Abstract
Background
Larch is an important timber tree species. The traditional methods of tree genetic breeding have been progressing slowly. It is necessary to carry out gene function analysis and genetically modified breeding research. The NAC transcription factor family is a plant-specific transcription factor family with various biological functions, as shown in recent research. However, there are few studies on the NAC gene among gymnosperm coniferous species.
Results
LoNAC3 with complete cds was identified and isolated from the cDNA of Larix olgensis based on transcriptome data. The cDNA length of LoNAC3 is 1185 bp, encoding 394 amino acids, with a conserved NAM domain located at the N-terminus, and subcellular localization in the nucleus. The results of real-time quantitative PCR analysis showed that at different growth stages and in different tissues of L. olgensis, the relative expression level of LoNAC3 was highest in the needles. After drought, salt, alkali stress and hormone treatment, expression was induced to different degrees. The expression level of LoNAC3 was significantly increased under drought and salt conditions. The relative expression level changed under methyl jasmonate (MeJA) and abscisic acid (ABA) treatment. By observing the phenotype of overexpressed LoNAC3 tobacco, it was found that overexpressed tobacco is shorter and blooms earlier than wild-type tobacco. Under abiotic stress, LoNAC3 overexpressed tobacco has lower germination rates and poorer growth status. Transgenic tobacco under stress treatment has a higher malondialdehyde (MDA) content than wild-type tobacco, while peroxidase (POD) activity is lower than wild-type tobacco.
Conclusions
Through the analysis of LoNAC3 sequence and promoter expression, it can be concluded that LoNAC3 is involved in the drought and salt stress response processes of L. olgensis, and is induced by ABA and MeJA expression. Overexpression of LoNAC3 leads to stunted tobacco growth and negatively regulates its tolerance to drought and salt stress through the reactive oxygen species pathway. The preliminary analysis of the expression pattern and function of the LoNAC3 can provide a theoretical basis and high-quality materials for genetic improvement of larch in later stages.
Similar content being viewed by others
Background
Abiotic stresses such as drought and salt can lead to a decrease in the yield and quality of agricultural and forestry crops [1]. Drought can lead to a decrease in plant enzyme activity, which affects the growth rate of plants, especially in the seedling stage [2], leading to physiological disorders in plants; Salt can cause changes in soil osmotic potential, plant water potential, and ion toxicity, thereby affecting the growth and development process of plants [3], because the impact of changing osmotic potential on plants is similar to that of drought stress. Research has found that drought can cause changes in plant enzyme activity, thereby affecting its physiological functions. It mainly affects two aspects of plant photosynthesis and respiration. Drought stress can directly lead to changes in chloroplast structure, reducing photosynthetic pigment content and leading to a decrease in photosynthetic rate [4]. Mild drought can increase crop respiration rate, which gradually decreases with increasing drought severity [5]. Non biological stresses such as drought and salt can directly lead to the accumulation of proline in plants, such as cell membrane lipid peroxidation, accumulation of malondialdehyde in cells, resulting in the destruction of biofilm structure and loss of function [6]. It can also cause protein denaturation, DNA strand breakage, and other harmful cellular effects, leading to cellular dysfunction [7].
NAC transcription factors are a class of plant specific transcription factors widely distributed in terrestrial plants, and are also one of the gene families with the most transcription factors in the plant genome. In the early stage, 117 transcription factors of the NAC family have been found in Arabidopsis [8]. With the deepening of sequencing work, 163 and 79 NAC family members were found in poplar and rice, respectively [9, 10]. In early research, the NAC family was divided into three subfamilies: ATAF, NAM and, OsNAC3 [11]. With the deepening of research and the completion of model plant genome sequencing work, researchers conducted a comprehensive analysis of the NAC family genes in rice and Arabidopsis, dividing NAC family proteins into two major groups including 18 subgroups [12]. NAC genes are generally composed of three exons and two introns. The first two exons form a highly conserved special structural domain at the N-terminus of NAC proteins, while the third exon forms a diverse transcriptional activation domain at the C-terminus, which is an important recognition feature of NAC protein structure [13]. The special structural domain of NAC protein at the N-terminus is generally composed of about 150 amino acids, which can be divided into five subdomains: A, B, C, D, and E. Among them, the three subdomains A, C, and D belong to highly conserved domains, and the remaining two subdomains with weak conservation are only conserved in some NAC subgroups [14]. The C-terminus of NAC protein has highly repetitive simple amino acids, containing a large amount of Ser, Thr, Glu, and some acidic amino acid residues [15]. Some studies have found that the C-terminal domain of NAC protein has the function of transcriptional activation domain in yeast and plant cells [16].
In the NAC gene family, the earliest discovered Petunia NAM gene is related to plant morphogenesis. Plants with NAM gene mutations may wither during the seedling stage due to the inability to form normal apical meristem tissue in the stem [14]. The NAC transcription factor family has been found to not only participate in plant growth, development, and morphogenesis, but also participate in various stress resistance responses, especially abiotic stress. The double mutant plants of Arabidopsis CUC1 and CUC2 were found to exhibit healing of stamens, sepals, and cotyledons [13]. The Arabidopsis CUC3 gene is involved in regulating the formation of stem tip meristem and cotyledon edges [17]. Overexpression or inhibition of the NAC1 gene in Arabidopsis leads to an increase and decrease in the number of lateral roots [18]. Overexpression of the soybean NAC gene GmNAC20 in Arabidopsis promotes the generation of lateral roots in plants [19]. Arabidopsis ANAC019, ANAC055, and ANAC072 can bind to the promoter sequences of drought response factors. Overexpression of Arabidopsis transgenic plants can improve drought resistance [20]. ATAF1 plays a role in drought resistance by participating in regulating the expression of related genes in osmotic stress response [21]. Arabidopsis ANAC096 can help plants survive under dehydration osmotic stress [22]. TsNAC1 can target an important proton transport protein to improve plant salt tolerance [23].
Previous studies have mainly focused on NAC transcription factors discovered in model plants. In recent years, with the progress of transcriptomics and molecular breeding technology, the NAC family transcription factors in coniferous tree species have gradually been discovered, and research on gene function is also gradually underway. Overexpression of the PwNAC2 in Arabidopsis can enhance the tolerance of plants to stress treatment [24]. The PtaVNS protein in Pinus taeda participates in the process of tracheid formation by regulating the transcription induction of genes required for SCW formation and PCD [25]. LoNAC1 and LoNAC2 in L. olgensis can be induced to express under drought and salt stress [26].
In this study, LoNAC3 gene in the NAC family of L. olgensis was identified through transcriptome data from the laboratory. The gene coding region sequence was cloned and isolated from the cDNA of L. olgensis, and the LoNAC3 promoter sequence was obtained from the gDNA of L. olgensis. Through analysis of sequence information and expression ability, as well as research on overexpressing LoNAC3 tobacco, the potential role of LoNAC3 gene in plant growth and development was preliminarily elucidated, enriching the NAC family genes of larch and providing a molecular basis for studying the growth and stress resistance mechanisms of larch.
Methods
Plant materials, growth conditions, and treatments
The seedlings of L. olgensis (Jixi species) and wild-type tobacco used in the experiment were preserved by the State Key Laboratory of Tree Genetics and Breeding of Northeast Forestry University.
Non-lignified (approximately 60 days), semi-lignified (approximately 120 days) and lignified (180 days) L. olgensis were used as experimental material. Roots, stems and needles were separated, wrapped in aluminium foil, labelled, rapidly frozen in liquid nitrogen and stored at −80 °C. The 60-day L. olgensis material was treated with solutions of PEG6000 (25% w/v), NaCl (250 mmol.L-1), NaHCO3 (50 mmol.L-1), 50 mg.L-1 Gibberellin A3 (GA3), abscisic acid (ABA), N-(phenylmethyl)-9 H-purin-6-amine(6-BA), indole-3-acetic acid(IAA), methyl jasmonate (MeJA), and water (control) at 0 h, 48 h, 72 h, and 84 h, respectively. Samples were taken uniformly at 96 h (three samples for each treatment and time) [26].
After disinfection, tobacco seeds are inoculated onto 1/2MS solid culture medium to germinate and obtain sterile seedlings. Select individual plants with good growth potential for tissue culture and propagate them as genetic transformation receptor materials [27].
RNA extraction and DNA extraction
Total RNA was extracted using Universal Plant Total RNA Extraction Kit (BIOTEKE, Beijing, China), and cDNA obtained through reverse transcription using total RNA as a template (ReverseScript RT reagent Kit, TaKaRa). Plant DNA was extracted using Universal Plant DNA Extraction Kit (Omega).
Isolation and sequence analysis of LoNAC3
Design primers based on transcriptome data (forward:5’ATGATTTATCAGTATGGGCCAG3’; reverse:5’TTACTTACGAGTTCCATAAATCTAT3’) and amplify PCR using cDNA from L.olgensis as a template. Purify the target DNA bands and sequence them to obtain the sequence information of LoNAC3. Upload the sequence information of the successfully aligned LoNAC3 gene to NCBI (GenBank: MW206689).
Perform sequence multiple comparison analysis on the amino acid sequence of LoNAC3 using BioEdit software. MEGA5.0 software was used to construct the Neighbour-Joining tree, and the structural domains of the sequences were predicted and analysed with the online tool NCBI CD-search. Protparam (https://web.expasy.org/cgi-bin/protparam/protparam) was used to predict and analyse the physicochemical properties of the protein encoded by LoNAC3 gene. GOR4 (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html) and SwissModel (https://www.swissmodel.expasy.org/) was used to predict the secondary structure and three-dimensional structure of the protein, respectively. Utilizing WOLF PSORT (https://wolfpsort.hgc.jp/) predict the subcellular localization of proteins.
Isolation and vector construction of LoNAC3 promoter
Primers were designed based on the NCBI Larix kaempferi genome (taxid: 54800, L. kaempferi GenBank assembly GCA_013171265.2 Nucleotide BLAST): Forword primers (5’-3’) GCCCCGTTTGTCGTAG; Reverse primers (5’-3’) CATGTCCTGTAAACAGAAAACAGGATTT. Using the DNA of L. olgensis as a template, a fragment of approximately 2000 bp before the start codon (ATG) of LoNAC3 was cloned and insert it into the BamHI and NcoI enzyme cleavage sites of the pCAMBIA1301 plant expression vector to obtain the recombinant plasmid.
Activity validation and bioinformatics analysis of LoNAC3 promoter
Transfer the constructed plasmid (LoNAC3pro:: GUS) into Agrobacterium GV3101.Cultivate Agrobacterium in LB containing 50 mg/L kan and Rif until OD600 = 0.8 (200 rpm, 28 ℃). Soak tobacco seedlings that have grown for a week in LoNAC3pro:: GUS Agrobacterium (OD600 = 0.5 ~ 0.6) suspended at 1/2MS, shake for 3 to 4 h (120 rpm, 25 ℃), and then incubate them in dark at 25 ℃ on 1/2MS medium for 2 days. Place them in the prepared GUS staining solution. After incubating at 37 ℃ for 12 h, decolorize the stained tobacco seedlings in 95% alcohol. Utilizing PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) predict the cis acting element of the promoter.
Analysis of LoNAC3 expression in L. olgensis
Primer Premier 5.0 software designed the qRT-PCR primers: Forword primers (5’-3’) GCACGATTGTTATTTTAGCC; Reverse Primers (5’-3’) ACACGAGAGTCTTTCGCATTCC, the primers specifically screened by gel electrophoresis detection. LoTubulin was selected as a reference gene (Forword primers:5’GCCGTGCTGCTGGATAATGAGG3’;Reverse primers:5’TGTCTGGAACTCAGTCACATCAACG3’) and amplification was performed automatically according to the Real Master Mix (SYBR Green) kit 20ul system and an ABI 7500 real-time PCR instrument was used. Three replicates were set up in the quantitative PCR instrument and the results analysed by the 2−ΔΔCt method [28].
Vector construction and genetic transformation of LoNAC3 gene
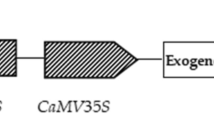
Insert the CDS sequence of the cloned LoNAC3 into the XbaI and HindIII enzyme cleavage sites of the VB191104 plant expression vector to obtain the recombinant plasmid. After successful sequencing of the connecting products, the plasmid was extracted and transferred to Agrobacterium GV3101. Using the Agrobacterium mediated leaf disc transformation method, tobacco leaves were infected with Agrobacterium GV3101 (VB191104-LoNAC3), and the infected transgenic leaves differentiated into resistant buds on a differentiation medium containing hygromycin (25 mg/L). After the isolated resistant buds were extracted from a culture medium containing hygromycin, they were transplanted to a culture medium containing hygromycin for rooting. After rooting, tobacco seedlings were transplanted into a nutrient soil substrate for cultivation [27].
Identification and phenotypic observationof overexpressed tobacco
Tobacco leaf DNA was extracted from the seedlings, and PCR identification was performed. Targeted bands were confirmed as positive seedlings. The seeds of the positive seedlings were collected and screened for hygromycin. After 2 generations of resistance screening, harvest T2 generation resistant tobacco seeds, and after sowing T3 generation on 1/2MS medium for 10 days, extract the entire RNA of each strain. Identify the RNA level through RT-qPCR, and select the three strains with the highest relative expression level for the next experiment.
Select the three overexpression strains and wild-type tobacco with the highest relative expression levels and sow them on 1/2MS medium. After normal growth for 7 days, move them to the soil to continue growing. Observe the growth status of the tobacco, record the height and diameter of each strain after stem extraction (45 days), calculate the growth rate, and observe the flowering time.
Stress resistance analysis of overexpressed tobacco
Sow each tobacco strain onto a 1/2MS medium containing 0.1 M NaCl (5.844 g/L) and 0.2 M (36.4344 g/L) mannitol, and record the seed germination rate. Sow wild-type seeds and overexpressed seeds on 1/2MS for 3 days. After the white heads of the seeds are exposed, transfer the seeds with the same conditions to 1/2MS medium containing 0.1 M NaCl and 0.2 M mannitol, place them vertically, and cultivate them in a constant temperature incubator at 25℃. After about 10 days of cultivation, measure the root length.
All strains of tobacco grown for fifteen days were subjected to stress treatment without watering or with water containing 0.2 M NaCl. After 15 days of treatment, the peroxidase (POD) activity and malondialdehyde (MDA) content of each strain of tobacco were measured. DAB and NBT were used to analyze in situ ROS accumulations, transgenic gametophytes were incubated in 1 mg/mL DAB solution (pH 3.8) or NBT solution (10 mM phosphate buffer, pH 7.8) for 6–8 h, then boiled ethanol (98%) for 10 min to remove chlorophyll. To further validate the role of LoNAC3 in abiotic stress, six genes (NtSOD, NtPOD, NtCAT, NtSOS, NtGST, NtAPX) related to ROS clearance, were selected to detect their relative expression levels (Table 1).
Results
Isolation and bioinformatics analysis of LoNAC3
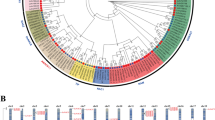
Using the cDNA of L. olgensis as a template, the PCR amplification product was located correctly with clear bands. The sequence information of LoNAC3 gene was obtained through gel recovery sequencing and uploaded to NCBI (GenBank: MW206689). The gene has a length of 1185 bp and encodes 394 amino acids. NCBI conserved structure analysis revealed the presence of a NAM conserved domain in LoNAC3, which is located at the N-terminus (from 12 to 139) of the gene. According to the subfamily classification of Ooka H et al. [12], LoNAC3 belongs to the OSNAC7 subgroup through evolutionary tree and homology analysis with Arabidopsis NAC family proteins (Fig. 1A). Phylogenetic analysis shows that LoNAC3 shares similar homology with Pinus taeda, Picea sitchensis, and Picea wilsonii (Fig. 1B). Motif analysis results also confirmed that the N-terminus is more conserved and the C-terminus exhibits diversity (Fig.S1), consistent with the structural characteristics of the NAC family.
According to the physical and chemical properties prediction on the Protparam website, the theoretical molecular weight of LoNAC3 protein is 45659.20 Da (1Da = 1 g.mol−1), and the predicted isoelectric point is 6.53. The negatively charged residues in the amino acid composition are dominant, and the instability coefficient is greater than 40. It is an unstable protein with a hydrophobicity of -0.851. The subcellular localization prediction results of WOLF PSORT show that LoNAC3 is mainly concentrated in the nucleus. GOR4 predicted its secondary structure and found that the LoNAC3 protein is mainly randomly curled, with 21.07% α-helices, 15.23% extension chains, 63.71% random coils, and lack β-turns, which has some similarity with the identified NAC gene of L. olgensis [26]. The SwissMode homologous modeling method was used to predict the tertiary structure of LoNAC3 (Fig. 1B), and the aligned model was A0A5S9HI04.1.A NAC transcription factor(gene: PtaVNS5, organism: Pinus taeda (Loblolly pine)), with a sequence identity of 95.41%.
Cloning and cis-acting elements analysis of the LoNAC3 promoter
Using NCBI genome data of L. kaempferi as a reference, primers were designed to clone the sequence information of the approximately 2000 bp fragment before the LoNAC3 start codon using the entire gDNA of L. olgensis as a template. Select the correctly sequenced LoNAC3pro::GUS Escherichia coli positive strain plasmids, transfer them into Agrobacterium receptive cells using heat shock method, apply plates, select single colonies for expansion culture, and use the bacterial solution as a template for PCR detection. The target bands are clear (Fig.S2), and the recombinant plasmid has been successfully transformed into Agrobacterium. By infecting tobacco, it was found that the LoNAC3 promoter can initiate GUS expression in tobacco, with promoter activity, which is weaker than the 35 S promoter. Under stress treatment, the LoNAC3 promoter exhibited stronger activity (Fig. 2B).
Analysis of LoNAC3 promoter and gene expression patterns. A The cis-acting elements in LoNAC3 promoter sequence. B The results of tobacco GUS staining. C Expression analysis of LoNAC3 in different growth stages of L. olgensis. D Expression analysis of LoNAC3 under different stress treatment. E Expression analysis of LoNAC3 under different hormone treatment
The prediction results on the PlantCARE website show that in addition to multiple common cis-acting element in promoter and enhancer regions (CAAT box), core promoter element around − 30 of transcription start (TATA box), there are also multiple MYB recognition sites on the LoNAC3 promoter. Analysis found that there are two cis-acting element involved in defense and stress responsiveness, as well as multiple cis-acting regulatory element involved in light responsiveness (G-Box) on the promoter. In addition, there are also multiple hormone related elements, three cis-acting element involved in the abscisic acid responsiveness, two cis-acting regulatory element involved in the MeJA-responsiveness and one gibberellin-responsive element (Fig. 2A).
Analysis of LoNAC3 gene expression patterns
Through the analysis of qRT-PCR results, it can be seen that LoNAC3 has the highest relative expression level in the leaves of L. olgensis at three time periods (Fig. 2C). Under abiotic stress treatment, the relative expression level of NaHCO3 showed little change, and the expression level was down regulated at 24 and 96 h; the changes in relative expression levels were relatively small at 12 h and 24 h under NaCl and PEG treatment, but were more significantly upregulated at 48 h and 96 h, reaching a tenfold increase (Fig. 2D). Under hormone treatment, the relative expression level of LoNAC3 induced by different hormones varied. The expression level of LoNAC3 was lower at several time points under IAA and 6-BA treatment than that of WT; The relative expression level of LoNAC3 gene under GA3 treatment reached a maximum of 1.99 times at 12 h; the relative expression levels of ABA and MeJA under two hormone treatments showed significant changes, reaching their maximum at 24 h, reaching 3.60 and 8.16 times, respectively (Fig. 2E).
Obtaining and phenotypic observation of overexpressed tobacco
The transgenic resistant buds obtained through Agrobacterium mediated leaf disc transformation grew well on a medium containing hygromycin (25mg.L−1) and were reinduced into seedlings (Fig. 3A-D). Extracting tobacco leaf DNA for PCR identification, there were clear and correctly positioned target bands (Fig. 3F), and the LoNAC3 gene was successfully integrated into the DNA of resistant tobacco seedlings. Collect the seeds of positive seedlings and screen them for hygromycin. The resistant seedlings grew well but the wild-type was unable to grow (Fig. 3E). After 2 generations of resistance screening, the T2 generation resistant tobacco seeds were harvested, and the T3 generation was seeded on 1/2MS medium. After 10 days, RNA was extracted from the entire tobacco plant of each strain. RNA levels were identified through RT-qPCR, and all positive strains were upregulated (Fig. 3H). Select OE3, OE4, and OE7 with the highest relative expression levels for the next experiment.
Obtaining and phenotypic observation of overexpressed tobacco. A Co-cultivation. B Induced resistant differentiated buds. C Rooted tissue culture seedlings. D Transplanted into soil. E Selection of transgenic tobacco seed resistance (25mg.L−1 Hyg). F PCR detection of VB191104-LoNAC3 transgenic tobacco, M: DL2000 Marker, 1: negative control (DNA of wild-type tobacco), 2: positive control (Agrobacterium tumefaciens of VB191104-LoNAC3), 3 ~ 11: DNA of OE1 ~ OE9(The original, unprocessed gel image can be found in the additional file.). G Phenotype of tobacco. H Relative expression levels of VB191104-LoNAC3 transgenic tobacco. I Plant height of tobacco with different asexual strains. J Ground diameter of tobacco with different asexual strains. Significance was determined by the least significant difference, asterisks indicate statistically significant differences from WT (*P < 0.05, **P < 0.01, ***P < 0.001)
Compared to the wild-type, overexpressed tobacco showed a situation of plant dwarfism (Fig. 3G). Throughout the growth process, the plant height of overexpressed tobacco was lower than that of wild-type, and the ground diameter of overexpressed tobacco was also smaller than that of wild-type (Fig. 3I-J). As the tobacco grew, the ground diameter difference gradually decreased. Within 45–90 days, the growth rate of plant height was lower than that of wild-type plants, while the growth rate of ground diameter was higher than wild-type plants, and flowering occurred earlier than wild-type plants.
Analysis of stress resistance of tobacco seeds
The germination rate of tobacco seeds sown on 1/2MS medium was almost 100%, while the germination rate of tobacco seeds sown on 1/2MS medium containing 0.2 M NaCl and 0.3 M mannitol was almost zero. Observing the germination rate of various tobacco strains sown on a 1/2MS medium containing 0.1 M NaCl (5.844 g.L−1) and 0.2 M mannitol (36.4344 g.L−1), it was found that the germination rate of overexpressed LoNAC3 tobacco was lower than that of wild-type tobacco (Fig. 4C-D), and wild-type tobacco seedlings showed better growth status (Fig. 4A-B). In addition, after stress treatment, the root length of overexpressed tobacco was shorter than that of wild-type tobacco (Fig. 4E-F).
Analysis of stress resistance in genetically modified tobacco seeds. A Sowing tobacco seeds in 1/2MS medium containing 0.2 M mannitol for 10 days. B Sowing tobacco seeds in 1/2MS medium containing 0.1 M NaCl for 10 days. C The germination rate of Tobacco in 1/2MS medium with 0.2 M mannitol. D The germination rate of Tobacco in 1/2MS medium with 0.1 M NaCl. E-F Root length of tobacco under stress treatment. Significance was determined by the least significant difference, asterisks indicate statistically significant differences from WT (*P < 0.05, **P < 0.01, ***P < 0.001)
Analysis of stress resistance of tobacco
Drought and salt stress can lead to the accumulation of reactive oxygen species, causing damage to plants. Under normal growth conditions, there was no significant difference in POD activity and MDA content between transgenic strains and wild-type tobacco. DAB and NBT staining showed only slight staining of plant leaves, but after treatment, transgenic strains showed weaker POD activity and lower MDA content (Fig. 5A). Different strains of tobacco also experienced varying degrees of deepening of staining, while overexpressing tobacco showed significantly deeper staining, indicating that it accumulated more hydrogen peroxide and superoxide ions (Fig. 5B). Similarly, under normal circumstances, the expression level of genes is low and there is no significant difference between different strains. After stress treatment, the expression level of genes increases, and the growth of most genes in transgenic tobacco is lower than that in wild-type tobacco (Fig. 5C).
Analysis of stress resistance in tobacco. A Determination of POD activity and MDA content in various tobacco strains before and after treatment. B DAB and NBT staining of tobacco leaves before and after treatment. C Detection of ROS-related gene expression levels before and after treatment. Significance was determined by the least significant difference, asterisks indicate statistically significant differences from WT (*P < 0.05, **P < 0.01, ***P < 0.001)
Discussion
Bioinformatics analysis of LoNAC3
Through bioinformatics analysis, LoNAC3 is a full-length gene sequence containing a NAM specific domain which located at the N-terminus. In terms of instability coefficient, LoNAC3 greater than 40 is an unstable protein, and the LoNAC3 protein may appear in stages. The predicted hydrophobicity of the protein is negative, suggesting that it is a hydrophilic protein. Through sequence alignment of NCBI, it was found that Picea wilsonii NAC transcription factor PwNAC30 with high homology to LoNAC3, negatively regulates the abiotic stress tolerance of transgenic Arabidopsis [29]. LoNAC3 belongs to the OSNAC7 subgroup through evolutionary tree and homology analysis with Arabidopsis NAC family proteins. ANAC012 (NST3) in the same subgroup negatively regulates the development of Arabidopsis wood fibers [30], and ANAC033 (SMB) is involved in the development of Arabidopsis root cap [31]. It is speculated that LoNAC3 may also be involved in the stress resistance and development processes related to L. olgensis. Further experimental analysis is needed for this conclusion.
Expression analysis of LoNAC3 gene and promoter
Through the analysis of qRT-PCR results, it can be seen that LoNAC3 can detect fluorescence signals in different parts of L. olgensis at three stages, indicating that the specific fragments of LoNAC3 have been successfully amplified in these parts, and LoNAC3 is expressed at all stages. LoNAC3 has the highest relative expression level in leaves, but overall, the difference in expression levels among different parts is not significant. Under abiotic stress, the relative expression level of LoNAC3 indicates that it is more likely to participate in drought and salt stress responses, and is more likely to participate in later regulation. Under several different hormone treatments, LoNAC3 was induced to varying degrees of expression. The relative expression levels varied significantly under MeJA and ABA hormone treatments, suggesting a greater correlation between LoNAC3 and these two hormones.
Due to the lack of genomic data information of L. olgensis, different from using RACE technology to clone promoter sequence information, this experiment designed primers based on NCBI L. kaempferi genomic data, cloned the LoNAC3 promoter sequence using L. olgensis gDNA as a template, and constructed the LoNAC3pro:: GUS plasmid. The LoNAC3 promoter can initiate GUS expression in tobacco and has promoter activity. The analysis of cis-acting elements in the promoter shows that there are three ABA responsive elements and two MeJA responsive elements on the LoNAC3 promoter.
ABA plays a crucial role in stress response and regulates various developmental processes such as seed maturation and dormancy, organ abscission, and leaf senescence in plants [32, 33]. Methyl jasmonic acid (MeJA) is a member of the jasmonic acid family. JAs not only play an important role in defense responses to biological stress [34], but also participate in a series of plant responses to abiotic stress [35,36,37]. Both hormones play important roles in various plant activities, and expression analysis results indicate that the LoNAC3 gene is likely associated with two hormone signals, and the response mechanism needs further research.
Preliminary analysis of LoNAC3 gene function
Although in a number of conifer species, progress has been made regarding somatic embryogenesis research, only a handful of somatic embryogenesis systems can be applied for practical production [38]. The stability of the larch somatic embryogenesis system is very poor, making it almost impossible to cultivate a large number of transgenic larch seedlings in the short term. In order to preliminarily analyze the function of the LoNAC3 gene, overexpressing LoNAC3 tobacco was used as plant material for experiments. This study found that transgenic tobacco exhibited stunted growth and poorer performance under stress. Under drought and salt stress, overexpressing LoNAC3 tobacco accumulates more reactive oxygen species and MDA content, while exhibiting weaker peroxidase activity.
Many reports indicate that NAC TFs are actively involved in the regulation of ROS metabolism and induction of important ROS scavenging enzymes. Under salt stress, overexpression of LpNAC17 in tobacco increases plant stress resistance through enhanced peroxidase activity [39]. MusaATAF2 overexpression in banana plants downregulates the ROS scavenging genes, leading to accumulation of H2O2 and inducing plant senescence [40]. MpSNAC67 is the main regulator of stress-induced aging in banana plants, triggering SA dependent ROS generation during senescence [41]. Transgenic MusaSNAC1 and MusaNAC68 lines displayed lower oxidative damage caused by ROS in terms of lower MDA content than control plants suggesting the drought protective function of MusaSNAC1 [42] and MusaNAC68 [43]. This study found that overexpressing LoNAC3 tobacco also exhibited weaker peroxidase activity under drought and salt stress, suggesting that LoNAC3 may participate in plant growth and development through a similar regulatory network.
Conclusion
This experiment cloned a gene with a NAM specific conserved domain named LoNAC3 from L. olgensis and obtained its promoter sequence information. Bioinformatics analysis identified LoNAC3 as a NAC family gene belonging to the OSNAC7 subgroup, and speculated that its function is related to the secondary wall. Through the analysis of LoNAC3 sequence and promoter expression, it can be concluded that LoNAC3 is involved in the drought and salt stress response processes of L. olgensis, and is induced by ABA and MeJA expression. Overexpression of LoNAC3 leads to stunted tobacco growth and negatively regulates its tolerance to drought and salt stress through the reactive oxygen species pathway.
Availability of data and materials
Data will be made available on request.
Data availability
Data is provided within the manuscript or supplementary information files. Data will be made available on request.
References
Zhang Y, Xu J, Li R, Ge Y, Li Y, Li R. Plants’ Response to Abiotic Stress: Mechanisms and Strategies. Int J Mol Sci. 2023;24(13). https://doi.org/10.3390/ijms241310915.
Schuppler U, He PH, John PC, Munns R. Effect of water stress on cell division and cell-division-cycle 2-like cell-cycle kinase activity in wheat leaves. Plant Physiol. 1998;117(2):667–78. https://doi.org/10.1104/pp.117.2.667.
van Zelm E, Zhang Y, Testerink C. Salt tolerance mechanisms of plants. Annu Rev Plant Biol. 2020;71:403–33. https://doi.org/10.1146/annurev-arplant-050718-100005.
M FEDERICO GIACOMO G. Stomatal, mesophyll conductance and biochemical limitations to photosynthesis as affected by drought and leaf ontogeny in ash and oak trees. Plant Cell Environ. 2005;28:834–49.
Reddya AR, Chaitanyaa KV, Vivekanandanb M. Drought-induced responses of photosynthesis andantioxidant metabolism in higher plants. J Plant Physiol. 2004;161(11):1189–202. https://doi.org/10.1016/j.jplph.2004.01.013.
Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48(12):909–30. https://doi.org/10.1016/j.plaphy.2010.08.016.
Baxter A, Mittler R, Suzuki N. ROS as key players in plant stress signalling. J Exp Bot. 2014;65(5):1229–40. https://doi.org/10.1093/jxb/ert375.
Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2020;290(5499):2105–10. https://doi.org/10.1126/science.290.5499.2105.
Hu R, Qi G, Kong Y, Kong D, Gao Q, Zhou G. Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. Bmc Plant Biol. 2010;10:145. https://doi.org/10.1186/1471-2229-10-145.
de Oliveira TM, Cidade LC, Gesteira AS, Coelho Filho MA, Soares Filho WS, Costa MGC. Analysis of the NAC transcription factor gene family in citrus reveals a novel member involved in multiple abiotic stress responses. Tree Genet Genomes. 2011;7(6):1123–34. https://doi.org/10.1007/s11295-011-0400-8.
Kikuchi K, Ueguchi-Tanaka M, Yoshida KT, Nagato Y, Matsusoka M, Hirano HY. Molecular analysis of the NAC gene family in rice. Mol Gen Genet. 2000;262(6):1047–51. https://doi.org/10.1007/pl00008647.
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. Dna Res. 2003;10(6):239–47. https://doi.org/10.1093/dnares/10.6.239.
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9(6):841–57. https://doi.org/10.1105/tpc.9.6.841.
Souer E, van Houwelingen A, Kloos D, Mol J, Koes R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85(2):159–70. https://doi.org/10.1016/s0092-8674(00)81093-4.
Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10(2):79–87. https://doi.org/10.1016/j.tplants.2004.12.010.
Hegedus D, Yu M, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D. Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol Biol. 2003;53(3):383–97. https://doi.org/10.1023/b:plan.0000006944.61384.11.
Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MACJ, de Vries SC. TheCUP-SHAPED COTYLEDON3 gene is required for Boundary and shoot Meristem formation in Arabidopsis. Plant Cell. 2003;15(7):1563–77. https://doi.org/10.1105/tpc.012203.
Xie Q, Frugis G, Colgan D, Chua NH. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000;14(23):3024–36. https://doi.org/10.1101/gad.852200.
Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, Zou HF, Lei G, Tian AG, Zhang WK, Ma B, Zhang JS, Chen SY. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011;68(2):302–13. https://doi.org/10.1111/j.1365-313X.2011.04687.x.
Tran LP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a Drought-Responsivecis-element in theearly responsive to dehydration stress 1 Promoter[W]. Plant Cell. 2014;16(9):2481–98. https://doi.org/10.1105/tpc.104.022699.
Lu P, Chen N, An R, Su Z, Qi B, Ren F, Chen J, Wang X. A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol Biol. 2006;63(2):289–305. https://doi.org/10.1007/s11103-006-9089-8.
Xu ZY, Kim SY, Hyeon DY, Kim DH, Dong T, Park Y, Jin JB, Joo SH, Kim SK, Hong JC, Hwang D, Hwang I. The Arabidopsis NAC Transcription Factor ANAC096 Cooperates with bZIP-Type transcription factors in dehydration and osmotic stress responses. Plant Cell. 2013;25(11):4708–24. https://doi.org/10.1105/tpc.113.119099.
Liu C, Wang B, Li Z, Peng Z, Zhang J. TisAC1key transcriptionifactorFactor in Abiotic Stress ResistancgrowthGrowth. Plant Physiol. 2018;176(1):742–56. https://doi.org/10.1104/pp.17.01089.
Zhang H, Cui X, Guo Y, Luo C, Zhang L. Wilsoniilsonii transcription factor NAC2 enhanced plant tolerance to abiotic stress and participated in RFCP1-regulated flowering time. Plant Mol Biol. 2018;98(6):471–93. https://doi.org/10.1007/s11103-018-0792-z.
Akiyoshi N, Nakano Y, Sano R, Kunigita Y, Ohtani M, Demura T. Involvement of VNS NAC-domain transcription factors in tracheid formation in Pinus taeda. Tree Physiol. 2020;40(6):704–16. https://doi.org/10.1093/treephys/tpz106.
Cao Q, An P, Zhang S, Wang J, Zhang H, Zhang L. Preliminary analysis of two NAC transcription factor expression patterns in Larix olgensis. J Forestry Res. 2022;33(2):601–9. https://doi.org/10.1007/s11676-021-01331-x.
An P, Wang C, Cao Q, Zhao Q, Qin R, Zhang L, Zhang H. Genetic transformation and growth index determination of the Larix olgensis LoHDZ2 transcription factor gene in tobacco. Sci Rep. 2021;11(1):20746. https://doi.org/10.1038/s41598-021-99533-0.
Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. https://doi.org/10.1093/nar/29.9.e45.
Liang K, Wang A, Yuan Y, Miao Y, Zhang L. Wilsoniilsonii NAC TranscrifactorFactor PwNAC30 NegatregulatesuabioticbstressStoleranceerantransgenicsgenic Arabidopsis. Plant Mol Biol Rep. 2020;38(4):554–71. https://doi.org/10.1007/s11105-020-01216-z.
Ko JH, Yang SH, Park AH, Lerouxel O, Han KH. ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J. 2007;50(6):1035–48. https://doi.org/10.1111/j.1365-313X.2007.03109.x.
Fendrych M, Van Hautegem T, Van Durme M, Olvera-Carrillo Y, Huysmans M, Karimi M, Lippens S, Guérin CJ, Krebs M, Schumacher K, Nowack MK. Programmed cell death controlled by ANAC033/SOMBRERO determines Root Cap Organ size in Arabidopsis. Curr Biol. 2014;24(9):931–40. https://doi.org/10.1016/j.cub.2014.03.025.
Chandler P. Gene expression regulated by Abscisic Acid and its relation to stress tolerance. Annu Rev Plant Physiol Plant mol biol. 1994;45(1):113–41.
Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–79. https://doi.org/10.1146/annurev-arplant-042809-112122.
Cao S, Zheng Y, Yang Z, Tang S, Jin P, Wang K, Wang X. Effect of methyl jasmonate on the inhibition of Colletotrichum acutatum infection in loquat fruit and the possible mechanisms. Postharvest Biol Tec. 2008;49(2):301–7. https://doi.org/10.1016/j.postharvbio.2007.12.007.
Hildmann T, Ebneth M, Pena-Cortes H, Sanchez-Serrano JJ, Willmitzer L, Prat S. General roles of abscisic and jasmonic acids in gene activation as a result of mechanical wounding. Plant Cell. 1992;4(9):1157–70. https://doi.org/10.1105/tpc.4.9.1157.
Mohamed HI, Latif HH. Improvement of drought tolerance of soybean plants by using methyl jasmonate. Physiol Mol Biol Pla. 2017;23(3):545–56. https://doi.org/10.1007/s12298-017-0451-x.
Bu Q, Hongling J, Chang-Bao LQZJ, Jiaqiang S, Xie Q, Chuanyou L. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-sig- naled defense responses. Cell Res. 2008;18:756–67.
Zhu J, Zhang K, Xiong H, Xie Y, Li R, Wu X, Yang Y, Wu H, Hao Z, Sun X. H2O2 significantly affects Larix kaempferi × Larix olgensis somatic embryogenesis. Int J Mol Sci. 2024;25:669. https://doi.org/10.3390/ijms25010669.
Wang Y, Cui Y, Liu B, Wang Y, Sun S, Wang J, Tan M, Yan H, Zhang Y. Lilium pumilum stress-responsive NAC transcription factor LpNAC17 enhances salt stress tolerance in tobacco. Fro Pla Sci. 2022. https://doi.org/10.3389/fpls.2022.993841.
Negi S, Tak H, Singh S, et al. MusaATAF2 like protein, a stress-related transcription factor, induces leaf senescence by regulating chlorophyll catabolism and H2O2 accumulation. Physiol Plant. 2022;174(1). https://doi.org/10.1111/ppl.13593.
Negi S, Bhakta S, Ganapathi T, et al. MpSNAC67 transcription factor of banana regulates stress induced senescence through salicylic acid dependent pathway. Environ Exp Bot. 2022;105104. https://doi.org/10.1016/j.envexpbot.
Negi S, Tak H, Ganapathi TR. A banana NAC transcription factor (MusaSNAC1) impart drought tolerance by modulating stomatal closure and H2O2 content. Plant Mol Biol. 2018. https://doi.org/10.1007/s11103-018-0710-4.
Negi S, Tak H, Ganapathi TR. Expression analysis of MusaNAC68 transcription factor and its functional analysis by overexpression in transgenic banana plants. Plant Cell Tiss Organ Cult. 2016;125:59–70. https://doi.org/10.1007/s11240-015-0929-6.
Acknowledgements
Not applicable.
Funding
This work was supported by the Science and Technology Innovation 2030-Agricultural Biological Breeding Major Project “Creation of Drought Resistant New Germplasm and Cultivation of New Varieties of Larch” (2023ZD040580204) and the grants from the Sci-Tech Innovation 2030 Agenda “Cultivation of New Varieties of High Quality and High Yield Pine Structural Timber” (No. 2023ZD0405903) .
Author information
Authors and Affiliations
Contributions
Q. C. and L. Z. conceived and designed the study. Q. C., T. Z. and L. L. performed the experiments. D. X., C. W. and Q. Z. analyzed the data. Q. C. and J. H. wrote the paper. H. Z. and L. Z. reviewed and edited the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Data is provided within the manuscript or supplementary information files. Data will be made available on request.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cao, Q., Hao, J., Zhang, T. et al. Isolation and functional analysis of the Larix olgensis LoNAC3 transcription factor gene. BMC Plant Biol 24, 881 (2024). https://doi.org/10.1186/s12870-024-05619-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-024-05619-y