Abstract
The NAC transcription factor family is plant-specific with various biological functions. However, there are few studies on the NAC gene involving coniferous species. Bioinformatics research and expression analysis of NAC genes in Larix olgensis can be used to analyse the function of the NAC gene in the future. Screening of excellent genetic materials and molecular breeding have been utilized to cultivate high-quality, stress-resistant larches. According to the transcriptome data for L. olgensis, the genes Unigene81490 and Unigene70699 with complete ORFs (open reading frames) were obtained by conserved domain analysis and named LoNAC1 and LoNAC2, respectively. The cDNAs of LoNAC1 and LoNAC2 were 1971 bp and 1095 bp in length, encoding 656 and 364 amino acids, respectively. The molecular weights of the proteins encoded by the two genes were predicted to be 72.61 kDa and 41.13 kDa, and subcellular localization analysis indicated that the proteins were concentrated in the nucleus. The results of real-time quantitative PCR analysis showed that at different growth stages and in different tissues of L. olgensis, the relative expression levels of the two NAC genes were highest in the stem, and the expression differences were more obvious in non-lignified tissues. After drought, salt and alkali stress and hormone treatment, expression was induced to different degrees. The expression levels of LoNAC1 and LoNAC2 in semi-lignified L. olgensis were higher than in the other two periods (non-lignified and lignified), and expression levels significantly increased under drought and salt stress. Relative expression levels changed under hormone treatment. It is speculated that these two genes may not only be related to drought and salt stress and secondary growth but may also be induced by hormones such as abscisic acid. Overall, LoNAC1 and LoNAC2 are genetic materials that can be used for molecular breeding of larch.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The NAC transcription factor family is plant-specific and widely distributed in terrestrial plants. It is also regarded as one of the gene families with the most transcription factors (Riechmann et al. 2000; Olsen et al. 2005). More than 8000 transcription factors of the NAC family have been found in plants, and 151 in Arabidopsis thaliana (L.) Heynh. alone (de Oliveira et al. 2011). This study found that the N-terminus of the NAC protein has a highly conserved domain, while the C-terminus has a transcription activation domain and presents diversity, which is an important recognition-related feature of the NAC protein structure (Aida et al. 1997). There are approximately 150 amino acids in the N-terminal domain of the NAC protein, which can be divided into five subdomains (Chen et al. 2019). The C-terminus has a simple amino acid sequence with high repetition and contains a greater number of Ser, Thr, and Glu and some acidic amino acid residues than the N-terminal domain (Olsen et al. 2005). By aligning the NAC protein sequences of Arabidopsis, researchers have shown that some common sequences can be found even at the C-terminus (Ooka et al. 2003).
In the NAC gene family, the Petunia NAM gene was the first one found to be related to plant morphogenesis (Souer et al. 1996). Researchers found that in Arabidopsis, the key factor regulating secondary wall thickening of xylem fibre cells was the specific expression of the NAC family NST1 and NST3/SND1 genes (Mitsuda et al. 2007). Huang et al. (2015) found that the genes NAC29 and NAC31 in rice are related to the regulation of cellulose synthesis. He et al. (2005) reported that the Arabidopsis transcription factor AtNAC2 was highly expressed in roots under high salt conditions, the lateral roots of plants overexpressing this gene were well-developed, and changes in auxin and ethylene were observed. Arabidopsis ANAC096 can help plants survive dehydration-related osmotic stress and is related to ABA-induced genes (Xu et al. 2013). Liu et al. (2018) noted that TsNAC1 can target an important proton transporter to improve salt tolerance. Researchers have studied the function of the NAC gene family but most of these have focused on plants such as Arabidopsis, tobacco, and rice. The function of NAC family of genes in coniferous species remains to be explored and verified.
Larix olgensis A. Henry, belonging to the larch genus of the pine family, is a fast-growing timber species as well as a species for soil and water conservation in China (Zhang 2012). With the advance of science and technology resulting in the genetic improvement of larch, molecular breeding has been combined with traditional breeding to improve the efficiency of genetic improvement and to accelerate the improvement process (Levee et al. 1997). In this study, two full-length NAC genes of L. olgensis were used, and their functions initially estimated from the expression levels of genes in different tissue parts and under drought and saline-alkali conditions and hormone treatment. This will provide the basis for the verification of the NAC gene in subsequent experiments and for the screening of genetic material by genetic engineering-based breeding.
Materials and methods
Plant materials
Seedlings of L. olgensis at different growth stages, non-lignified (approximately 60 days), semi-lignified (approximately 120 days) and lignified (180 days), were used as experimental material. Roots, stems and needles were separated, wrapped in aluminium foil, labelled, rapidly frozen in liquid nitrogen and stored at − 80 °C. The 180-day L. olgensis material was treated with solutions of PEG6000 (25% w/v), NaCl (250 m mol/L), and NaHCO3 (50 m mol/L). Samples were taken at 0 h, 12 h, 24 h, 48 h and 96 h (three samples for each treatment and time). Gibberellin A3 (GA3), abscisic acid (ABA), N-(phenylmethyl)-9H-purin-6-amine(6-BA), indole-3-acetic acid(IAA), 2,4-dichlorophenoxyacetic acid (2,4-D) and methyl jasmonate (MeJA), each at 50 mg/L, were used to treat samples (at approximately 90 days) for 2 h, 4 h, 8 h, 12 h, 24 h, 48 h, 72 h, and 96 h (Lv et al. 2014; Zhao 2019). Untreated L. olgensis seedlings were used as controls.
Prediction of gene sequence structure and function
Through the National Center for Biotechnology Information (NCBI) online tool Blastx, more than 10 candidate sequences of NAC genes obtained in the laboratory were compared, and the structural domains of the sequences were predicted and analysed with the online tool CD-search (Marchler-Bauer et al. 2015). Unigene81490 (LoNAC1) and Unigene70699 (LoNAC2) were selected with the NAC family conserved structural domain and complete open reading frame. MEGA5.0 software (Tamura et al. 2011) was used to construct the neighbour-joining tree. Amino acid sequences corresponding to full-length genes in Blastx and to several full-length genes with the highest similarity in the evolutionary tree were combined, and these were used to perform multiple sequence comparisons by BioEdit software. Protparam (https://web.expasy.org/cgi-bin/protparam/protparam) was used to predict and analyse the physicochemical properties of the protein encoded by the full-length NAC gene. GOR4 (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html) was used to predict the secondary structure, and WOLF PSORT (https://wolfpsort.hgc.jp/) and SwissModel (https://www.swissmodel.expasy.org/) to predict the subcellular localization and three-dimensional structure of the protein, respectively.
Analysis of LoNAC1 and LoNAC2 expression in L. olgensis
Total RNA was extracted using Universal Plant Total RNA Extraction Kit (BIOTEKE, Beijing, China), and cDNA obtained by reverse transcription of total RNA was used as a template (ReverseScript RT reagent Kit, TaKaRa). Primer Premier 5.0 software designed the qRT-PCR primers (Table 1), the primers specifically screened by gel electrophoresis detection. LoTublin was selected as a reference gene and amplification was performed automatically according to the Real Master Mix (SYBR Green) kit instructions and an ABI 7500 real-time PCR instrument was used. Three replicates were set up in the quantitative PCR instrument and the results analysed by the 2−ΔΔCt method (Relative Expression = 2−((Ct gene−Ct LoTublin)sample−(Ct gene−Ct LoTublin)control) = 2−(ΔCt sample−ΔCt control) = 2−ΔΔCt) (Pfaffl 2001).
Results
Identification of LoNAC1 and LoNAC2
The NCBI online tool blastx was used to compare the genes from the L. olgensis transcriptome database obtained in the laboratory. Two genes, Unigene81490 and Unigene70699, with the NAC family special conserved domain (NAM), were obtained and named LoNAC1 and LoNAC2, respectively. The cDNAs were 1974 bp and 1098 bp in length, encoding 657 and 365 amino acids, respectively (Fig. 1).
Predicted physicochemical properties of LoNAC1 and LoNAC2 proteins
According to the prediction of physical and chemical properties, the theoretical molecular weights of the LoNAC1 and LoNAC2 proteins are 72.61 kDa and 41.13 kDa (1 kDa = 1000 Da = 1000 g/mol), respectively, and the predicted isoelectric points are 4.99 and 4.79, respectively. The LoNAC1 and LoNAC2 proteins contain 63 and 40 positively charged amino acids, respectively, and 91 and 61 negatively charged amino acids, respectively. The instability coefficient of the LoNAC1 gene is 38.13, the instability coefficient of the LoNAC2 gene is 50.12, and the protein hydrophobicity of the two sequences is − 0.460 and − 0.667, respectively. Subcellular localization analysis predicted that the proteins were concentrated in the nucleus.
Using the GOR4 webpage to predict the secondary structure of the two genes (Fig. 2) showed that they also have certain similarities in secondary structure. LoNACl and LoNAC2 proteins are mainly composed of random coils. LoNAC1 and LoNAC2 contain 21.31% and 26.58% α-helices, 18.57% and 16.44% extension chains, and 60.12% and 56.99% random coils, respectively, and lack β-turns. SwissModel homology modelling was used to predict the tertiary structure of the two proteins. As shown in Table 2 and Fig. 3, the N-terminus of LoNAC1 and LoNAC2 proteins have a highly conserved domain (NAM), which is an important recognition-related feature of the NAC protein structure and the prediction results for the tertiary structure of the two proteins were very similar.
Sequence alignment and evolution tree analysis of LoNAC1 and LoNAC2 proteins
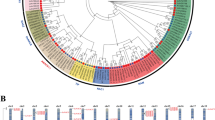
LoNAC1 and LoNAC2 were translated to obtain the amino acid sequence and the sequence was compared with the sequences of NAC family proteins of Arabidopsis thaliana by MEGA5.0 software to construct a phylogenetic tree (Fig. 4). Figure 4 shows that LoNAC1 and LoNAC2 of L. olgensis and AtNAC053, AtNAC078, AtNAC082, and AtNAC103 of Arabidopsis thaliana are clustered on the same branch. It is speculated that these genes are relatively similar in evolutionary kinship. Using BioEdit software, LoNAC1 and LoNAC2 were multiplexed and compared with the Arabidopsis NAC members AtNAC053, AtNAC078, AtNAC082, and AtNAC103 (Fig. 5). There is a highly conserved domain at the N-terminus of the amino acid, and this domain can be divided into multiple subdomains. The C-terminus shows diversity, a highly variable transcription activation region, which is consistent with the characteristics of NAC family transcription factors.
Analysis of the tissue expression patterns of LoNAC1 and LoNAC2
Different seedling growth stages, i.e., non-lignified (approximately 60 days), semi-lignified (approximately 120 days) and lignified (180 days) (Fig. 6) stored at − 80 °C was used to extract plant RNA and reverse transcribe it to the corresponding cDNA. The gene expression in different tissues (roots, stems, needles) was measured at different growth stages by qRT-PCR (Fig. 7). In the needles of L. olgensis, both genes had the highest relative expression levels during the semi-lignification period, and the expression levels at different stages were in the order: semi-lignified > lignified > non-lignified; among the stems, the relative expression of genes was also highest during the semi-lignification period, and the relative expression levels at different stages were in the order: semi-lignified > non-lignified > lignified. In the roots, the relative expression level was again highest in the semi-lignification stage, and the expression levels at different stages were in the order: semi-lignified roots > lignified roots > non-lignified roots, similar to the needles. In the three tissues, the two genes had the highest relative expression levels during the semi-lignification period. The relative expression levels in the roots were significantly different among the three growth stages (Fig. 7).
Expression analysis of LoNAC1 and LoNAC2 under abiotic stress
Under drought stress, the expression levels of LoNAC1 and LoNAC2 were upregulated at five time periods (Fig. 8). The relative gene expression levels in L. olgensis seedlings for 96 h reached the highest levels, increasing 12.7-fold (LoNAC1) and 12.8-fold (LoNAC2), respectively. Under salt stress, LoNAC1 was down regulated at 12 h and 24 h, while LoNAC2 was down regulated at 12 h. In addition, both expression levels were up regulated and the relative expression level reached the highest level at 96 h, approximately 10 times that before treatment. Under alkali stress, the relative expression levels of LoNAC1 at 12 h and 24 h and LoNAC2 at 24 h and 48 h were lower than in the controls, and expression was inhibited. Expression was induced at the remaining time periods, and the relative expression level was highest 96 h after treatment, reaching 2–3 times the value of the controls.
Analysis of the expression patterns of LoNAC1 and LoNAC2 under hormone induction
RT-PCR was used to measure gene expression in L. olgensis treated with six hormones (Fig. 9). At the same time, according to the results, the relative expression levels of LoNAC1 and LoNAC2 changed under different hormone induction conditions. Under IAA treatment, both LoNAC1 and LoNAC2 genes were up regulated at most time periods; LoNAC1 expression reached a maximum of approximately eightfold at 96 h, and LoNAC2 of approximately threefold at 96 h. Under MeJA treatment, LoNAC1 was only slightly up regulated at 96 h and LoNAC2 slightly up regulated at 4 h and 72 h. In addition, the expression of both genes was down regulated. Under 2,4-D treatment, LoNAC1 and LoNAC2 were both up regulated; LoNAC1 expression reached a maximum of 7.5-fold at 72 h, and LoNAC2 5.7-fold at 12 h. Under ABA treatment, both genes were up regulated, except for a slight down regulation at 4 h and 48 h, and their relative expression levels reached a maximum at 96 h, increasing 7.0- and 5.4-fold, respectively. Under GA3 treatment, both genes were up regulated, except for a slight down regulation at 24 h, and their relative expression levels reached a maximum at 2 h. Under 6-BA treatment, LoNAC1 was down regulated at 2 h, 4 h, 8 h, 12 h and 24 h, but was most highly down regulated at 8 h. LoNAC2 was up regulated, except for the down regulation observed at 8 h, and the relative expression reached a maximum at 48 h.
Discussion
Bioinformatics analysis of LoNAC1 and LoNAC2
Bioinformatics analysis showed that the full-length sequences of both the LoNAC1 and LoNAC2 genes contain special NAM domains. The coefficient of instability of LoNAC1 is less than 40, and that of LoNAC2 greater than 40, i.e., the LoNAC1 protein is stable and the LoNAC2 protein unstable. It is speculated that the LoNAC1 protein may be present for a long duration in L. olgensis, while the LoNAC2 protein may appear at certain stages. Both proteins had negative hydrophobicity results and are presumed to be hydrophilic proteins. The evolutionary tree and homology analysis of the amino acids encoded by Arabidopsis NAC family genes showed that LoNAC1 and LoNAC2 are clustered on the same branch with AtNAC053, AtNAC078, AtNAC082, and AtNAC103, and it is speculated that these genes are close in evolutionary kinship. Their structures and functions may be similar.
Under the condition of protein toxicity, AtNAC053 and AtNAC078 work together to activate the expression of many factors so that the plant produces sufficient protein homeostasis factors, such as the 26S proteasome to regulate the protein toxicity stress response. The AtNAC053 and AtNAC078 proteins play a role as central regulators in this process (Gladman et al. 2016). However, the induction of AtNAC103 expression in Arabidopsis depends on bZIP60, which participates in the growth and development of Arabidopsis and in the endoplasmic reticulum stress response (Sun et al. 2018). It is believed that the functions of LoNAC1 and LoNAC2 are similar and in the phylogenetic tree analysis, the affinity of LoNAC1 and LoNAC2 reached 97%. These two genes may cooperate in some regulatory mechanisms. Due to the differences in gene functions between different plants and the influence of distant and close relationships, this conclusion requires further study.
Analysis of LoNAC1 and LoNAC2 qRT-PCR results
Abiotic stress restricts the growth and development of plants, leading to a reduction in the yield and quality of agricultural and forestry crops. Research indicates that environmental stresses have caused almost half crop losses globally. Among environmental stress factors, drought is considered the most important factor restricting the development of global agriculture (Boyer 1982). As soil salinity becomes an increasingly serious problem, saline-alkali stress has also emerged as one of the factors that restrict crop growth. Plant hormones are a class of substances that regulate growth and development and are related to processes of plant environmental adaptation. They not only independently but also cooperatively regulate seed maturation, dormancy, and germination, vegetative and reproductive growth, and plant adaptation to abiotic and biological stresses during growth. Previous studies have found that NAC genes participate in both the stress response of plants(Lu et al. 2007; Mao et al. 2014), secondary growth (Wang et al. 2015) and also regulate plant cells and tissue death effects (Tran et al. 2009; Ma et al. 2018), which are related to hormone synthesis and regulatory networks (Fujita et al. 2004; Gao et al. 2010; Mao et al. 2017). NAC is a family of transcription factors with various biological functions discovered in recent research.
According to the analysis of the qRT-PCR results, during the non-lignification period, the distribution of LoNAC1 and LoNAC2 in different tissues varied greatly. There was little difference in the lignification period, and the two genes had the highest relative expression levels in the semi-lignification period, indicating that the genes may participate in the secondary growth of L. olgensis. Under drought and salt stresses, the relative expression levels of LoNAC1 and LoNAC2 were quite different, while under alkaline stress, the differences were relatively small. These results indicate that the genes LoNAC1 and LoNAC2 respond to drought and salt stress. Under hormone treatment, both LoNAC1 and LoNAC2 were induced to different degrees, and the relative expression levels in the 2,4-D, ABA and GA3 treatments were significantly different, suggesting that the expression of the two genes was highly correlated with these three hormones.
2,4-D is a representative, artificially synthesized plant hormone that is an auxin analogue and widely used as a growth regulator for some crops (Hu et al. 2019). Studies have shown that 2,4-D can delay senescence in citrus plants. The levels of many endogenous hormones changed, and defence-related genes and proteins were up regulated, which improved the ability of defence under adversity. Some NAC family genes were over expressed (Ma et al. 2014). ABA plays a vital role in stress responses and regulates various developmental processes such as seed maturation and dormancy, organ shedding, and leaf senescence (Erik and Stokstad 2010). There also exist ABA-dependent regulatory pathways that respond to various abiotic stresses such as drought, high salt stress, and cold stress (Yamaguchi-Shinozaki and Shinozaki 2005).
This study found that some NAC genes are related to the corresponding pathways of 2,4-D and ABA stress. For example, in 2,4-D-treated citrus plants, some NAC family genes were up regulated which improved the ability of defence under adversity (Hu et al. 2019). OsNAC52 may respond to ABA to increase drought tolerance in transgenic plants (Gao et al. 2010). In this study, analysis of the qRT-PCR results show that the expression of two NAC genes was significantly up regulated under drought and salt stress, and these genes were induced by treatment with the hormones 2,4-D and ABA. It may also be related regulatory networks in L. olgensis. The promoters of the two genes are currently unknown but will be identified by RACE technology in the future, and promoter sequence analysis will be completed to establish the regulatory network of these two genes.
Conclusion
LoNAC1 and LoNAC2 have special conserved NAM domains and it is believed that their functions are similar to those of NAC family genes. Analysis of the qRT-PCR results show that the two genes were induced by 2,4-D, ABA, and GA3 and participated in the secondary growth process of L. olgensis and in the response to drought and salt stress. These genes can be used as excellent genetic material for molecular breeding.
References
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9(6):841–857
Boyer JS (1982) Plant productivity and environment. Science 218(4571):443–448
Chen S, Lin X, Zhang D, Li Q, Zhao X, Chen S (2019) Genome-wide analysis of NAC gene family in Betula pendula. Forests 10(9):741
de Oliveira TM, Cidade LC, Gesteira AS, Coelho Filho MA, Soares Filho WS, Costa MGC (2011) Analysis of the NAC transcription factor gene family in citrus reveals a novel member involved in multiple abiotic stress responses. Tree Genet Genomes 7(6):1123–1134
Erik S (2010) Agriculture: biotech crops good for farmers and environment academy finds. Science 328(5976):295–295
Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LP, Yamaguchi-Shinozaki K, Shinozaki K (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39(6):863–876
Gao F, Xiong A, Peng R, Jin X, Xu J, Zhu B, Chen J, Yao Q (2010) OsNAC52, a rice NAC transcription factor, potentially responds to ABA and confers drought tolerance in transgenic plants. Plant Cell Tissue Organ Culture (PCTOC) 100(3):255–262
Gladman NP, Marshall RS, Lee K, Vierstra RD (2016) The proteasome stress regulon is controlled by a pair of NAC transcription factors in Arabidopsis. Plant Cell 28(6):1279–1296
He X, Mu R, Cao W, Zhang Z, Zhang J, Chen S (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44(6):903–916
Hu C, Zhao H, Shi J, Li J, Nie X, Yang G (2019) Effects of 2,4-dichlorophenoxyacetic acid on cucumber fruit development and metabolism. Int J Mol Sci 20(5):1126
Huang D, Wang S, Zhang B, Shang-Guan K, Shi Y, Zhang D, Liu X, Wu K, Xu Z, Fu X, Zhou Y (2015) A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 27(6):1681–1696
Levee V, Lelu MA, Jouanin L, Cornu D, Pilate G (1997) Agrobacterium tumefaciens-mediated transformation of hybrid larch (Larix kaempferi T L. decidua) and transgenic plant regeneration. Plant Cell Rep 16(10):680–685
Liu C, Wang B, Li Z, Peng Z, Zhang J (2018) TsNAC1 is a key transcription factor in abiotic stress resistance and growth. Plant Physiol 176(1):742–756
Lu PL, Chen NZ, An R, Su Z, Qi BS, Ren F, Chen J, Wang XC (2007) A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol Biol 63(2):289–305
Lv J, Rao J, Zhu Y, Chang X, Hou Y, Zhu Q (2014) Cloning and expression of lipoxygenase genes and enzyme activity in ripening persimmon fruit in response to GA and ABA treatments. Postharvest Biol Technol 92:54–61
Ma Q, Ding Y, Chang J, Sun X, Zhang L, Wei Q, Cheng Y, Chen L, Xu J, Deng X (2014) Comprehensive insights on how 2,4-dichlorophenoxyacetic acid retards senescence in post-harvest citrus fruits using transcriptomic and proteomic approaches. J Exp Bot 65(1):61–74
Ma X, Zhang Y, Turečková V, Xue G, Fernie AR, Mueller-Roeber B, Balazadeh S (2018) The NAC transcription factor SlNAP2 regulates leaf senescence and fruit yield in tomato. Plant Physiol 177(3):1286–1302
Mao X, Chen S, Li A, Zhai C, Jing R (2014) Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS ONE 9(1):e84359
Mao C, Lu S, Lv B, Zhang B, Shen J, He J, Luo L, Xi D, Chen X, Ming F (2017) A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol 174(3):1747–1763
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43(D1):D222–D226
Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19(1):270–280
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10(2):79–87
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10(6):239–247
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45
Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, Creelman R, Pilgrim M, Broun P, Zhang JZ, Ghandehari D, Sherman BK, Yu G (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290(5499):2105–2110
Souer E, van Houwelingen A, Kloos D, Mol J, Koes R (1996) The No Apical Meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85(2):159–170
Sun L, Wang X, Zhang Y, Sun W, Liu J, Song Z (2018) The resistance analysis of Arabidopsis thaliana transcription factor NAC-103 in the ER stress response. Mol Plant Breed 16(05):1475–1481
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Tran LP, Quach TN, Guttikonda SK, Aldrich DL, Kumar R, Neelakandan A, Valliyodan B, Nguyen HT (2009) Molecular characterization of stress-inducible GmNAC genes in soybean. Mol Genet Genom 281(6):647–664
Wang F, Lin R, Feng J, Chen W, Qiu D, Xu S (2015) TaNAC1 acts as a negative regulator of stripe rust resistance in wheat, enhances susceptibility to Pseudomonas syringae, and promotes lateral root development in transgenic Arabidopsis thaliana. Front Plant Sci 6:108
Xu ZY, Kim SY, Hyeon DY, Kim DH, Dong T, Park Y, Jin JB, Joo SH, Kim SK, Hong JC, Hwang D, Hwang I (2013) The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-Type transcription factors in dehydration and osmotic stress responses. Plant Cell 25(11):4708–4724
Yamaguchi-Shinozaki K, Shinozaki K (2005) Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci 10(2):88–94
Zhang L (2012) Research on Larix olgensis families' variation under drought stress. Northeast Forestry University, Doctoral theses, p. 106
Zhao Z (2019). Clonging and functionnal analysis of PeNAC10 gene in Phyllostachys edulis. Zhejiang A&F University, Doctoral theses, p. 89
Author information
Authors and Affiliations
Contributions
QC and LZ conceived and designed the study. QC, PA and SZ performed the experiments. QC wrote the paper. JW, HZ and LZ reviewed and edited the manuscript. All authors read and approved the manuscript.
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Project funding: This project was supported by the National Science and Technology Major Project (2018ZX08020003-001-001), the National Natural Science Foundation of China (Grant No. 31700595), and the Fundamental Research Funds for the Central Universities (2572019BA13), Heilongjiang Touyan Innovation Team Program.
The online version is available at http://www.springerlink.com
Corresponding editor: Yu Lei
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cao, Q., An, P., Zhang, S. et al. Preliminary analysis of two NAC transcription factor expression patterns in Larix olgensis. J. For. Res. 33, 601–609 (2022). https://doi.org/10.1007/s11676-021-01331-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-021-01331-x