9