Abstract
Background
Malaria remains an important public health problem in Latin America, and the development of insecticide resistance in malaria vectors poses a major threat to malaria elimination efforts. Monitoring of insecticide susceptibility and the determination of the mechanisms involved in insecticide resistance are needed to effectively guide the deployment of appropriate vector control measures. Here, molecular assays have been developed to screen for mutations associated with insecticide resistance on the voltage-gated sodium channel (VGSC) and acetylcholinesterase-1 (Ace-1) genes in four malaria vectors from Latin America.
Methods
Degenerate primers were designed to amplify a partial fragment on the VGSC and Ace-1 genes. Wild-caught individuals for Anopheles albimanus (also historical samples and individuals from a laboratory strain), Anopheles darlingi, Anopheles vestitipennis and Anopheles pseudopunctipennis were used to optimize the PCR assays. All samples were sequenced to validate the PCR results and DNA alignments were constructed for each gene using the unique haplotypes observed.
Results
Primers designed successfully amplified the VGSC gene in An. albimanus, An. darlingi, An. vestitipennis and An. pseudopunctipennis, and the Ace-1 gene in both An. albimanus and An. darlingi. DNA sequencing revealed that compared with Anopheles gambiae, there were a total of 29, 28, 21 and 24 single nucleotide polymorphisms (SNPs) on the VGSC gene for An. albimanus (308 bp), An. darlingi (311 bp), An. pseudopunctipennis (263 bp) and An. vestitipennis (254 bp), respectively. On the 459 bp fragment of the Ace-1 gene, a total of 70 SNPs were detected in An. darlingi and 59 SNPs were detected in An. albimanus compared with An. gambiae. The SNPs detected on the VGSC gene were all synonymous. On the Ace-1 gene, non-synonymous substitutions were identified on three different codons. All species showed the homozygous wild-type kdr allele (coding for leucine) at codon 995 (formerly reported as codon 1014) on the VGSC gene, but one sample was heterozygous at codon 280 (formerly reported as codon 119) on the Ace-1 gene, coding for both the resistant (serine) and susceptible (glycine) amino acids.
Conclusions
New molecular assays to amplify and screen the regions of the VGSC and Ace-1 genes associated with insecticide resistance are reported for An. albimanus, An. darlingi, An. vestitipennis, and An. pseudopunctipennis. The development of these PCR assays presents an important advance in the analysis of target-site resistance in malaria vectors in the Americas, and will further facilitate the characterization of insecticide resistance mechanisms in these species.
Similar content being viewed by others
Background
Impressive gains have been made in malaria control in recent years, however, malaria remains as an endemic disease in Latin America. In 2017, a total of 976,800 confirmed cases were reported in the Americas by the World Health Organization (WHO) [1]. Currently, Plasmodium vivax, Plasmodium falciparum and Plasmodium malariae are the only malaria species reported in the Americas, with the vectors Anopheles albimanus and Anopheles darlingi present over a wide geographical range [1, 2]. In addition, there is ongoing malaria transmission in areas with Anopheles vestitipennis and Anopheles pseudopunctipennis as principal or secondary vectors [2, 3].
The main vector control measures used in the Americas are long-lasting insecticide-treated bed nets (LLINs) and indoor residual spraying (IRS) [1]. These measures rely on the use of insecticides, mainly pyrethroids, which are often the most cost-effective and, until recently, the only class of insecticides approved by WHO for use on LLINs [1, 4]. Given this reliance on chemical interventions, monitoring of insecticide resistance is critical. Between 2010 and 2016, a total of 55 countries with ongoing malaria transmission reported the development of pyrethroid resistance in malaria vectors [5]. In Latin America, insecticide resistance has been detected in An. albimanus populations from Peru, Ecuador, Dominican Republic, Panama, Mexico, Honduras, and Guatemala [3, 5,6,7,8,9,10,11,12]. Moreover, pyrethroid resistance has been reported in An. darlingi populations from Brazil, Bolivia, Peru, and Colombia [5, 13]. In the case of An. vestitipennis and An. pseudopunctipennis, resistance to DDT was reported across Latin America during 1980s [14, 15]. However, resistance to pyrethroids, carbamates and organophosphates was observed only in populations of An. pseudopunctipennis from Guatemala and Honduras during the same period. Despite the limited information collected between 2005 and 2019, recent surveillance data suggest that An. vestitipennis and An. pseudopunctipennis remain susceptible to all classes of insecticides in Guatemala (unpublished results, Norma Padilla), but resistance to pyrethroids was detected in a population of An. pseudopunctipennis from Peru [7].
Insecticide resistance in mosquitoes can be mediated by changes in insect behavior, modification in the composition of the exoskeleton or digestive tract linings, increases in enzymatic activity (metabolic resistance), or single nucleotide polymorphisms (SNPs) which produce amino acid changes on insecticide target-sites (target-site resistance) [16]. The most studied mechanisms in Anopheles are those related to metabolic and target-site resistance. Metabolic resistance arises from increases in the levels of major detoxification enzyme families, principally cytochrome P450s, carboxylesterases and glutathione-S-transferases [16,17,18]. In the case of target-site resistance, non-synonymous SNPs at codon 995 on the voltage-gated sodium channel (VGSC) gene can confer resistance to pyrethroids and DDT (referred to as ‘knockdown resistance’, or kdr) [19, 20]. The most common amino acid changes on the VGSC gene are from leucine to phenylalanine, serine, or cysteine [21]. Similarly, non-synonymous substitutions at codon 280 on the acetylcholinesterase-1 (Ace-1) gene produce an amino acid change from leucine to serine resulting in cross resistance to organophosphates and carbamates [22]. Based on the new Anopheles gambiae coding numbering [23, 24], codon 995 corresponds to what had previously been referred to as position 1014 (in reference to its position in the house fly Musca domestica) and codon 280 corresponds to what had previously been referred to as position 119 (in reference to its position in the fish Torpedo californica).
According to the World Health Organization’s Global Plan for Insecticide Resistance Management (GPIRM), the emergence and spread of insecticide resistance poses a major threat to malaria elimination efforts worldwide [25, 26]. The plan stresses the need to fill knowledge gaps on mechanisms of insecticide resistance in order to develop more comprehensive resistance management strategies. Managing insecticide resistance requires not only the detection of resistant phenotypes, but also the identification of the mechanisms underlying the resistance. However, the characterization of the insecticide resistance mechanisms in malaria vectors in the Americas has lagged behind those in Africa. As such, the lack of molecular tools to describe target-site resistance in malaria vectors in the Americas has impeded the development of comprehensive resistance diagnostics, which are critical for the development of resistance management plans.
A preliminary analysis of historical samples of An. albimanus collected across Latin America during the 1990s when insecticide resistance was frequent in field populations reported for the first time the presence of kdr mutations (L995F and L995C) on the VGSC gene [27]. Recently, non-synonymous SNPs at codon 280 (G280S) and duplication events on the Ace-1 gene in field populations of An. albimanus from Peru were associated with cross-resistance to carbamates and organophosphates [28]. At present, molecular assays to screen the regions of the VGSC and Ace-1 genes associated with insecticide resistance have only been described for An. albimanus. Herein, new molecular assays have been developed to screen for non-synonymous SNPs on regions of the VGSC and Ace-1 genes associated with insecticide resistance in four malaria vectors from Latin America: An. albimanus, An. darlingi, An. vestitipennis and An. pseudopunctipennis.
Methods
Primer design
Degenerate primers were designed to amplify and sequence the regions associated with insecticide resistance on the VGSC and Ace-1 genes. The AKDRF2 and AADKDRR2 primers were designed to amplify the kdr region (including codon 995) on the VGSC gene for An. albimanus (309 bp, between exons 22 and 23) and An. darlingi (312 bp, between exons 20 and 21). These primers were designed based on the published sequences of An. albimanus [GenBank: KF137581.1, APCK01001913] and An. darlingi [GenBank: ADMH02001922]. The AAKDRF and AAKDRR primers previously described by Lol and colleagues [27] were used to amplify the kdr region in An. vestitipennis (263 bp) and An. pseudopunctipennis (254 bp). For the Ace-1 gene, ACE1DAF and ACE1DAR primers were designed to amplify a partial fragment of exon 4 for An. albimanus and exon 2 for An. darlingi, both fragments include the codon 280. The primers for the Ace-1 gene were designed based on the sequences of An. albimanus [AALB002313-RA] and An. darlingi [ADAC000377-RA] available on VectorBase. The primer sequences are presented in Table 1.
Samples and DNA extraction
For the optimization of the conventional PCR assays, wild-caught An. albimanus (including DNA from historical samples of this species used in previous population genetic studies [29, 30]), An. darlingi, An. vestitipennis, and An. pseudopunctipennis were used as DNA templates, as well as individuals from the insecticide-susceptible An. albimanus Sanarate laboratory strain (Table 2). Genomic DNA was extracted from mosquitoes collected during 2014–2017 and the Sanarate strain using DNAzol (Invitrogen), according to the manufacturer’s instructions with modifications. Briefly, each mosquito was grounded with 100 µL of DNAzol and resuspended in 100 µL of 1× TE buffer.
PCR conditions and sequencing of the VGSC and Ace-1 genes
The amplification of the kdr region of the VGSC gene for An. albimanus and An. darlingi was carried out in a 50 µL reaction mix containing 1× Colorless GoTaq® Flexi Buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 2.5 µM of each primer (AKDRF2 and AADKDRR2), 1.5 U of GoTaq® Hot Start Polymerase (Promega), and 2.5 to 10 ng of DNA template. The cycling conditions were 95 °C for 3 min, followed by 40 cycles of 95 °C for 45 s, 45 °C for 45 s, and 72 °C for 1 min with a final extension step at 72 °C for 5 min in a 2720 Thermal Cycler (Applied Biosystems). In the case of An. vestitipennis and An. pseudopunctipennis, the same region was amplified using the AAKDRF and AAKDRR primers with the same reaction mix and cycling conditions as mentioned above with the exception that the annealing temperature was 40 °C for An. vestitipennis and 45 °C for An. pseudopunctipennis. For the Ace-1 gene, a 25 µL reaction mixture was optimized containing 1× AccuStart™ II Gel Track™ PCR SuperMix (Quantabio), 0.4 µM of each primer (ACE1DAF and ACE1DAR), and 11 to 27 ng of DNA template. The cycling conditions were 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 61 °C for 30 s, and 72 °C for 1 min with a final extension step at 72 °C for 10 min in a Bio-Rad T100 Thermocyler (Bio-Rad).
All PCR products were visualized on a 2% agarose gel stained with ethidium bromide and observed under UV light. Then, 50 µL of PCR product for the VGSC gene were purified with 10 µL of a mix containing 1× MULTI-CORE™ buffer (Promega), 1 U of Exonuclease I (Biolabs), and 1 U of TSAP (Promega). Each sample was incubated at 37 °C for 30 min and 80 °C for 20 min in a thermocycler. Purified samples for the VGSC gene were directly sequenced by Macrogen Inc. (Maryland, USA) with the same primers used for the PCR amplification. For the Ace-1 gene, 25 µL of PCR product were purified using a Multiscreen PCR 96-well plate (Millipore), according to the manufacturer’s instructions. Sequencing of the Ace-1 gene was perfomed on an ABI 3130xl Genetic Analzyer (Applied Biosystems) in the laboratory at the U.S. Centers for Disease Control and Prevention (Atlanta, USA) using the same primers as in the PCR amplification.
Data analysis
Consensus sequences were obtained from the partial DNA sequences for the VGSC and Ace-1 genes using the SeqMan Pro tool of DNASTAR LaserGene 11.0 suite. The identity of DNA sequences were confirmed by a BLAST against the databases of GenBank and VectorBase. The intron positions for the VGSC gene in An. pseudopunctipennis and An. vestitipennis were determined by comparing the sequences obtained against the reference sequences of An. gambiae [VectorBase: AGAP004707-RA], An. albimanus [VectorBase: AALB007478-RA], and An. darlingi [VectorBase: ADAC011160-RA]. A DNA alignment for the unique haplotypes for each species was then constructed for each gene using Clustal Omega available on the EMBL-EBI website (https://www.ebi.ac.uk/Tools/msa/clustalo/). The An. gambiae sequences for the VGSC [VectorBase: AGAP004707-RA] and Ace-1 [VectorBase: AGAP001356-RA] genes were included in the DNA alignments as a reference sequence to detect SNPs. From the DNA sequences without introns, amino acid sequences were deduced and changes were detected compared to the amino acid sequences in An. gambiae. The consensus sequences obtained for the VGSC and Ace-1 genes were submitted to GenBank and the accession numbers are indicated in Table 3.
Results
PCR results for the VGSC and Ace-1 genes are presented in Figs. 1 and 2, respectively. Specific and efficient amplification was achieved for samples of An. albimanus, An. darlingi, An. pseudopunctipennis, and An. vestitipennis from different regions across Latin America with the described primers. In addition, DNA sequencing confirmed that the PCR products corresponded to the target areas of the VGSC and Ace-1 genes.
The DNA alignment of the fragments of the VGSC gene showed that when different species of malaria vectors from Latin America were compared with An. gambiae (Fig. 3), a total of 29 SNPs were detected in An. albimanus, 28 in An. darlingi, 24 in An. vestitipennis, and 21 in An. pseudopunctipennis. Moreover, sequences obtained for An. vestitipennis and An. pseudopunctipennis were submitted to GenBank since there were no sequences available for these species. For An. albimanus and An. darlingi, sequences obtained from all samples were identical to the sequences reported in VectorBase [An. albimanus: AALB002313-RA; An. darlingi ADAC00377-RA]. The genotypes observed at codon 995 on the VGSC gene were TTG (An. albimanus, An. pseudopunctipennis, and An. vestitipennis) and TTA (An. darlingi), both of which code for leucine, which is associated with susceptibility to pyrethroids and DDT. All other SNPs detected on the VGSC gene for the analyzed species were synonymous.
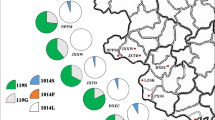
DNA alignment of the kdr region of VGSC gene of malaria vectors from Latin America. The alignment was constructed with partial DNA sequences for each species, and the aminoacid (AGa) and DNA (AG) sequences for An. gambiae were included as reference sequences [VectorBase: AGAP004707-RA]. Identical positions are indicated by an asterisk, primer positions are enclosed by an orange box, and the numbers above the amino acid sequence represent the codon position. Codon 995, which is associated with pyrethroid and DDT resistance (commonly reported as codon 1014, in reference to its position in the house fly Musca domestica), is highlighted in light blue, and it encodes the wildt-ype kdr allele (L995). AA, An. albimanus [VectorBase: AALB007478-RA]; AD, An. darlingi [VectorBase: ADAC011160-RA]; AP, An. pseudopunctipennis [GenBank: KX863726]; AV, An. vestitipennis [GenBank: KX907013]
The DNA alignment of the fragments of the Ace-1 gene from An. darlingi and An. albimanus are presented in Fig. 4. The alignment revealed 70 SNPs in An. darlingi and 59 SNPs in An. albimanus, as compared with An. gambiae. Based on these SNPs, five different haplotypes were detected for An. darlingi and three haplotypes were detected for An. albimanus in addition to the reference haplotypes in VectorBase for each species. At position 280 on the Ace-1 gene, An. darlingi exhibited three potential genotypes: GGG, GGS (S = C/G), and GGT, all of which code for glycine which is associated with susceptibility to carbamate and organophosphates. However, An. albimanus showed the genotypes GGC (coding for glycine; associated with susceptibility) and RGC (R = A/G), which codes for both the resistant (AGC, serine) and susceptible (GGC, glycine) amino acids. Two additional non-synonymous substitutions were detected at codons 221 (threonine to alanine) and 216 (threonine to serine) on the Ace-1 gene in all samples of An. albimanus and An. darlingi as compared with An. gambiae; these substitutions have not been previously reported before in any insect or associated with insecticide resistance [31].
DNA alignment of the Ace-1 gene for malaria vectors from Latin America. The alignment was constructed with partial DNA sequences for each species, and the aminoacid (AGa) and DNA (AG) sequences for An. gambiae were included as reference sequences [VectorBase: AGAP001356-RA]. Identical positions are indicated by an asterisk, primer positions are enclosed by an orange box, and the numbers above the amino acid sequence represent the codon position. Codon 280, which is associated with carbamate and organophosphate resistance (commonly reported as codon 119, in reference to its position in the fish Torpedo californica), is highlighted in light blue, and it encodes both homozygous susceptible (G280) and heterozygous resistant (G280S) alleles. Codons 216 and 221 are highlighted in red and yellow, respectively, and both codons result in an aminoacid change for all species from Latin American compared with the reference sequence of An. gambiae. Codon 215 is underlined and represents the choline binding site. Sequence AD1 corresponds to the sequence from VectorBase for An. darlingi [ADAC000377-RA]. Sequences AD2 [GenBank: MK477198], AD3 [GenBank: MK477199], and AD6 [GenBank: MK477202] are from Bolivia. Sequences AD4 [GenBank: MK477200] and AD5 [GenBank: MK477201] are from Guatemala. Sequence AA1 corresponds to the sequence from VectorBase for An. albimanus [AALB002313-RA]. Sequences AA2 [GenBank: MK477203], AA3 [GenBank: MK477204], and AA4 [GenBank: MK477205] are from Ecuador, Venezuela and Nicaragua, respectively. AA, An. albimanus; AD, An. darlingi
Discussion
Malaria case incidence rates decreased in Latin America by 31% between 2000 and 2015, and several countries have nearly eliminated the malaria transmission [32]. However, in recent years, many Latin American countries are increasing the use of insecticide-based vector control as part of the global push to eliminate the burden of malaria [1, 33]. This has increased the selection pressure of insecticides, favoring the emergence of insecticide resistance in malaria vectors. The continued intensive use of insecticides in commercial agriculture has also contributed to the resistance selection pressure, as was previously observed for An. albimanus populations from Guatemala during the 1980s [34].
Based on GPIRM recommendations, the routine monitoring of insecticide susceptibility is beginning to be included in the annual activities of National Malaria Control Programmes (NMCP) in some Latin American countries [5, 7]. However, the molecular characterization of the mechanisms underlying the resistance that has been detected is currently hindered by the lack of specific molecular assays for the principal malaria vectors of the region. Knowledge of these mechanisms is increasingly needed to better understand resistance patterns and improve vector control measures in ways that best manage and mitigate the effects of resistance. This is an important component of an Integrated Vector Management (IVM) framework, and will ultimately help elucidate the impact of insecticide resistance on malaria transmission [26]. Here, molecular assays have been developed to screen the regions of the VGSC and Ace-1 genes associated with insecticide resistance in four malaria vectors from Latin America.
The PCR primers designed successfully amplified the target regions of both genes of interest in all samples, suggesting that results are easily replicable. Moreover, these molecular assays present the advantage of being applicable across multiple species, resulting in a potential reduction in time and costs to process samples. The DNA sequencing results showed high sequence quality in the region of interest, allowing SNPs to be identified with a high degree of accuracy. Interestingly, different susceptible genotypes were observed at key codons of the VGSC and Ace-1 genes associated with insecticide resistance. As a consequence, these differences could limit the applicability of the allele-specific probes previously developed for African and Asian Anopheles. Additionally, nucleotide sequence variation in the region used to design the primes could also reduce the efficiency and specificity of the allele-specific probes developed for other Anopheles species. Therefore, these findings highlighting the value of this first step in characterizing important target site resistance regions in advance of developing allele-specific assays.
The molecular assays described in this paper will complement the bioassays that are required to detect resistant phenotypes. In addition to further elucidating the role of target-site mechanisms, additional research is also needed to develop molecular diagnostic tools to detect metabolic mechanisms of resistance in populations of malaria vectors from Latin America.
Conclusions
This study reports molecular assays that amplify the regions of the VGSC and Ace-1 genes associated with insecticide resistance in An. albimanus, An. darlingi, An. vestitipennis and An. pseudopunctipennis. These assays present an important advance in the analysis of target-site mutations in field populations of malaria vectors in Latin America, as now that these targets have been successfully amplified in these species, allele-specific diagnostic assays can be developed.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Abbreviations
- WHO:
-
World Health Organization
- LLINs:
-
long-lasting insecticide-treated bed nets
- IRS:
-
indoor residual spraying
- SNPs:
-
single nucleotide polymorphisms
- VGSC :
-
voltage-gated sodium channel
- kdr :
-
knockdown resistance
- Ace-1 :
-
acetylcholinesterase-1
- GPIRM:
-
Global Plan for Insecticide Resistance Management
- bp:
-
base pair
- DNA:
-
deoxyribonucleic acid
- PCR:
-
polymerase chain reaction
- UV:
-
ultraviolet
- BLAST:
-
basic local alignment search tool
- NMCP:
-
National Malaria Control Programmes
- IVM:
-
Integrated Vector Management
References
WHO. World malaria report 2018. Geneva: World Health Organization; 2018.
Sinka ME, Rubio-Palis Y, Manguin S, Patil AP, Temperley WH, Gething PW, et al. The dominant Anopheles vectors of human malaria in the Americas: occurrence data, distribution maps and bionomic precis. Parasit Vectors. 2010;3:72.
WHO. World malaria report 2015. Geneva: World Health Organization; 2015.
WHO. World malaria report 2014. Geneva: World Health Organization; 2014.
WHO. Global report on insecticide resistance in malaria vectors: 2010–2016. Geneva: World Health Organization; 2018.
Vargas VF, Córdova PO, Alvarado AA. Determinación de la resistencia a insecticidas en Aedes aegypti, Anopheles albimanus y Lutzomyia peruensis procedentes del Norte Peruano. Rev Peru Med Exp Salud Publica. 2006;23(4):259–64.
Quiñones ML, Norris DE, Conn JE, Moreno M, Burkot TR, Bugoro H, et al. Insecticide resistance in areas under investigation by the international centers of excellence for malaria research: a challenge for malaria control and elimination. Am J Trop Med Hyg. 2015;93(3 Suppl):69–78.
Cáceres L, Rovira J, García A, Torres R. Determinación de la resistencia a insecticidas organofosforados, carbamatos y piretroides en tres poblaciones de Anopheles albimanus (Diptera: Culicidae) de Panamá. Biomedica. 2011;31:419–27.
Penilla RP, Rodriguez AD, Hemingway J, Torres JL, Arredondo-Jimenez JI, Rodriguez MH. Resistance management strategies in malaria vector mosquito control. Baseline data for a large-scale field trial against Anopheles albimanus in Mexico. Med Vet Entomol. 1998;12(3):217–33.
Penilla RP, Rodríguez AD, Hemingway J, Trejo A, López AD, Rodríguez MH. Cytochrome P450-based resistance mechanism and pyrethroid resistance in the field Anopheles albimanus resistance management trial. Pestic Biochem Physiol. 2007;89(2):111–7.
Dzul FA, Penilla RP, Rodríguez AD. Susceptibility and insecticide resistance mechanisms in Anopheles albimanus from the southern Yucatan Peninsula, Mexico. Salud Publica Mex. 2007;49:302–11.
Mackenzie-Impoinvil L, Weedall GD, Lol JC, Pinto J, Vizcaino L, Dzuris N, et al. Contrasting patterns of gene expression indicate differing pyrethroid resistance mechanisms across the range of the New World malaria vector Anopheles albimanus. PLoS ONE. 2019;14(1):e0210586.
Fonseca-González I, Quiñones ML, McAllister J, Brogdon WG. Mixed-function oxidases and esterases associated with cross-resistance between DDT and lambda-cyhalothrin in Anopheles darlingi Root 1926 populations from Colombia. Mem Inst Oswaldo Cruz. 2009;104:18–26.
Acosta M. Niveles básicos de susceptibilidad del Anopheles pseudopunctipennis al DDT y al Dieldrin en la región occidental del Perú. Rev Peru Med Exp Salud Publica. 1958;12:99–106.
WHO. Vector resistance to pesticides: fifteenth report of the WHO Expert Committee on Vector Biology and Control. Geneva: World Health Organization; 1992.
Ranson H, N’Guessan R, Lines J, Moiroux N, Nkuni Z, Corbel V. Pyrethroid resistance in African anopheline mosquitoes: what are the implications for malaria control? Trends Parasitol. 2011;27(2):91–8.
Ffrench-Constant RH, Daborn PJ, Le Goff G. The genetics and genomics of insecticide resistance. Trends Genet. 2004;20(3):163–70.
Ranson H, Claudianos C, Ortelli F, Abgrall C, Hemingway J, Sharakhova MV, et al. Evolution of supergene families associated with insecticide resistance. Science. 2002;298(5591):179–81.
Martinez-Torres D, Chandre F, Williamson MS, Darriet F, Berge JB, Devonshire AL, et al. Molecular characterization of pyrethroid knockdown resistance (kdr) in the major malaria vector Anopheles gambiae s.s. Insect Mol Biol. 1998;7(2):179–84.
Ranson H, Jensen B, Vulule JM, Wang X, Hemingway J, Collins FH. Identification of a point mutation in the voltage-gated sodium channel gene of Kenyan Anopheles gambiae associated with resistance to DDT and pyrethroids. Insect Mol Biol. 2000;9(5):491–7.
Silva APB, Santos JMM, Martins AJ. Mutations in the voltage-gated sodium channel gene of anophelines and their association with resistance to pyrethroids—a review. Parasit Vectors. 2014;7:450.
Weill M, Malcolm C, Chandre F, Mogensen K, Berthomieu A, Marquine M, et al. The unique mutation in ace-1 giving high insecticide resistance is easily detectable in mosquito vectors. Insect Mol Biol. 2004;13(1):1–7.
The Anopheles gambiae Genomes Consortium. Genetic diversity of the African malaria vector Anopheles gambiae. Nature. 2017;552:96.
Lynd A, Oruni A, van’t Hof AE, Morgan JC, Naego LB, Pipini D, et al. Insecticide resistance in Anopheles gambiae from the northern Democratic Republic of Congo, with extreme knockdown resistance (kdr) mutation frequencies revealed by a new diagnostic assay. Malar J. 2018;17(1):412.
WHO. Global plan for insecticide resistance management in malaria vectors. Geneva: World Health Organization; 2012.
WHO. Framework for a national plan for monitoring and management of insecticide resistance in malaria vectors. Geneva: World Health Organization; 2017.
Lol JC, Castellanos ME, Liebman KA, Lenhart A, Pennington PM, Padilla NR. Molecular evidence for historical presence of knock-down resistance in Anopheles albimanus, a key malaria vector in Latin America. Parasit Vectors. 2013;6:268.
Liebman KA, Pinto J, Valle J, Palomino M, Vizcaino L, Brogdon W, et al. Novel mutations on the ace-1 gene of the malaria vector Anopheles albimanus provide evidence for balancing selection in an area of high insecticide resistance in Peru. Malar J. 2015;14:74.
De Merida AM, Palmieri M, Yurrita M, Molina A, Molina E, Black WC 4th. Mitochondrial DNA variation among Anopheles albimanus populations. Am J Trop Med Hyg. 1999;61(2):230–9.
Molina-Cruz A, De Merida AM, Mills K, Rodriguez F, Schoua C, Yurrita MM, et al. Gene flow among Anopheles albimanus populations in Central America, South America, and the Caribbean assessed by microsatellites and mitochondrial DNA. Am J Trop Med Hyg. 2004;71(3):350–9.
Guo D, Luo J, Zhou Y, Xiao H, He K, Yin C, et al. ACE: an efficient and sensitive tool to detect insecticide resistance-associated mutations in insect acetylcholinesterase from RNA-Seq data. BMC Bioinform. 2017;18(1):330.
WHO. World malaria report 2016. Geneva: World Health Organization; 2016.
Herrera S, Ochoa-Orozco SA, Gonzalez IJ, Peinado L, Quiñones ML, Arevalo-Herrera M. Prospects for malaria elimination in Mesoamerica and Hispaniola. PLoS Negl Trop Dis. 2015;9(5):e0003700.
Brogdon WG, Beach RF, Stewart JM, Castanaza L. Microplate assay analysis of the distribution of organophosphate and carbamate resistance in Guatemalan Anopheles albimanus. Bull World Health Organ. 1988;66(3):339–46.
Acknowledgements
We thank Maria E. Castellanos for assistance with primer design. We especially thank Pedro Peralta and Alfonso Salam from the CES and our collaborators from the Vector Control Programs from Guatemala and Bolivia, who done the mosquito collections.
Funding
Partial funding for this work was provided by the US Agency for International Development (USAID) under Amazon Malaria Initiative, CDC Cooperative Agreement Guatemala No. 5U01GH001003, Secretaria Nacional de Ciencia y Tecnologia de Guatemala FINDECYT/FODECYT 037-2017, and the Center for Health Studies, Universidad del Valle de Guatemala. The findings and conclusions in this paper are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Author information
Authors and Affiliations
Contributions
JCL performed the primers design, molecular assays, data analysis and drafted the manuscript. DC performed primer design and molecular assays. LI performed molecular assays and sequencing. CGR performed all An. darlingi collections. AL assisted with the data analysis, interpretation of results and contributed to the manuscript. NRP conceived and guided the study, participated in the interpretation of results, and contributed to the drafting of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Lol, J.C., Castañeda, D., Mackenzie-Impoinvil, L. et al. Development of molecular assays to detect target-site mechanisms associated with insecticide resistance in malaria vectors from Latin America. Malar J 18, 202 (2019). https://doi.org/10.1186/s12936-019-2834-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12936-019-2834-7