Abstract
Background
The lifestyle transition from autotrophy to heterotrophy often leads to extensive degradation of plastomes in parasitic plants, while the evolutionary trajectories of plastome degradation associated with parasitism in hemiparasitic plants remain poorly understood. In this study, phylogeny-oriented comparative analyses were conducted to investigate whether obligate Loranthaceae stem-parasites experienced higher degrees of plastome degradation than closely related facultative root-parasites and to explore the potential evolutionary events that triggered the ‘domino effect’ in plastome degradation of hemiparasitic plants.
Results
Through phylogeny-oriented comparative analyses, the results indicate that Loranthaceae hemiparasites have undergone varying degrees of plastome degradation as they evolved towards a heterotrophic lifestyle. Compared to closely related facultative root-parasites, all obligate stem-parasites exhibited an elevated degree plastome degradation, characterized by increased downsizing, gene loss, and pseudogenization, thereby providing empirical evidence supporting the theoretical expectation that evolution from facultative parasitism to obligate parasitism may result in a higher degree of plastome degradation in hemiparasites. Along with infra-familial divergence in Loranthaceae, several lineage-specific gene loss/pseudogenization events occurred at deep nodes, whereas further independent gene loss/pseudogenization events were observed in shallow branches.
Conclusions
The findings suggest that in addition to the increasing levels of nutritional reliance on host plants, cladogenesis can be considered as another pivotal evolutionary event triggering the ‘domino effect’ in plastome degradation of hemiparasitic plants. These findings provide new insights into the evolutionary trajectory of plastome degradation in hemiparasitic plants.
Similar content being viewed by others
Background
Parasitic plants rely on their host to alleviated competition with other plant species for essential resources (e.g., light, water, soil nutrients), thereby enhancing their adaptive capacity in unfavorable environmental conditions; consequently, parasitism is generally perceived as a life strategy that evolves under natural selection and environmental pressures [1]. To date, approximately 4,750 parasitic species have been documented in angiosperms and it is estimated that the lifestyle transition from photosynthetic autotrophy to parasitism may have independently evolved at least 12 or 13 times in the angiosperm tree of life [1, 2]. Based on their photosynthetic capacity, parasitic plants are divided into two categories: hemiparasites and holoparasites [3,4,5]. Hemiparasites retain varying degrees of photosynthetic capacity while obtaining nutrients (primarily water and mineral elements) from their hosts. Contrarily, holoparasites exhaustively lose their photosynthetic capacity, with severely or entirely degraded leaves; therefore, they must completely rely on the host plants for energy and nutrients [3, 5, 6].
Chloroplasts, which originated from the endosymbiosis of photosynthetic autotrophic cyanobacteria within primitive eukaryotic cells, serve as vital centers for energy acquisition and carbohydrate production in plants [7, 8]. Due to their distinct origin, chloroplasts possess autonomous genomes and transcription/translation systems that synergistically interact with the nuclear genomes for plastid protein synthesis [8, 9]. Although the chloroplast genome (plastome) is highly conserved in most photosynthetic autotrophic angiosperms in terms of genome size, structure, and gene content, the lifestyle transition from autotrophy to heterotrophy might relax the purifying selection pressures that play a crucial role in maintaining the stability of plastomes [10], resulting in varying degrees of gene loss, pseudogenization, size reduction and structural rearrangement in parasitic plastomes [10,11,12,13,14,15,16]. Previous studies have demonstrated that different levels of reliance on host plants for energy and nutrient requirement can exert various intensities of selective pressures on the plastomes of parasitic plants; as a result, holoparasites exhibit higher levels of plastome degradation compared to their hemiparasitic relatives [13,14,15,16,17,18,19].
Hemiparasitic plants are further classified into facultative root-parasites and obligate stem-parasites based on their feeding modes and nutritional dependence on the hosts [20]. Considering the relatively higher degrees of trophic reliance of obligate stem-parasites on their hosts [21], purifying selection of plastid genes tends to be gradually relaxed during the transition from facultative parasitism to obligate parasitism. Therefore, obligate stem-hemiparasites are expected to have higher levels of plastome degradation than facultative root-hemiparasites [14, 22]. Surprisingly, this theoretical prediction has not yet been empirically validated, as previous comparative studies on the plastomes of Santalales hemiparasites revealed that obligate stem-parasites did not consistently exhibit a higher level of gene loss or pseudogenization than facultative root-parasites [23–25,]. Additionally, a previous study suggested that plastome degradation in hemiparasitic plants may follow a ‘domino effect’, where initial losses of plastid genes trigger chain reactions leading to subsequent gene losses [23]. However, the evolutionary mechanisms underlying the enigmatic ‘domino effect’ of plastome degradation in hemiparasitic plants remain elusive. These unresolved issues leave critical gaps in the exploration of evolutionary trajectories of plastome degradation in hemiparasitic plants.
Previous studies have demonstrated multiple independent evolutionary transitions from facultative root-parasitism to obligate stem-parasitism in Santalales [1, 24, 25], and proposed that plastome degradation in Santalales hemiparasites may have evolved in a lineage-specific manner [24,25,26]. This suggests that the degree of plastome degradation may exhibit substantial variation among phylogenetically distant facultative root-parasites, potentially surpassing the differences in plastome degradation observed between closely related root-parasites and obligate stem-parasites. Consequently, previous studies [24, 25, 27] on the retrogressive evolution of hemiparasitic Santalales plastomes may be flawed, as comparative analyses of plastomes based solely on lifestyle variations (facultative root-parasitism vs. obligate stem-parasitism) without considering phylogenetic relationships could lead to biased inferences.
The hemiparasitic lineage Loranthaceae, which includes approximately 76 genera and more than 1,000 species, is the largest family within Santalales [20, 28, 29]. Within Loranthaceae, three monotypic genera, Nuytsia, Atkinsonia, and Gaiadendron, are facultative root-parasites, while the remaining genera are obligate stem-parasites [20]. Given the monophyletic origin of obligate stem-parasitism from facultative root-parasitism in Loranthaceae [20, 29, 30], this family provides an ideal system to investigate whether obligate stem-parasites have a higher degree of plastome degradation than closely related facultative root-parasites. To gain a better understanding of the evolutionary trajectories of plastome degradation in hemiparastic plants, this study aims to (1) verify the theoretical prediction that the lifestyle transition from facultative root-parasitism to obligate stem-parasitism may lead to further degradations of hemiparasitic plastomes [14, 27] and (2) explore the plausible events that could have triggered the ‘domino effect’ in the plastome degradation among Loranthaceae hemiparasites. To achieve these objectives, we sequenced the plastomes of 22 Loranthaceae hemiparasites and combined them with publicly available data, yielding an extensive dataset comprising the plastomes of 48 species from 14 genera within this family. Under the framework of plastome phylogeny, we analyzed plastome degradation in each clade to determine whether there was lineage-specific gene loss, pseudogenization, or plastome structural variation within Loranthaceae. Additionally, we compared the plastome features of obligate stem-parasites with those of facultative root-parasites to explore differences in the degree of plastome degradation.
Materials and methods
Plant materials, illumina sequencing, plastome assembly and annotation
In total, 22 species from eight genera of Loranthaceae were collected from the wild (voucher specimens were deposited in the herbarium of Yunnan University and taxonomically identified by Dr. Min Deng), within voucher information being presented in Table S1. The genomic DNA of each sample was extracted from approximately 100 mg of silica gel-dried leaf tissues using a modified CTAB method [31]. The purified genomic DNA was use to prepare a paired-end Illumina sequencing library (with an average insert size of approximately 350 bp) for each sample, employing the TruSeq DNA Sample Prep Kit (Illumina, Inc., San Diego, CA, USA) following the manufacturer’s guidelines. Low-coverage genome sequencing was performed on an Illumina HiSeq 2500 platform to generate approximately four Gbp raw reads for each sample.
Raw Illumina reads were filtered using an NGS QC tool kit [32] to remove adaptors and reads with ambiguous bases. The complete plastome of each sample was de novo assembled using NOVOPlasty v.2.7.0 [33] with the k-mer size set at 31, and the large subunit of the RuBisCO gene (rbcL) of Taxillus sutchuenensis (MG999457) was utilized as the seed for plastome assembly. These assembled plastomes were annotated using PGA [34], and the boundaries of start/stop codons and introns/extrons of protein-coding genes were checked using Geneious v10.2 [35]. The tRNAscan-SE 1.21 [36]. was used to verify the transfer RNA (tRNA) genes using default parameters. All annotated plastomes were deposited in the NCBI GenBank database and their accession numbers are listed in Table S1. In addition to these newly sequenced plastomes, publicly available complete plastomes of 26 Loranthaceae species (Table S2) were obtained from the NCBI GenBank database for phylogenetic and comparative analyses. These publicly available plastomes were reannotated using the same method.
Phylogenetic analyses
In total, 48 species of Loranthaceae hemiparasites were sampled for phylogenetic analysis. Erythropalum scandens (Erythropalaceae), an autotrophic relative of Loranthaceae [1, 28, 30] was selected as the outgroup. Based on the plastome dataset, 55 plastid protein-coding genes (PCGs) commonly shared by these species were extracted from each plastome using Geneious v10.2 [35]. These PCGs were aligned separately using MAFFT v7.3.1 [37], and each CDS matrix was concatenated into a supermatrix (3,8153 bp) using Phylosuite [38]. Based on the concatenated matrix, maximum likelihood (ML) and Bayesian inference (BI) approaches were employed to infer the intrafamilial phylogenetic relationships of Loranthaceae. ML analysis was implemented by IQ-TREE V2.1.2 [39, 40] using UFBoot2 with the best-fit model TVM + F + R2 determined by Modelfinder [41]; 1,000 replications were adopted to calculate the standard bootstrap percentage for each node. MrBayes v3.2.7a [42] was used for the BI analysis. According to the Akaike information criterion (AIC), the best-fitting partitioning schemes and models were selected for each gene with default values using PartitionFinder v2.1.1 [43]. . The Markov chain Monte Carlo algorithm was run for five million generations with every 5,000 generations for tree sampling. Trees resulting from the first 25% of generations were discarded as “burn-in”, and the remaining trees were used to build the 50% majority-rule consensus tree and estimate the posterior probability values (PP). FigTree 1.4.6 [44] was used to present and edit the phylogenetic trees.
Comparative analyses of Loranthaceae plastomes
Genomic features, including genome size, gene content, and GC percentage of the whole plastome, LSC regions, SSC regions, and IR regions, were compared according to the sequence and annotation using Geneious v10.2 [35]. The boundaries of LSC, SSC, and IRs of each plastome were compared to check IR expansion/contraction in Loranthaceae plastomes using Geneious v10.2 [35] by comparison with their autotrophic relative, Erythropalum scandens. The microsyntenic structure of Loranthaceae plastomes was investigated using the multiple genome alignment tool, Mauve v2.3.1 [45], after removing one copy of the IR region for each plastome. Gene loss and pseudogenization events detected in each plastome were mapped to a phylogenetic tree to trace the evolutionary trajectory of plastome degradation associated with the evolution of the hemiparasitic lifestyle in Loranthaceae.
Results
Plastome features of Loranthaceae hemiparasites
The plastomes of these 48 Loranthaceae hemiparasites exhibited a typical quadripartite structure, consisting of a pair of inverted repeat regions (IRs, 20,118–26,801 bp) separated by a large single copy (LSC, 67,837–79,519 bp) and a small single copy (SSC, 5,059–7,719 bp) regions (Table 1). The lengths of these plastomes varied from 115,635 bp (Dendrophthoe pentandra) to 139,027 bp (Nuytsia floribunda), exhibiting a genomic size difference of 24,004 bp. The guanine and cytosine (GC) content of these Loranthaceae hemiparasites ranged from 35.9 to 37.8% (Table 1), and exhibited variations across the four regions. Specifically, the IR regions displayed the highest GC content (42.0–43.0%), followed by the LSC region (33.4–35.5%) and the SSC region (24.1–26.9%) (Table 1). The plastome of the autotrophic Erythropalum scandens harbored a total of 114 genes, comprising 79 protein-coding genes (PCGs), 30 tRNAs, four rRNAs, and one ycf15 pseudogene. In contrast, the plastomes of Loranthaceae species exhibited varying degrees of gene loss and pseudogenization, encompassing 92–100 plastid genes including 59–66 PCGs, 25–28 tRNAs, four rRNAs, and one to seven pseudogenes (Table 1). Among Loranthaceae taxa sampled in this study, the facultative root-parasite Nuytsia floribunda possessed a relatively larger plastome size (139,027 bp) compared to obligate stem-parasites ranging from 115,635 − 129,570 bp (Table 1), as well as a higher number of potentially functional plastid genes (98 vs.91–96) (Table 1).
Phylogenetic relationships
ML and Bayesian inference BI analyses generated identical tree topologies (Fig. 1), with most nodes being fully supported (BS = 100%, PP = 1.00). Consistent with previous studies [20, 29, 30], the plastome phylogeny resolved all Loranthaceae obligate stem-parasites as a well-supported monophyletic lineage (BS = 100%, PP = 1.00), which was sister to the facultative root-parasite Nuytsia floribunda (tribe Nutsiea). Within the clade of obligate stem-parasites, a close relationship between the two tribes, Elytranthaea and Loranthaea was recovered (BS = 100%, PP = 1.00). For those six subtribes of the tribe Loranthaea (Scurrulinae, Dendrophthoinae, Amyeminae, Tapinanthinae, Emelianthinae, and Loranthae) sampled in this study, our phylogenetic analyses resolved them as three successively diverging branches, corresponding to (1) the subtribe Loranthae (BS = 100%, PP = 1.00), (2) Dendrophthoinae (except for Helixanthera parasitica) + Dendrophthoinae + Amyeminae + Tapinanthinae + Emelianthinae (BS = 100%, PP = 1.00), and (3) Scurrulinae + Helixanthera parasitica (BS = 100%, PP = 1.00). As the genus Helixanthera did not aggregate into a single branch, both ML and BI phylogenies failed to resolve the subtribe Dendrophthoinae as monophyletic. Within the tribe Elytranthaea, the monophyly of the two genera Elytranthe and Macrosolen was not supported by the plastome phylogeny.
Phylogeny of Loranthaceae reconstructed by analyzing 55 plastid protein-coding genes (PCGs) using Bayesian inference (A) and Maximum likelihood (B) methods. Numbers on each node indicate bootstrap (BS) percentage /posterior probability (PP), with the asterisk (*) indicating full support in both two analyses (BS = 100; PP = 1.00).
Phylogeny-oriented comparative analysis of Loranthaceae plastomes
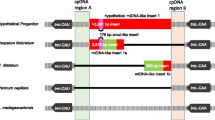
Except for a slight fragment translocation with a sequence length of 25 bp in Phyllodemis delavayi, microsynthetic analysis showed that no additional structural rearrangements occurred in these Loranthaceae plastomes (Fig. 2). Despite obvious variations in genome size (Table 1), these Loranthaceae plastomes exhibited a high level of conservatism with respect to their LSC/IRa, IRa/SSC, SSC/IRb, and IRb/LSC junctions (Fig. 3). Although Loranthaceae hemiparasites shared same SSC/IRb and IRb/LSC boundaries with the autotrophic relative Erythropalum sandens, the LSC/IRa boundaries of Loranthaceae plastomes moved into the rpl2 gene (Fig. 3). Within Loranthaceae, the SSC/IRa boundary of the facultative root-parasite Nuytsia floribunda moved into the rpl32 gene, whereas the SSC/IRa boundaries of all obligate stem-parasites were further contracted into the trnL-UAG gene (Fig. 3).
To trace the evolutionary trajectory of plastome degeneration associated with the formation of hemiparasitic lifestyle in Loranthaceae, the gene loss and pseudogenization events detected in each species were mapped to a phylogenetic tree (Fig. 4). As ycf15 was annotated as a pseudogene in all analyzed plastomes, this mutation may have occurred in the most recent common ancestor (MRCA) of Loranthaceae and Erythropalaceae (Fig. 4). Contrary to their autotrophic relative, Erythropalum scandens, all plastid NADH dehydrogenase-like complex genes, except for ndhB, two ribosomal protein genes (rps15 and rps16), and two tRNA genes (trnG-UCC and trnV-UAC), were absent from Loranthaceae plastomes, which may represent a molecular synapomorphy of the family (Fig. 4). Compared to the facultative root-parasite Nuytsia floribunda, additional deletion of rpl32 and ndhB genes might have taken place in the stem lineage ancestor of obligate stem-parasites (Fig. 4). Among Loranthaceae obligate stem-parasites, further loss of the infA gene is commonly observed in the tribe Elytranthaea (Fig. 4). Within the tribe Loranthaea, the pseudogenization of pasI and infA might have occurred in the stem lineage ancestor of the subtribe Loranthinae, and the losses of trnA-UGC and trnL-GAU likely occurred in the MRCA of the subtribes Dendrophthoinae (except for Helixanthera parasitica), Dendrophthoinae, Amyeminae, Tapinanthinae, and Emelianthinae (Fig. 4). Furthermore, the pseudogenization of plastid rpl16 gene could have taken place in the MRCA of the subtribes Scurrulinae and Helixanthera parasitica (Fig. 4).
Following the lineage-specific gene loss and pseudogenization events that occurred in the deep nodes of Loranthaceae tree topology, extensive gene loss or pseudogenization occurred independently in most shallow branches, such as the loss of trnA-UGC and trnI-GAU genes in Tolypanthus maclurei, Phyllodemis delavayi, Scurrula pulverulenta, Moquiniella rubra, Helicanthes elasticus, and Loranthus odoratus (Fig. 4). Additionally, independent deletion of trnA-UGC was observed in Taxillus caloreas, Loranthus guizhouensis, Loranthus grewinghii, Loranthus europaeus, and Loranthus tankae; independent loss of rpl36 was detected in Dendrophthoe pentandra, Helicanthes elasticus, and Plicosepalus acacia; and the independent loss of rpl33 was identified in Plicosepalus acacia, Plicosepalus curviflorum, and Cecarria abtusifolia (Fig. 4). Moreover, species-specific pseudogenization of psbZ and matK was detected in Cecarria abtusifolia (Fig. 4).
Discussion
Characterization of plastome degradation in Loranthaceae hemiparasites
A growing body of evidence suggests that the lifestyle transition from autotrophy to heterotrophy often results in rampant plastome degradation in parasitic plants, including the physical and functional loss of plastid genes, genome downsizing, and structural mutations [13, 14, 16, 23]. Consistent with this, the current study found that the plastomes of 48 Loranthaceae hemiparasites exhibited varying degrees of plastome size shrinkage, structural alteration, gene loss, and pseudogenization compared to their autotrophic relative, Erythropalum scandens. The findings indicate that although the hemiparasitic plants enable photosynthesis, their plastomes have experienced diverse modifications owing to the evolution of heterotrophic lifestyles.
Compared to their autotrophic relative, Erythropalum scandens, extensive gene loss and pseudogenization events were detected in the plastomes of Loranthaceae hemiparasites. Among them, Cecarria obtusifolia (116,508 bp), Dendrophthoe pentandra (115,635 bp), Helixanthera sampsonii (1206,58 bp), Loranthus europaeus (120,994 bp), Plicosepalus curviflorus (120,181 bp), Scurrula delavayi (119,914 bp), Scurrula pulverulenta (119,811 bp), and Taxillus caloreas (120,663 bp) possessed relatively smaller plastomes sizes and retained less functional plastid genes (91–93), suggesting that the plastome size shrinkage observed in Loranthaceae hemiparasites can be partially attributed to the physical or functional losses of plastid genes. Additionally, the deletion of 10 plastid ndh genes (ndhA, ndhC–K), two ribosomal protein genes (rps15 and rps16), and two tRNAs (trnG-UCC and trnV-GAC) was observed in all Loranthaceae hemiparasites. Deletion of seven ndh genes (ndhA, ndhD–I) and rps15 in the SSC region, and three ndh genes (ndhD, ndhJ, and ndhK) and trnG-UCC in the LSC region, is respected to result in a more pronounced contraction of these two single copy regions compared to the deletion of two genes (trnV-GAC and rps16) in the IR regions. Consequently, these structural alterations lead to shifts of LSC/IRa and SSC/IRa boundaries in Loranthiaceae plastomes.
Compared to Erythropalum scandens, gene loss and pseudogenization events occurring in the plastomes of Loranthaceae hemiparasites involved the following PCGs: ndhA–K, infA, rpl16, rpl32, rpl33, rpl36, rps15, rps16, psaI, psbZ, matK, ycf1, ycf2, ycf15, and rpoC2, as well as six tRNAs. Among them, only three PCGs (rps16, ycf2, and ycf15) and four tRNAs (trnA-UCC, trnH-GUG, trnI-GAD, and trnV-GAC) were located in the IR regions, whereas the remaining genes were located in the LSC and SSC regions. Although the plastomes of Loranthiaceae hemiparasites possessed lower GC content in the LSC and SSC regions than Erythropalum scandens, there were slight differences in the GC content in the IR regions. The higher levels of gene loss/pseudogenization and lower GC content in the LSC and SSC regions, in contrast to the lower levels of gene loss/pseudogenization and higher GC content in the IR regions observed in the plastomes of Loranthaceae hemiparasites, provide robust evidence for the theoretical prediction that loss/pseudogenization of plastid genes in parasitic plants may occur simultaneously with a decrease in GC content [16].
Previous studies have demonstrated that parasitic plants with low GC content in their plastomes tend to accumulate inversions and structural mutations around the IR regions [13, 14]. Although varying degrees of plastid gene loss occurred in Loranthiaceae hemiparasites, significant synteny was observed among their plastomes, suggesting that they did not undergo dynamic structural rearrangement after the formation of hemiparasitic lifestyle. Notably, compared with their autotropic relative, Erythropalum scandens (38.0%), the GC content of Loranthaceae plastome (35.9–37.8%) was only slightly decreased. The findings provide additional empirical evidence that a dramatic decrease in GC content is more likely to trigger the structural rearrangement of the parasitic plastomes [13, 14]. Due to a higher degree of plastid gene loss and reduction in GC content observed in the plastomes of holoplastic plants compared to hemiparasitic plants, the plastomes of holoparasites are more prone to independent structural rearrangement, as previous study demonstrated [46,47,48,49].
The gene contents of Loranthaceae plastomes and their autotrophic relative were compared based on the phylogenetic tree topologies. The results indicated that a total of 10 ndh genes (ndhA, ndhC–K), two ribosomal protein genes (rps15 and rps16), and two tRNA genes (trnG-UCC and trnV-UAC) might have been deleted in the stem lineage ancestor of Loranthaceae. The deleted ndh genes encode 10 subunits of the plastid NDH complex that mediates photosystem I cyclic electron transport and facilitates chlororespiration in plant cells [50]. The functional and physical loss of most plastid ndh genes has been commonly observed in a wide spectrum of parasitic angiosperm lineages and is regarded as an early response to the evolution of heterotrophic lifestyle [13, 14, 16]. Notably, this mutation has also been detected in a wide spectrum of photoautotrophic angiosperms [48, 51,52,53,54], and Frailey et al. proposed that the plastid ndh genes may have undergone negative selection in these photoautotrophic lineages [49]. Additionally, Lin et al. observed that several ndh genes encoded by the chloroplast and nuclear genomes may have been lost concomitantly in some epiphytic autotrophic orchids and proposed that this mutation may increase the possibility of evolving a heterotrophic lifestyle [19]. On this basis, we speculate that the massive reduction in the NDH pathway in the stem lineage ancestor of Loranthaceae might have played a pivotal role in triggering the transition from photoautotrophic to hemiparasitic lifestyles.
In addition to the loss of 10 plastid ndh loci, the deletion of two plastid ribosomal protein-encoding genes (rps15 and rps16) was inferred to have occurred in the stem lineage ancestor of Loranthaceae, and further loss/pseudogenization of other plastid ribosomal protein-encoding genes (rpl16, rpl32, rpl33, and rpl36) was commonly observed in Loranthaceae obligate stem-parasites. Notably, loss/pseudogenization of these plastid ribosomal protein-encoding genes has been detected not only in parasitic plants [55, 56], but also in a wide range of autotrophic angiosperms [57,58,59,60]. Although the rps15, rpl33, and rpl36 genes are not essential for chloroplast gene translation, the remaining genes play essential roles [61]. Hence, the loss of these plastid ribosomal protein-encoding genes in Loranthaceae may be compensated for by other plastid rpl/rps genes or by nuclear-encoded rpl/rps genes.
Plastid infA is another commonly reduced gene in the plastomes of Loranthaceae hemiparasites. While this gene was intact and retained in the facultative root-parasite Moquiniella rubra, the remaining Loranthaceae species (obligate stem-parasites) exhibited either loss or pseudogenization of this gene. This suggests that, after the evolution of a hemiparasitic lifestyle, plastid infA gene was recurrently deleted or pseudogenized in Loranthaceae obligate stem-parasites. Similarly, parallel losses of plastid infA genes have been observed in Santalales [24,25,26, 62] and a majority of holoparasitic plants [14, 16], as well as in diverse lineages of photoautotrophic angiosperms [16, 63]. Previous studies proposed that the plastid infA gene may have been constantly transferred to the nucleus [64]. Accordingly, horizontal transfer of the infA gene from plastomes to nuclear genomes may have occurred independently in Loranthaceae obligate stem-parasites.
In addition to the abovementioned protein-encoding genes, the deletion of six plastid tRNA genes was observed in the plastomes of Loranthaceae hemiparasites. It is well known that some heterotrophic plants generally contain only a fraction of tRNAs [14, 56, 65]. Despite the essential roles of tRNAs in plastid translation, the partial loss of these genes in plants may not represent a lethal mutation. Because of their relatively small size, tRNAs are easily transferred from the cytosol to plastid organelles [16], and photosynthetic plants most likely possess a specific transport mechanism that transports tRNAs to plastids from the cytosol [60, 65]. Accordingly, the loss of these tRNAs in Loranthaceae hemiparasites may not have a serious impact on their survival and is therefore tolerable.
New insights into evolutionary trajectories of plastome degradation in hemiparasitic plants
Along with the lifestyle transition from autotrophy to heterotrophy, parasitic plants developed numerous survival strategies to adapt to various environments, which might have led to diverse plastome degradation trajectories [15,16,17, 56, 66]. Among hemiparasitic plants, facultative root-parasites show a slight degree of trophic reliance on the host compared to obligate stem-parasites, which require host support water and mineral elements to complete their life cycle after germination [20, 21]. Theoretically, the varying nutritional dependence of facultative root-parasites and obligate stem-parasites on their hosts may influence the reductive evolution of their plastomes [14, 22]. Although this theoretical prediction seems plausible, it has not been validated by empirical studies, leaving a critical gap in the exploration of the evolutionary trajectory of plastome degradation in hemiparasitic plants.
In the current study, the plastome phylogeny resolved all obligate stem-parasties as a well-supported monophyletic lineage sister to the facultative root-parasite, Nuytia floribunda, confirming the monophyletic origin of obligate stem-parasitism from facultative root-parasitism in Loranthaceae proposed by previous studies [20, 29, 30]. Compared with their autotrophic relative, Erythropalum scandens, the plastomes of Loranthaceae hemiparasites exhibit various degrees of plastome downsizing and gene loss/pseudogenization. Among them, the plastome size of the early diverged facultative root-parasite, Nuytsia floribunda, was 139,027 bp, retaining 98 functional genes, which represents the largest plastome size and highest gene content in the Loranthaceae hemiparasites sampled in this study. Contrarily, the plastomes of all obligate stem parasites possessed relatively small genome sizes (115,635–129,570 bp) and retained relatively few functional plastid genes. Comparative analyses of Loranthaceae plastomes under the phylogenetic framework also showed that the stem lineage ancestor of obligate stem-parasites might have undergone more gene deletions (losses of rpl32 and ndhB genes) than the facultative root-parasite Nuytsia floribunda (pseudogenization of ndhB gene). Taken together, these results provide empirical evidence supporting the theoretical expectation that the evolution of obligate stem-parasitism from facultative root-parasitism might have caused higher levels of plastome degradation in hemiparasites [14, 22].
Nevertheless, the taxonomic sampling in this study is still subject to certain limitations. Specifically, the Loranthaceae family has been documented to include three monotypic genera (Nuytsia, Atkinsonia, and Gaiadendron), each hosting a facultative root-parasite [20]. Among them, Gaiadendron exhibits a closer phylogenetic relationship with the clade of obligate stem-parasites compared to Nuytsia and Atkinsonia [29]. Given that this study only included one facultative root-parasite (Nuytia floribunda), comparing its plastome with those of obligate stem-parasites may not provide an accurate reflection of the evolutionary trajectory of plastome degradation associated with the lifestyle transition in hemiparasites. From this perspective, our data may not provide robust support for the theoretical expectation that the transition from facultative root-parasitism to obligate stem-parasitism could lead to increased levels of plastome degradation in hemiparasites [14, 22]. Therefore, the further validation is necessary to determine whether the lifestyle transitions resulting in increased nutritional reliance on host plants constitute pivotal evolutionary events that trigger the ‘domino effect’ in plastome degradation of hemiparasitic plants.
Within the clade of obligate stem-parasites, only one gene (infA), was lost in the plastomes of the early divergent tribe Elytranthaea. Within the tribe Loranthaea, further lineage-specific gene loss and pseudogenization occurred in the stem lineage ancestors of the subtribe Loranthinae, the MRCA of the subtribes Dendrophthoinae (except for Helixanthera parasitica), Dendrophthoinae, Amyeminae, Tapinanthinae, and Emelianthinae, as well as the MRCA of the tribes Scurrulinae and Helixanthera parasitica. Additionally, independent gene loss and pseudogenization were observed in some shallow nodes of the Loranthaea tribe. As a result, taxa within shallow branches exhibit a heightened degree of physical and functional loss of plastid genes compared to taxa within deep clades (e.g., the tribe Elytranthaea). This suggests that plastome degradation in Loranthaceae hemiparasites is likely a gradual process that intensifies with lineage and species divergence. Considering the occurrence of lineage-specific gene loss/pseudogenization at deep nodes and additional independent genes loss/pseudogenization in shallow branches, it is plausible to hypothesize that cladogenesis may act as an essential evolutionary force triggering the cascading degradation (the ‘domino effect’) of plastome degradation in hemiparasitic plants.
Phylogenetic implications
The infra-familial relationships of Loranthaceae recovered in this study were largely congruent with previous studies [20, 29, 30, 67,68,69] but received full support (BS = 100; PP = 1.00) for most nodes. Notably, the plastome phylogeny failed to resolve the genus Helixanthera as monophyletic, given that Helixanthera parasitica formed a sister relationship with the subtribe Scurrulinae clade, which is consistent with the findings of Su et al. [70] and Liu et al. [29]. This relationship is supported by morphological evidence that the flowers of Helixanthera parasitica and the two genera (Scurrula and Taxillus) of the subtribe Scurrulinae are 5-merous, which is different from the 4-merous flowers of Helixanthera terrestris and Helixanthera sampsonii [29]. Collectively, this line of evidence suggests that the current taxonomic delineation of the genus Helixanthera is likely problematic. To reasonably resolve this taxonomic issue, further studies based on multidisciplinary methods are required.
Consistent with a previous study [29], the plastome phylogeny showed that the two genera (Elytranthe and Macrosolen) of the tribe Elytranthaea were not monophyletic, as the species of the two genera were interwoven in the tree topology. Notably, both genera share similar vegetation, flower, and fruit morphology, and the diagnostic characteristics used to discriminate between them is that the inflorescences of Macrosolen have relatively larger bracts. Accordingly, Barlow argued that this trait may not be robust enough to distinguish these two genera from each other and proposed treating Macrosolen and Elytranthe as congeneric [71]. The current study provides a phylogenomic evidence supporting this taxonomic proposal.
Conclusion
The plastome features of 48 Loranthaceae hemiparasites were characterized in this study. Under a phylogenetic framework, comparative analyses of these Loranthaceae plastomes revealed that all obligate stem-parasites exhibited a higher degree of plastid degeneration than the closely related facultative root-parasite, Nuytia floribunda, providing empirical evidence for the theoretical expectation that the evolution of obligate stem-parasitism from facultative root-parasitism may have caused a higher level of plastome degradation in hemiparasites [14, 22]. Phylogeny-oriented comparative analyses of Loranthaceae plastomes have also revealed that plastome degradation in Loranthaceae is likely a gradual process that intensifies with lineage and species divergence. Therefore, cladogenesis can be a key evolutionary force that triggered the ‘domino effect’ in plastome degradation of hemiparasitic plants. These findings provide new insights into the evolutionary trajectory of plastome degradation in hemiparasitic plants.
Data availability
The complete plastome sequences generated in this study can be accessed at the NCBI GenBank database, by searching their corresponding accession numbers (OR909691, OR909692, OR909704, OR909697, OR909702, OR909693, OR909707, OR909710, OR909711, OR909708, OR909709, OR909701, OR909703, OR909705, OR909695, OR909690, OR909699, OR909706, OR909700, OR909698, OR909694, OR909696).
Abbreviations
- bp:
-
Base Pair
- BS:
-
Bootstrap
- CTAB:
-
Cetyl Trimethylammonium Bromide
- DNA:
-
Deoxyribonucleic Acid
- GbP:
-
Giga Base Pairs
- GC:
-
Guanine and Cytosine
- IR:
-
Inverted Repeat
- LSC:
-
Large Single Copy
- MCMC:
-
Markov Chain Monte Carlo
- ML:
-
Maximum Likelihood
- NCBI:
-
National Center for Biotechnology Information
- PCGs:
-
Protein-Coding Genes
- PP:
-
Posterior Probability
- rRNA:
-
Ribosomal RNA
- SSC:
-
Small Single Copy
- tRNA:
-
Transfer RNA
References
Nickrent DL. Parasitic angiosperms: how often and how many? Taxon. 2020;69:5–27.
Westwood JH, Yoder JI, Timko MP, dePamphilis CW. The evolution of parasitism in plants. Trends Plant Sci. 2010;15:227–35.
Heide-Jørgensen H. Parasitic flowering plants. The Netherlands: Brill; 2008.
Těšitel J. Functional biology of parasitic plants: a review. Plant Ecol Evol. 2016;149:5–20.
Twyford AD. Parasitic plants. Curr Biol. 2018;28:R857–9.
Nickrent DL. The parasitic plant connection. (updated on 15 May 2018). Carbondale, IL: Southern Illinois University; 1997.
Mcfadden GI. Origin and evolution of plastids and photosynthesis in eukaryotes. Cold Spring Harb Perspect Biol. 2014;6:a016105.
Dyall SD, Brown MT, Johnson PJ. Ancient invasions: from endosymbionts to organelles. Science. 2004;04:253–7.
Clegg MT, Gaut BS, Learn GHJr, Morton BR. Rates and patterns of chloroplast DNA evolution, Proc. Natl. Acad. Sci. U.S.A. 1994;91: 6795–6801.
Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 2011;76:273–97.
Neuhaus HE, Emes MJ. Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:111–40.
Bungard RA. Photosynthetic evolution in parasitic plants: insight from the chloroplast genome. BioEssays. 2004;26:235–47.
Wicke S, Müller KF, de Pamphilis CW, Quandt D, Wickett NJ, Zhang Y, Renner SS, Schneeweiss GM. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 2013;25:3711–25.
Wicke S, Müller KF, dePamphilis CW, Quandt D, Bellot S, Schneeweiss GM. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc Natl Acad Sci U S A. 2016;113:9045–50.
Barrett CF, Wicke S, Sass C. Dense infraspecific sampling reveals rapid and independent trajectories of plastome degradation in a heterotrophic orchid complex. New Phytol. 2018;218:1192–204.
Wicke S, Naumann J. Molecular Evolution of Plastid genomes in parasitic flowering plants. Adv Bot Res. 2018;2018:315–47.
Molina J, Hazzouri KM, Nickrent D, Geisler M, Meyer RS, Pentony MM, Flowers JM, Pelser P, Barcelona J, Inovejas SA, Uy I, Yuan W, Wilkins O, Michel CI, Locklear S, Concepcion GP, Purugganan MD. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia Lagascae (Rafflesiaceae). Mol Biol Evol. 2014;31:793–803.
Cai L, Arnold BJ, Xi Z, Khost DE, Patel N, Hartmann CB, Manickam S, Sasirat S, Nikolov LA, Mathews S, Sackton TB, Davis CC. Deeply altered Genome Architecture in the Endoparasitic Flowering Plant Sapria Himalayana Griff (Rafflesiaceae). Curr Biol. 2021;31:1002–11.
Lin CS, Chen JJW, Chiu CC, Hsiao HCW, Yang CJ, Jin XH, Leebens-Mack J, de Pamphilis CW, Huang YT, Yang LH, Chang WJ, Kui L, Wong GK, Hu JM, Wang W, Shih MC. Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J. 2017;90:994–1006.
Vidal-Russell R, Nickrent DL. Evolutionary relationships in the showy mistletoe family (Loranthaceae). Am J Bot. 2008;95:1015–29.
Těšitel J, Plavcová L, Cameron DD. Interactions between hemiparasitic plants and their hosts: the importance of organic carbon transfer. Plant Signal Behav. 2010;5:1072–6.
Petersen G, Cuenca A, Seberg O. Plastome Evolution in Hemiparasitic Mistletoes. Genome Biol Evol. 2015;7:2520–32.
Krause K. Piecing together the puzzle of parasitic plant plastome evolution. Planta. 2011;234:647–56.
Chen X, Fang D, Wu C, Liu B, Liu Y, Sahu SK, Song B, Yang S, Yang T, Wei J, Wang X, Zhang W, Xu Q, Wang H, Yuan L, Liao X, Chen L, Chen Z, Yuan F, Chang Y, Lu L, Yang H, Wang J, Xu X, Liu X, Wicke S, Liu H. Comparative Plastome Analysis of Root- and stem-feeding parasites of Santalales Untangle the footprints of Feeding Mode and Lifestyle transitions. Genome Biol Evol. 2020;12:3663–76.
Guo XR, Liu CK, Zhang GF, Su WH, Landis JB, Zhang X, Wang HC, Ji YH. The complete plastomes of Five Hemiparasitic plants (Osyris wightiana, Pyrularia edulis, Santalum album, Viscum liquidambaricolum, and V. ovalifolium): Comparative and evolutionary analyses within Santalales. Front. Genet. 2020;11:597.
Guo XR, Liu CK, Wang HC, Zhang GF, Yan HJ, Jin L, Su WH, Ji YH. The complete plastomes of two flowering epiparasites (Phacellaria Glomerata and P. compressa): gene content, organization, and plastome degradation. Genomics. 2021;113:447–55.
Shin HW, Lee NS. Understanding plastome evolution in Hemiparasitic Santalales: complete chloroplast genomes of three species, Dendrotrophe varians, Helixanthera Parasitica, and Macrosolen cochinchinensis. PLoS ONE. 2018;13:e0200293.
Nickrent DL, Anderson F, Kuijt J. Inflorescence evolution in Santalales: integrating morphological characters and molecular phylogenetics. Am J Bot. 2019;106:402–14.
Liu B, Le CT, Barrett RL, Nickrent DL, Chen Z, Lu L, Vidal-Russell R. Historical biogeography of Loranthaceae (Santalales): diversification agrees with emergence of tropical forests and radiation of songbirds. Mol Phylogenet Evo. 2018;124:199–212.
Nickrent DL, Malécot V, Vidal-Russell R, Der JP. A revised classification of Santalales. Taxon. 2010;59:538–58.
Doyle JJ, Doyle JL. A Rapid DNA isolation Procedure for small quantities of Fresh Leaf tissue. Phytochem Bull. 1987;19:11–5.
Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE. 2012;7:e30619.
Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45(4):e18.
Qu XJ, Moore MJ, Li DZ, Yi TS. PGA: a software package for rapid, accurate, and fexible batch annotation of plastomes. Plant Methods. 2019;15:50.
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9.
Chan PP, Lowe TM. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 2019;1962:1–14.
Katoh K, Standley DM. MAFFT multiple sequence alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80.
Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. PhyloSuite: an Integrated and Scalable Desktop Platform for Streamlined Molecular Sequence Data Management and Evolutionary Phylogenetics studies. Mol Ecol Resou. 2020;20:348–55.
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4.
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9.
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the Ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–22.
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large Model Space. Sys Biol. 2012;61:539–42.
Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: New methods for selecting Partitioned models of Evolution for Molecular and Morphological phylogenetic analyses. Mol Biol Evol. 2017;34:772–3.
Rambaut A. FigTree v1.4.5, a graphical viewer of phylogenetic trees. 2014.
Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–403.
Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, Boore JL, Jansen RK. The complete chloroplast genome sequence of Pelargonium xhortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 2006;23:2175–90.
Naumann J, Der JP, Wafula EK, Jones SS, Wagner ST, Honaas LA, Ralph PE, Bolin JF, Maass E, Neinhuis C, Wanke S, dePamphilis CW. Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae). Genome Biol Evol. 2016;8:345–63.
Bellot S, Cusimano N, Luo S, Sun G, Zarre S, Gröger A, Temsch E, Renner SS. Assembled plastid and mitochondrial genomes, as well as nuclear genes, place the parasite family Cynomoriaceae in the Saxifragales. Mol Biol Evol. 2016;8(7):2214–30.
Frailey DC, Chaluvadi SR, Vaughn JN, Coatney CG, Bennetzen JL. Gene loss and genome rearrangement in the plastids of five hemiparasites in the family Orobanchaceae. BMC Plant Biol. 2018;6:18–30.
Yamori W, Shikanai T. Physiological functions of cyclic electron transport around photosystem I in sustaining photosynthesis and plant growth. Annu Rev Plant Biol. 2016;67:81–106.
Blazier JC, Guisinger-Bellian MM, Jansen RK. Recent loss of plastid encoded ndh genes within Erodium (Geraniaceae). Plant Mol Biol. 2011;76:1–10.
Peredo EL, King UM, Les DH. The plastid genome of Najas flexilis: adaptation to submersed environments is accompanied by the complete loss of the NDH complex in an aquatic angiosperm. PLoS ONE. 2013;8:e68591.
Li HT, Yi TS, Gao LM, Ma PF, Zhang T, Yang JB, Gitzendanner MA, Fritsch PW, Cai J, Luo Y, Wang H, van der Bank M, Zhang SD, Wang QF, Wang J, Zhang ZR, Fu CN, Yang J, Hollingsworth PM, Chase MW, Soltis DE, Soltis PS, Li DZ. Origin of angiosperms and the puzzle of the jurassic gap. Nat Plants. 2019;5:461–70.
Sun Y, Moore MJ, Lin N, Adelalu KF, Meng A, Jian S, Yang L, Li J, Wang H. Complete plastome sequencing of both living species of Circaeasteraceae (Ranunculales) reveals unusual rearrangements and the loss of the ndh gene family. BMC Genomic. 2017;18:592.
McNeal JR, Kuehl JV, Boore JL, de Pamphilis CW. Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 2007;7:57.
Su HJ, Barkman TJ, Hao W, Jones SS, Naumann J, Skippington E, Wafula EK, Hu JM, Palmer JD, dePamphilis CW. Novel genetic code and record-setting AT-richness in the highly reduced plastid genome of the holoparasitic plant Balanophora. Proc. Natl. Acad. Sci. U.S.A. 2018;116:934–943.
Sabir J, Schwarz E, Ellison N, Zhang J, Baeshen NA, Mutwakil M, Jansen R, Ruhlman T. Evolutionary and biotechnology implications of plastid genome variation in the inverted–repeat lacking clade of legumes. Plant Biotechnol J. 2014;12:743–54.
Park S, Jansen RK, Park S. Complete plastome sequence of Thalictrum coreanum (Ranunculaceae) and transfer of the rpl32 gene to the nucleus in the ancestor of the subfamily Thalictroideae. BMC Plant Biol. 2015;15:40.
Zhang X, Sun Y, Landis JB, Lv Z, Shen J, Zhang H, Lin N, Li L, Sun J, Deng T, Sun H, Wang H. Plastome phylogenomic study of Gentianeae (Gentianaceae): widespread gene tree discordance and its association with evolutionary rate heterogeneity of plastid genes. BMC Plant Biol. 2020;20:340.
Silva GMD, Lopes ADS, Pacheco TG, Machado KLDG, Silva MC, de Oliveira JD, de Baura VA, Balsanelli E, de Souza EM, de Oliveira PF, Rogalski M. Genetic and evolutionary analyses of plastomes of the subfamily Cactoideae (Cactaceae) indicate relaxed protein biosynthesis and tRNA import from cytosol. Braz J Bot. 2021;44:97–116.
Tiller N, Bock R. The translational apparatus of plastids and its role in plant development. Mol Plant. 2014;7:1105–20.
Guo X, Zhang G, Fan L, Liu C, Ji Y. Highly degenerate plastomes in two hemiparasitic dwarf mistletoes: Arceuthobium chinense and A. Pini (Viscaceae). Planta. 2021;253:125.
Ahmed I, Biggs PJ, Matthews PJ, Collins LJ, Hendy MD, Lockhart PJ. Mutational dynamics of aroid chloroplast genomes. Genome Biol Evol. 2012;4:1316–23.
Millen RS, Olmstead RG, Adams KL, Palmer JD, Lao NT, Heggie L, Kavanagh TA, Hibberd JM, Gray JC, Morden CW, Calie PJ, Jermiin LS, Wolfe KH. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 2001;13:645–58.
Morden CW, Wolfe KH, dePamphilis CW, Palmer JD. Plastid translation and transcription genes in a nonphotosynthetic plant: intact, missing and pseudogenes. EMBO J. 1991;10:3281–8.
Li X, Yang JB, Wang H, Song Y, Corlett RT, Yao X, Li DZ, Yu WB. Plastid NDH pseudogenization and Gene Loss in a recently derived lineage from the Largest Hemiparasitic Plant Genus Pedicularis (Orobanchaceae). Plant Cell Physiol. 2021;62:971–84.
Nickrent DL, Su HJ, Lin RZ, Devkota MP. Examining the needle in the Haystack: Evolutionary relationships in the Mistletoe Genus Loranthus (Loranthaceae). Syst Bot. 2021;46:403–15.
Su HJ, Liang SL, Nickrent DL. Plastome variation and phylogeny of Taxillus (Loranthaceae). PLoS ONE. 2021;16:e0256345.
Darshetkar AM, Pable AA, Nadaf AB, Barvkar VT. Understanding parasitism in Loranthaceae: insights from plastome and mitogenome of Helicanthes Elastica. Gene. 2023;20:861–147238.
Su HJ, Hu JM, Anderson FE, Der JP, Nickrent DL. Phylogenetic relationships of Santalales with insights into the origins of holoparasitic Balanophoraceae. Taxon. 2015;64:491–506.
Barlow BA. Loranthaceae. In: C. Kalkman, P.F. Stevens, D.W. Kirkup, W.J.J.O. de Wilde, H.P. Nooteboom, editors, Flora Malesiana Series 1. National Herbarium of the Netherlands, Leiden. 1997; p209–401.
Funding
The current study is financially supported by the National Natural Science Foundation of China (32370395) and Yunnan Revitalization Talent Support Program “Top Team” Project (202305AT350001).
Author information
Authors and Affiliations
Contributions
YJ, XG, and MD conceived and designed the research framework. LT, TW, and LH collected and analyzed the data. LT, and YJ wrote the original draft manuscript. YJ, XG, MD, and GZ revised and edited the final manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Ethical approval and consent to participate
Collection of all plant samples in this study completely followed the Regulations on the Protection of Wild Plants of the People’s Republic of China, the IUCN Policy Statement on Research Involving Species at Risk of Extinction and the Convention on the Trade in Endangered Species of Wild Fauna and Flora. All the methods included in this study are in accordance with the relevant guidelines.
Consent to publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tang, L., Wang, T., Hou, L. et al. Comparative and phylogenetic analyses of Loranthaceae plastomes provide insights into the evolutionary trajectories of plastome degradation in hemiparasitic plants. BMC Plant Biol 24, 406 (2024). https://doi.org/10.1186/s12870-024-05094-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-024-05094-5