Abstract
Background
Cucumber downy mildew, caused by P. cubensis, is an important leaf disease that can severely affect cucumber production. In recent years, cucumber target spot, caused by C. cassiicola, has been reported in both Asia and Europe and is now considered as a major disease disrupting cucumber production. Single-disease-resistant cucumber varieties have been unable to satisfy production needs.
To explore the molecular mechanisms of cucumber resistance to these two diseases, cucumber cultivars D9320 (resistant to downy mildew and target spot) and D0401 (susceptible to downy mildew and target spot) were used as experimental materials in this study. We used transcriptome sequencing technology to identify genes related to disease resistance and verified using transgenic technology.
Results
We screened out the cucumber resistance-related gene CsERF004 using transcriptome sequencing technology. Induction by pathogens, salicylic acid (SA), and ethylene (ET) resulted in the up-regulation of CsERF004. Three treatments, namely, inoculation with C. cassiicola alone, inoculation with P. cubensis alone, and simultaneous inoculation with both pathogens, all resulted in the significant and sustained up-regulation of CsERF004 in the resistant cultivar D9320, during the early stage of infection. In the susceptible cultivar D0401, CsERF004 expression was also significantly up-regulated at the later stage of infection but to a lesser extent and for a shorter duration than in the resistant cultivar D9320. The CsERF004 gene encodes a protein localizes to the nucleus. The over-expression of CsERF004 in the susceptible cultivar D0401 resulted in the significant up-regulation of the CsPR1 and CsPR4 genes and increased the levels of SA and ET, which enhanced the resistance of cucumber to downy mildew and target spot.
Conclusions
Analyses of the CsERF004 expression pattern in disease-resistant and susceptible cucumber cultivars and transgenic validation indicate that CsERF004 confers resistance to P. cubensis and C. cassiicola. The findings of this study can help to better understanding of mechanisms of response to pathogens and in establishment the genetic basis for the development of cucumber broad-spectrum resistant cultivars.
Similar content being viewed by others
Background
Cucumber (Cucumis sativus L.) is a staple vegetable that produces tender edible fruits. In production, cucumber is susceptible to a variety of infections that severely affect its yield and quality. Two major diseases that affect cucumber production are cucumber downy mildew caused by Pseudoperonospora cubensis (Berk. & M.A.Curtis, Rostovzev) [1] and cucumber target spot caused by Corynespora cassiicola (Berk & Curt, Wei) [2]. Disease epidemics involving these pathogens generally spread quickly, disrupting cucumber production. Downy mildew and target spot, reduce cucumber yields by 50% [3] and 60–70% [4], respectively. The production and quality of cucumber have been affected resulting in economic losses. There is some controversy about genetic control of resistance to downy mildew and target spot. Some studies reported that resistance to downy mildew is controlled by multiple genes [5,6,7,8,9], while others believe it to result from a single recessive gene [10,11,12,13]. Resistance to target spot is generally believed to be controlled by a single gene. However, whether this gene is dominant or recessive has been discussed. Several studies have shown that resistance is controlled by a single dominant gene [14], while others indicate that it is controlled by a single recessive gene [15,16,17]. The discrepancies in the above results may be related to the use of different experimental materials. The use of single-disease-resistant cucumber varieties has already proven insufficient to prevent losses in cucumber production due to these diseases. Therefore, the study of broad-spectrum disease resistance mechanisms can be important to establish the genetic basis for the development of multi-disease-resistant cucumber varieties.
Unlike mammals, plants are not equipped with mobile cells and do not possess an adaptive immune system. However, during co-evolution with pathogens, plants developed an innate immune system [18] consisting of two levels. The first level involves pathogen-associated molecular pattern-triggered immunity (PTI), which can prevent some pathogens from entering the cell via oxygen bursts and callose deposition [19,20,21]. The second level consists of the specific recognition of resistance proteins and avirulence proteins, triggering the hypersensitive response and inhibiting the growth of pathogenic bacteria via programmed cell death in a process known as effector-triggered immunity (ETI) [22]. Transcription factors can activate or inhibit the expression of genes related to disease resistance; thus, they play an important role in disease resistance in plants [23]. The AP2/ERF-like transcription factors compose a large family of proteins that is divided into five subfamilies based on the number of AP2/ERF domains; they include AP2, RAV, DREB, ERF, and others [24]. The ERFs are a major subfamily of the AP2/ERF transcription factor family and occur widely in plants. ERF-subfamily transcription factors have been isolated from Arabidopsis thaliana [25], tobacco [26], soybean [27], rice [28], maize [29], and tomato [30]. ERFs can regulate the expression of the PR genes in combination with the GCC-box and DRE/CRT cis-acting elements in the promoter region of the gene [31, 32]. PR genes play an important role in plant resistance to various infections [33]. Expression of PR-1a, PR4, PR5, and PR10 genes has been associated with resistance to pathogen infection and has been shown to improve plant disease resistance [34,35,36,37]. ERF transcription factors are involved in a variety of plant hormone signaling pathways [38, 39], and are the connecting factors of signal cross-linking pathway under stress, thus playing an important role in plant growth and development as well as in resistance to biotic and abiotic stresses [40,41,42,43,44]. Over-expression of ERF genes in rice [44] and soybean [23] has been shown to increase plant disease resistance.
To date, a total of 103 genes that encode complete AP2/ERF domains have been identified in the cucumber genome [45]. However, our understanding of the function of the cucumber ERF gene subfamily is limited because only a single gene, CsERF (Csa7G448110), has been reported to regulate the expression of the bitter gene Bi [46]. Furthermore, ERF has not been reported that regulates to cucumber disease resistance. In the present study, we cloned the CsERF004 gene, analysed its expression patterns after inoculation with only C. cassiicola, only P. cubensis or both pathogens, and explored its role in disease resistance using transgenic validation in cucumber, thereby laying the foundation for cultivating multi-resistant cucumber varieties.
Results
Screening of disease-related genes
The raw data were uploaded to the National Center for Biotechnology Information (NCBI) (GenBank accession no.SRX2468535). Transcriptome sequencing after the three treatments identified 61 up-regulated and 276 down-regulated genes in the resistant cultivar D9320 and 427 up-regulated and 763 down-regulated genes in the susceptible cultivar D0401 (Fig. 1). A total of 61 genes were up-regulated in the D9320 genotype under all three inoculation conditions; 16 of them were also up-regulated in the D0401 genotype and were not analysed further. The remaining genes that were up-regulated in the D9320 genotype were either down-regulated or not differentially expressed in the D0401 genotype. According to annotations in the genome database, 10 of these genes may be associated with disease resistance (Table 1). The over-expression of ERF transcription factors can enhance resistance to two or three diseases [41, 44]. The purpose of our study was to explore the mechanism of resistance to two diseases in cucumber. Therefore, the CsERF004 gene (Csa7M432080.1) was selected to study the mechanism of resistance to two diseases in cucumber.
Differentially expressed genes venn diagrams of cucumber cultivars D9320 and D0401 under pathogen stress. D9320: resistant cultivar. D0401: susceptible cultivar. P/CK represents a set of differentially expressed genes of comparison between the P. cubensis-infected sample and the control sample at 48 h. C/CK represents a set of differentially expressed genes of comparison between the C. cassiicola-infected sample and the control sample at 48 h. PC/CK represents a set of differentially expressed genes of comparison between the two pathogens-infected sample and the control sample at 48 h. Sterilized water was used as a control. Red arrows indicate up-regulated genes in infected samples, and green arrows represent down-regulated genes in infected samples
Gene cloning and bioinformatics analysis
The CsERF004 gene was cloned from the resistant cucumber cultivar D9320 by PCR using cDNA extracted from cucumber leaves as a template; its CDS was 591 bp in length and encoded 196 amino acids. The protein encoded by this gene has one AP2/ERF domain, and belongs to the ERF transcription factor B-6 family. (Fig. 2).
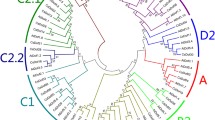
Phylogenetic analysis and sequence alignment of CsERF004. a Phylogenetic analysis of CsERF004 with ERF proteins; the phylogenetic tree was constructed using the amino acid sequences of the AP2/ERF domains in each ERF protein. The GenBank accession numbers are as follows: AtRAP2.11 (AAC49777.1), NcRAP2–11 (JAU24088.1), CaRAP2–11 (XP_010493057.1), BnRAP2–11-like (XP_013667343.1), AtESE3 (NP_197901.1), AtERF1 (NP_567530.4), NcERF1a (JAU16327.1), AtEREBP-2 (CAB45963.1), AtERF2 (NP_199533.1), Pti4 (NP_001275437.1), AtRAP2.2 (NP_850582.1), AtRAP2.12 (NP_175794.1), GmAP2-EREBP (NP_001238300.1), GmERF4 (NP_001238595.1), AtERF4 (NP_188139.1), AtRAP2.5 (AAC49771.1), and NcERF4 (JAU43921.1). b The conserved AP2/ERF domain of the B-6 group proteins. The conserved AP2/ERF domain is marked with a black horizontal solid line
Analysis of the CsERF004 gene expression pattern under pathogen stress
After inoculation with either P. cubensis or C. cassiicola as well as after inoculation of both pathogens, the expression of the CsERF004 gene was differed between the resistant cultivar D9320 and the susceptible cultivar D0401.
After inoculation with P. cubensis, the expression level of the CsERF004 gene in the D9320 genotype was significantly increased at 8 h, and reached a peak at 48 h (approximately 10-fold), and then decreased slightly, but yet remained higher than that of the control, (Fig. 3a). The expression level of the CsERF004 gene in the D0401 genotype was only significantly increased at 12 h (Fig. 3b).
Relative expression levels of the CsERF004 gene in two cucumber varieties and after three treatments. a and b represent the relative expression levels of CsERF004 in the resistant cultivar D9320 and susceptible cultivar D0401, respectively. W: water control. P: inoculation of P. cubensis. C: inoculation of C. cassiicola. PC: inoculation of both pathogens. There were three biological replicates per treatment and three technical replicates per sample. Student’s t-test was used for comparison between two samples. “**” indicates that the difference between the corresponding column value and the adjacent control column value is significant
After inoculation with C. cassiicola, the expression level of CsERF004 gene in the D9320 genotype was significantly increased at 4 h, and reached a peak at 96 h (approximately 9.5-fold), and then decreased slightly, but yet remained higher than that of the control (Fig. 3a). The expression level of CsERF004 gene in the D0401 genotype was significantly increased at 12 h (1–1.5-fold) and 120 h (approximately 2.5-fold) (Fig. 3b).
After inoculation with both pathogens, the expression level of CsERF004 gene in the D9320 genotype was significantly increased at 8 h, and reached a peak at 96 h (approximately 8.5-fold), and then decreased slightly, but yet remained higher than that of the control (Fig. 3a). The expression level of CsERF004 gene in the D0401 genotype was significantly increased at 12 h (1–1.5-fold) and 120 h (approximately 1.5-fold) (Fig. 3b).
The above results show that the expression of CsERF004 gene was significantly up-regulated in the resistant cultivar D9320 during the early stage of infection, and this expression was high and long-lasting. In the susceptible cultivar D0401, the expression of CsERF004 gene was also significantly up-regulated at the later stage of infection, although this induction was comparatively lower and shorter in duration than that in D9320.
Gene expression patterns are often associated with gene function [47]. Differences in the expression of the CsERF004 gene in response to P. cubensis and C. cassiicola were observed between the resistant cultivar D9320 and the susceptible cultivar D0401, indicating that the CsERF004 gene might be closely associated with resistance to downy mildew and target spot.
Analysis of the CsERF004 gene expression pattern in response to hormone induction
Plant resistance to diseases involves hormone signal transduction pathways. ERF transcription factors are involved in a variety of hormone responses [38, 39]. In the present study, the resistant cucumber cultivar D9320 was treated with methyl jasmonate (MeJA), salicylic acid (SA) and ethylene(ET), respectively. After MeJA treatment, CsERF004 expression did not significantly change (Fig. 4). After SA treatment, CsERF004 was significantly up-regulated and reached its peak expression at 12 h, with an expression level 6.5–7-fold higher than that in the control (Fig. 4). After ET treatment, the expression of CsERF004 also was significantly up-regulated and peaked at 12 h, with an expression level 7–7.5-fold higher than that in the control (Fig. 4). These findings show that CsERF004 can be up-regulated by SA and ET, suggesting that CsERF004 may be involved in the SA and ET signalling pathways.
Relative expression levels of CsERF004 induced by different hormones in the resistant cultivar D9320. There were three biological replicates per treatment and three technical replicates per sample. Sterilized water was used as a control. Student’s t-test was used to compare two samples. “**” indicates that the difference between the corresponding column value and the adjacent control column value is significant
Subcellular localization analysis
The CsERF004-GFP fusion expression vector and an empty hGFP vector were individually introduced into Arabidopsis protoplasts. The subcellular localization of the CsERF004-GFP fusion protein was observed under a laser scanning confocal microscope. The CsERF004-GFP fusion protein was enriched in the nuclei of Arabidopsis cells (Fig. 5), indicating that CsERF004 is a nuclear-localized protein.
Over-expression of CsERF004 improves the resistance of cucumber to downy mildew and target spot
The over-expression vector PCXSN-CsERF004 was successfully transferred into the susceptible cultivar D0401 using the cucumber genetic transformation technology. In the T0 generation, lines E4, E7, and E9 (with the highest expression levels) were inoculated with C. cassiicola and P. cubensis. After 7 days, the symptoms of disease in the leaves of the transgenic plants were significantly less severe than those of the susceptible D0401 plants (Fig. 6). T1 generation plants over-expressing CsERF004 were used to analyse disease resistance. After inoculation with P. cubensis, the disease index decreased from 90.7 in wild-type plants to 58.7, 62.7, and 60.0 in the E4, E7, and E9 lines, respectively. After inoculation with C. cassiicola, the disease index decreased from 85.3 in wild-type plants to 49.3, 52.0 and 56.0 in the E4, E7, and E9 lines, respectively (Table 2). These results show that the over-expression of CsERF004 can improve the resistance of cucumber to downy mildew and target spot.
The ERF genes regulate the expression of PR genes, thereby improving plant disease resistance [33,34,35,36,37]. In the present study, the expression levels of the CsPR1 and CsPR4 genes were significantly up-regulated after CsERF004 over-expression. The relative expression of the CsPR4 gene increased to more than 20-fold that of the control (Fig. 7), and the relative expression of the CsPR1 gene increased to more than 6-fold that of the control (Fig. 7). Furthermore, we predicted the cis-acting elements of the CsPR1 and CsPR4 promoter regions. The CsPR4 promoter region contained two core CCGAC sequences of DRE/CRT cis-acting elements Additional file 1: Table S1. The CsPR1 promoter region contained one CCGCC sequence and one CCGTC sequence Additional file 1:Table S1. LeERF2 has been shown to bind to the CCGCC motif [48]. The over-expression of the CsERF004 gene may enhance cucumber disease resistance by directly regulating defense genes such as CsPR1 and CsPR4.
Expression levels of the CsERF004, CsPR1, and CsPR4 genes in the T1-generation CsERF004-over-expressing plants. E4, E7, and E9: T1-generation plants; WT: control. There were three biological replicates per treatment and three technical replicates per sample. Student’s t-test was for comparisons between two samples. “**” indicates that the difference between the corresponding column value and the adjacent control column value is significant
CsERF004 may require the salicylic acid and ethylene signalling pathways to enhance disease resistance in cucumber
SA and ET are important hormones in plant defense responses, and play roles against biotrophic and necrotrophic pathogens, respectively [49]. To further explore the mechanism by which CsERF004 over-expression enhances the resistance of cucumber to different pathogens, gas chromatography and enzyme-linked immunosorbent assay (ELISA) were performed to determine the ET and SA contents in T1 generation plants that over-expressed CsERF004. Fig. 8 shows that both the ET and SA contents in CsERF004-over-expressing plants were significantly higher than those of wild-type plants. The plants over-expressing CsERF004 had more than 2-fold more ethylene than wild-type plants (Fig. 8a) and SA levels of approximately 1.5-fold those in the wild-type plants (Fig. 8b). The over-expression of the CsERF004 gene significantly increased the contents of SA and ET, indicating that CsERF004 may depend upon the SA and ET signalling pathways to improve cucumber disease resistance.
Discussion
CsERF004 is associated with resistance to downy mildew and target spot in cucumber
Gene expression patterns are often associated with gene function [47]. Under the stress of P. cubensis, C. cassiicola, or the combination of both pathogens, CsERF004 was significantly up-regulated in the resistant cultivar in the early stage of pathogen stress, with high and long-lasting expression (Fig. 3a). In the susceptible cultivar, CsERF004 was significantly up-regulated at 12 h and 120 h after inoculation, although the expression was relatively low and short in duration (Fig. 3b). The expression pattern of CsERF004 was related to plant disease resistance, which indicated that CsERF004 plays a role in cucumber resistance to downy mildew and target spot.
Over-expression of CsERF004 enhances cucumber resistance to downy mildew and target spot
Previous studies have shown that the up-regulation of ERF enhances plant resistance to single or multiple diseases. Furthermore, the over-expression of the BrERF11 gene induces the expression of NtPR-1a/c, NtPR3, and NtPR-1b and enhanced the resistance of tobacco to Ralstonia solanacearum [50]. The over-expression of GmERF3 enhanced tobacco resistance to bacterial wilt, brown spot, and tobacco mosaic virus [41]. The over-expression of AaERF1 induced the expression of the PDF1.2 gene and positively regulated Artemisia annua resistance to Botrytis cinerea [51]. Chen and Guo introduced the tobacco OPBP1 gene into rice to enhance its resistance to Magnaporthe grisea and Rhizoctonia solani [44]. Other studies have shown that ERF negatively regulates plant disease resistance. McGrath et al. [52] and Onate-Sanchez et al. [53] over-expressed AtERF4 and AtERF14, respectively, in Arabidopsis, and both reduced the resistance of Arabidopsis to Fusarium wilt. The results of the present study showed that the disease symptoms in CsaERF004-over-expressing plants were significantly less severe than those of D0401 plants (Fig. 6). Furthermore, the disease index of plants inoculated with downy mildew decreased from 90.7 in wild-type plants to 58.7, 62.7, and 60.0 in the transgenic plants (Table 2). After inoculation with C. cassiicola, the disease index decreased from 85.3 in wild-type plants to 49.3, 52.0, and 56.0 in the transgenic plants (Table 2). Therefore, the over-expression of CsERF004 could improve the resistance of cucumber to downy mildew and target spot. P. cubensis is an oomycete, and C. cassiicola is a fungus, indicating that CsERF004 plays an important role in resistance of cucumber to fungal and oomycete infections. Previous studies have shown that the over-expression of ERFs not only increases the resistance of plants to fungi, bacteria, and viruses but also improves the resistance of plants to the oomycete family.
CsERF004 positively regulates the expression of CsPR1 and CsPR4 and enhances cucumber disease resistance
ERF transcription factors are capable of binding to the GCC-box or DRE/CRT cis-acting elements of the gene promoter region to regulate gene expression [31, 32, 48, 54, 55]. Previous studies have shown that the over-expression of ERFs in plants enhances plant disease resistance by increasing the expression of PR genes. The over-expression of the GbERF2 transcription factor gene raised the expression levels of PR-1b, PR2 and PR4 to enhance the resistance of tobacco to Alternaria longipes [56]. The over-expression of the BrERF11 gene up-regulated the expression of NtPR-1a/c, NtPR3, and NtPR-1b and enhanced the resistance of tobacco to Ralstonia solanacearum [50]. PR1 and PR4 play important roles in plant resistance to pathogens. PR4 has antibacterial activity against Magnaporthe grisea and Fusarium solani [35, 57]. The over-expression of PR-1a in tobacco enhances tobacco resistance to downy mildew and black shank [34]. The results of the present study showed that CsPR1 and CsPR4 could be significantly up-regulated after the over-expression of the CsERF004 gene, which agrees with the findings of previous studies. Compared to that in wild-type plants, the expression of CsPR4 increased to more than 20-fold, whereas that of CsPR1 increased to more than 6-fold (Fig. 7). The expression of CsPR4 was higher than that of CsPR1, which may be related to the cis-acting elements in the promoter regions of CsPR4 and CsPR1. The CsPR4 gene promoter region contains two core CCGAC sequences of DRE/CRT cis-acting elements, and CsERF004 may regulate expression of CsPR4 by binding to these CCGAC sequences. The CsPR1 promoter region has neither a DRE/CRT cis-acting element nor a GCC-box (GCCGCC) cis-acting element and has one CCGCC sequence and one CCGTC sequence. Zhang et al. showed that LeERF2 binds to the CCGCC box [48]. Wang et al. reported that the third, fourth, and sixth bases of the conserved GCCGCC sequence of the GCC box are essential for ERF protein binding and that other base alterations may affect their binding efficiency [55]. Therefore, the low binding efficiency of CsERF004 and the CsPR1 promoter may have affected the expression level of CsPR1. CsERF004 may positively regulate the expression of CsPR1 and CsPR4, thereby enhancing disease resistance in cucumber.
CsERF004 may require the salicylic acid and ethylene signalling pathways to enhance disease resistance in cucumber
Plants are equipped with different disease resistance signalling pathways that are triggered in response to different infections. The SA disease resistance signalling pathway is mainly involved in resistance to biotrophic pathogens, whereas the ET and JA disease resistance pathways are involved in the resistance to necrotrophic pathogens [49]. P. cubensis is a biotrophic pathogen, deriving nutrients from living host tissues [58], and C. cassiicola is a necrotrophic pathogen, deriving nutrients from dead or dying cells [59]. There is crosstalk among the SA, ET, and JA signalling pathways, and ERF transcription factors serve as the link among them [38, 39]. GmERF089 is up-regulated during ET, SA, and JA stress [60]. The expression of GmERF5 is significantly up-regulated during ET and SA stress and is down-regulated by JA treatment [23]. The results of the present study show that SA and ET, but not JA, could significantly up-regulate the CsERF004 gene (Fig. 4). The response of the CsERF004 gene to SA and ET may involve specific signalling pathways in cucumber. The SA and ET contents increased in plants over-expressing CsERF004, suggesting that CsERF004 over-expressing enhances the resistance of cucumber to downy mildew and target spot and that this resistance may be dependent on the SA and ET signalling pathways.
Implications in plant breeding
Cucumber is a widely cultivated vegetable crop in worldwide. Cucumber downy mildew has occurred in 70 countries [61]. Cucumber target spot has been reported in America, China, Japan and Korea [62]. The two diseases have affected cucumber production. Single-disease-resistant cucumber cultivars have been unable to meet production needs. Therefore, cultivating multi-resistant cucumber cultivars is very important in cucumber production.
This study utilized transcriptome sequencing techniques, screened out the genes related to plant disease resistance. Using transgenic techniques, we analysed gene function and it was confirmed that CsERF004 is involved in resistance to cucumber downy mildew and target spot. Conventional breeding methods were utilized to cultivate multi-resistant cultivars, which required the identification of multiple resistance genes aiming at different pathogens. The many genes involved may lead to excessively long breeding periods. Identifying the genes related to multi-resistance and utilizing transgenic techniques to support breeding will possible shorten the cultivation period for multi-resistant cultivars.
Conclusions
In the present study, the CsERF004 gene, encoding a member of the ERF transcription factor subfamily, was isolated from cucumber and was found to be significantly up-regulated under the stress of P. cubensis and C. cassiicola infection in resistant varieties and responsive to SA and ET treatments. In terms of the molecular mechanism of cucumber disease resistance, CsERF004 may promote resistance to P. cubensis and C. cassiicola in an SA-and-ET-pathway-dependent manner. Based on the analysis of CsERF004 gene expression patterns and transgenic validation, CsERF004 is involved in resistance to cucumber downy mildew and target spot. Results could be useful in the development of new resistant cultivars and in understanding the mechanisms of response to pathogens in cucumber.
Methods
Plant material
In this study, the homozygous cucumber lines D9320 and D0401 were used as experimental materials. The homozygous cucumber lines D9320 (resistant to downy mildew and target spot) and D0401 (susceptible to downy mildew and target spot) were identified inoculation with P. cubensis and C. cassiicola, 10 plants per replicate and three replicates tested per treatment [63]. The materials were provided by cucumber research group of Northeast Agricultural University, Harbin, China.
Pathogen inoculum preparation and inoculation method
C. cassiicola isolate of monoconidial cultures (accession number Cc-Tj) was obtained from the Associate Researcher Huizhe Wang (Tianjin Kerun Cucumber Research Institute, Tianjin, China). P. cubensis isolate of monosporangial cultures (accession number Pc-hrb6) was obtained from the Professor Yanju Zhang (Northeast Agricultural University, Harbin, China).
Inoculation with C. cassiicola [17, 64]: The C. cassiicola inoculum was prepared by gently scraping the surface of potato dextrose agar (PDA) medium in sterile water, after which mycelial and conidial suspensions were filtered through four layers of sterile gauze. The conidial suspensions were counted using a haemocytometer and adjusted to 1 × 105 conidia·mL−1 using sterile tap water. The first true leaves were inoculated with 30 droplets (approximately 10 μL) of the conidial suspension.
Inoculation with P. cubensis [65]: The P. cubensis inoculum was prepared by gently scraping the surface of diseased leaves in sterile water, after which sporangia were filtered through four layers of sterile gauze. The sporangial suspensions were counted using a haemocytometer and adjusted to 2.0 × 103 sporangia·mL−1 using sterile tap water. The first true leaves were inoculated with 30 droplets (approximately 10 μL) of sporangial suspension.
Simultaneous inoculation with P. cubensis and C. cassiicola: The first true leaves were inoculated with 15 droplets of the sporangial suspension (2.0 × 103 sporangia·mL−1) and 15 droplets of the conidial suspension (1.0 × 105 spores·mL−1), each approximately 10 μL in volume.
The culture conditions were as follows: 26 °C/18 °C day and night, and a 12-h/12-h light cycle. Sterile water was used as a control. The three inoculation treatments and the water controls were sampled at 2, 4, 8, 12, 24, 48, 96, and 120 h after inoculation, respectively, frozen in liquid nitrogen, and then stored at −80 °C. Ten plants were inoculated at every time point, and three experimental replicates were performed.
Resistance was assessed by inoculating the T0 generation of the E4, E7, and E9 lines with either C. cassiicola or P. cubensis, and the leaf disease symptoms were photographed (Nikon D5500, Japan) after 7 days. Seeds of the T1 generation were obtained via the self-cross of the T0 generation lines. A total of 15 individuals from the T1 generation (lines E4, E7, and E9) were selected for inoculation, and 15 D0401 plants were inoculated as controls. After inoculation, the disease index was recorded. The severity scale was based on a previous report [17], and the disease index was calculated using the following formula: disease condition index [66] = Σ (number of disease-level plants × representative levels) × 100/(total number of plants × highest representative value)
Hormone induction treatment
At the two-true-leaf stage of the cucumber cultivar D9320, the leaves were sprayed with 100 μM MeJA (dissolved in was 0.01% ethanol), 1 mM SA (dissolved in sterile water), or 1 mM ET (2 mL 40% EtOH and 1 g NaHCO3 dissolved in 200 mL sterile water); sterilized water was used as a control. The ET treatment was conducted in a sealed glass box. Sampling was performed at 6, 12, and 24 h after spraying, and the leaves were then frozen in liquid nitrogen and stored at −80 °C. Ten plants were treated at every time point, and three experimental replicates were performed. Hormone treatment was performed as described elsewhere [23, 67].
Transcriptome analysis
In this study, the homozygous cucumber lines D9320 and D0401 were used as experimental materials. Samples were taken 48 h after the three inoculation treatments and water control treatments, then frozen in liquid nitrogen and stored at −80 °C. Total RNA was extracted from the cucumber leaves using TRIzol™ (Invitrogen, USA) [48]. RNA samples that met the quality control requirements were sent to Shenzhen Genomics. Then, a cDNA library was obtained by PCR amplification and sequenced on the Illumina HiSeq 2000 platform with 100 cycles of paired-end (2 × 101 bp) sequencing. The raw data (raw reads) were filtered with the FASTQ_Quality_Filter tool from the FASTX-toolkit. The filtered data were used for further analysis. After preprocessing the RNA-Seq data, the reads were mapped to the cucumber genome database (http://cmb.bnu.edu.cn/Cucumis_sativus_v20/) [68]. Differentially expressed genes were identified based on a p value ≤0.01 and | log2 ratio ≥ 1 |.
Gene cloning and bioinformatics analysis
The full-length CDS of the cucumber gene ID Csa7M432080.1 (CsERF004) was searched in the cucumber genome database, and specific primers for cloning the full-length CDS were designed using Primer Premier 5.0 (Table 3). PCR was used to obtain the full-length CDS of CsERF004. The PCR conditions were as follows: denaturation at 94 °C for 30 min, annealing at 50 °C for 30 s, extension at 72 °C for 10 min for a total of 30 cycles; and a final extension at 72 °C for 10 min. The amplified products were stored at 4 °C.
The analysis of the conserved domains in the CsERF004 protein was performed using the NCBI CDD database for conserved protein domains (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). A phylogenetic tree was constructed using MAGE5.10 software. The sequence alignment of the conserved domains was performed using DNAMAN software. All sequences data were obtained from the National Center for Biotechnology Information (NCBI) Additional file 2:Table S2.
Promoter sequence analysis
BLAST sequence alignment was performed in the cucumber genome database (http://www.icugi.org/) to obtain the complete sequence of the gene; the sequence 2000 bp upstream of the transcription initiation region was considered the promoter region. The online tool PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and the PLACE database (http://www.dna.affrc.go.jp/htdocs/PLACE/) were used to perform promoter sequence analysis.
qRT-PCR analysis
Samples were taken 2, 4, 8, 12, 24, 48, 96, and 120 h after inoculation or water control treatment. Samples from the three hormone induction treatments and water controls were taken 6, 12, and 24 h after spraying. Total RNA was extracted from cucumber leaves using the TRIzol (Invitrogen, USA) method [69]. The first-strand cDNA was reverse-transcribed using a Toyobo reverse transcription kit ReverTra Ace qPCR RT-Kit (ToYoBo, Japan). The quality and concentration of the cDNA were determined using a SMA3000 UV spectrophotometer (Beijing, China). After dilution, the cDNA was stored at −20 °C until use.
The qRT-PCR primers were designed using the online tool GenScript Real-time PCR (TaqMan) Primer Design (https://www.genscript.com/ssl-bin/app/primer) (Table 3). qRT-PCR was performed using SYBR® Green Master Mix (ToYoBo, Japan) in an iQ5 (Bio-Rad) thermocycler. CsEF1α (XM_004138916) [70] was used as the housekeeping gene. Three biological replicates per treatment and three technical replicates per sample were analysed. The relative gene expression was calculated using the 2−ΔΔCT relative quantitative analysis method [71], and variance and significance were analysed with DPS 7.05 data processing system software. Significant differences between the treatment and the control were determined using Student’s t- test.
Subcellular localization
A CsERF004 and GFP gene fusion vector was constructed, primers with enzyme digestion sites for BamHI and SmaI were designed (Table 3), and the CsERF004 open reading frame was amplified without its stop codon. The fusion expression vector pSASY-T3-CsERF004 and the transient expression vector pGII-eGFP were digested with BamHI and SmaI, and the product recovered from the gel was ligated to obtain the fusion expression vector 35S–CsERF004-eGFP. The A. thaliana (Columbia ecotype) were obtained from the European Arabidopsis Stock Centre (NASC, Nottingham, UK). The A. thaliana protoplasts were extracted and transformed as described elsewhere [72]. Subcellular localization was observed using confocal laser scanning microscopy (Leica, Germany).
Genetic transformation of the susceptible cultivar D0401
The full-length CDS of the CsERF004 gene obtained using PCR was ligated into the PCXSN-1250 vector, which had been digested with XcmI [73], to yield the over-expression vector PCXSN-CsERF004. PCXSN-CsERF004 and PCXSN-1250 plasmids were transferred into Agrobacterium tumefaciens LBA4404 (BioVector NTCC Inc., Beijing, China) using the freeze-thaw method [74].
The susceptible cultivar D0401 was transformed with the over-expression vector PCXSN-CsERF004 using the cucumber genetic transformation technology [75] described by Zhang et al. and grown on a concentration of 1 mg/L glufosinate.
Expression analysis and physiological index determination of CsERF004-over-expressioning plants
The expression of CsPR1 and CsPR4 was analysed by qRT-PCR in CsERF004-over-expressing plants. The ET content was determined by gas chromatography [76]. SA extraction was performed as described elsewhere [77], and a plant SA ELISA kit (Shanghai, China enzyme biotechnology Co., Ltd.) was used to determine the SA content.
Abbreviations
- cDNA:
-
Complementrary DNA
- ET:
-
Ethylene
- MeJA:
-
Methyl jasmonate
- PCR:
-
Polymerase chain reaction
- PDA:
-
Potato dextrose agar medium
- PR1:
-
Pathogenesis-related protein 1-like
- PR4:
-
Pathogenesis-related protein 4A
- qRT-PCR:
-
Real-time quantitative reverse transcription-polymerase chain reaction
- RNA:
-
Ribonuncleic acid
- SA:
-
Salicylic acid
References
Zhang Y, Pu Z, Qin Z, Zhou X, Liu D, Dai L, et al. A study on the overwintering of cucumber downy mildew oospores in China. J Phytopathol. 2012;160(9):469–74.
Teramoto A, Martins MC, Ferreira LC, Cunha MG. Reaction of hybrids, inhibition in vitro and target spot control in cucumber. Hortic Bras. 2011;29(3):342–8.
Holmes GJ, Ojiambo PS, Hausbeck MK, Quesada-Ocampo L, Keinath AP. Resurgence of cucurbit downy mildew in the United States: a watershed event for research and extension. Plant Dis. 2015;99:428–41.
Yang SJ, Gu XF, Zhang SP, Miao H, Li BJ. Research progress on cucumber target leaf spot (Corynespora cassiicola). China Veg. 2012;4:1–9.
Kozik EU, Klosinska U, Call AD, Wehner TC. Heritability and genetic variance estimates for resistance to downy mildew in cucumber accession ames 2354. Crop Sci. 2013;53:177–82.
Zhang S, Liu MM, Miao H, Zhang SQ, Yang YH, Xie BY. Chromosomal mapping and QTL analysis of resistance to downy mildew in Cucumis sativus. Plant Dis. 2013;97:245–51.
Pang X, Zhou XH, Wan HJ, Chen JF. QTL mapping of downy mildew resistance in an introgression line derived from interspecific hybridization between cucumber and Cucumis hystrix. J Phytopathol. 2013;161:536–43.
Szczechura W, Staniaszek M, Klosinska U, Kozik EU. Molecular analysis of new sources of resistance to Pseudoperonospora cubensis (Berk.et Curt.) Rostovzev in cucumber. Russ J Genet. 2015;51(10):974–9.
Wang YH, VandenLangenberg K, Wehner TC, Kraan PAG, Suelmann J, Zheng XY, et al. QTL mapping for downy mildew resistance in cucumber inbred line WI7120(PI330628). Theor Appl Genet. 2016;129(8):1493–505.
Van Vliet GJA, Meijsing WD. Inheritance of resistance to Pseudoperonospora cubensis rost. In cucumber (Cucumis sativus L.). Euphytica. 1974;23(2):251–5.
Van Vliet GJA, Meijsing WD. Relation in the inheritance of resistance to Pseudoperonospora cubensis rost. And Sphaerotheca fuligineapoll in cucumber (Cucumis sativus L.). Euphytica. 1977;26(3):793–6.
Fanourakis NE, Simon PW. Analysis of genetic linkage in the cucumber. J Hered. 1987;78(4):238–42.
Meng PQ, Cai LJ, Zhang GH, Du SL. Study on molecular markers linked to cucumber downy mildew resistance gene. J Changjiang Veg. 2014;08:12–4.
Call AD, Wehner TC. Gene list 2010 for cucumber. Cucurbit Genet Cooperative. 2010;28–29:105–41.
Wang HZ, Li SJ, Guan W. Identification and genetic analysis of resistance to cucumber target spot. China Cucurbits and Veg. 2010;1:24–5.
Yang LM, Koo DH, Li YH, Zhang XJ, Luan FS, Havey MJ, et al. Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J. 2012;71(6):895–906.
Wen C, Mao A, Dong C, Liu H, Yu S, Guo YD, et al. Fine genetic mapping of target leaf spot resistance gene cca-3 in cucumber, Cucumis sativus L. Theor Appl Genet. 2015;128(12):2495–506.
Jones JD, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–9.
Zipfel C, Robatzek S. Pathogen-associated molecular pattern-triggered immunity: Veni, Vidi…? Plant Physiol. 2010;154(2):551–4.
Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323(5910):95–101.
Chen SY, Chronis D, Wang XH. The novel GrCEP12 peptide from the plant-parasitic nematode Globodera rostochiensis suppresses flg22-mediated PTI. Plant Sign Behav. 2017; DOI: 10.4161/psb.25359.
Cheng X, Tian CJ, Li AN, Qiu JL. Advances on molecular mechanisms of plant-pathogen interactions. Hereditas. 2012;34(2):134–44.
Dong L, Cheng Y, Wu J, Cheng Q, Li W, Fan S, et al. Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J Exp Bot. 2015;66(9):2635–47.
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration-and cold-inducible gene expression. Biochem Bioph res co. 2002;290(3):998–1009.
Jofuku KD, Den Boer BG, Van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6(9):1211–25.
Ohme-Takagi M, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell. 1995;7(2):173–82.
Hernandez-Garcia CM, Bouchard RA, Rushton PJ, Jones ML, Chen X, Timko MP, et al. High level transgenic expression of soybean (Glycine max) GmERF and Gmubi gene promoters isolated by a novel promoter analysis pipeline. BMC Plant Biol. 2010;10(1):237–52.
Zhang H, Zhang J, Quan R, Pan X, Wan L, Huang R. EAR motif mutation of rice OsERF3 alters the regulation of ethylene biosynthesis and drought tolerance. Planta. 2013;237(6):1443–51.
Fountain JC, Raruang Y, Luo M, Brown RL, Guo B, Chen ZY. Potential roles of WRKY transcription factors in regulating host defense responses during Aspergillus flavus infection of immature maize kernels. Physiol Mol Plant P. 2015;89:31–40.
Gu YQ, Yang C, Thara VK, Zhou J, Martin GB. Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell. 2000;12(5):771–85.
Caarls L, Pieterse CM, Van Wees S. How salicylic acid takes transcriptional control over jasmonic acid signaling. Front Plant Sci. 2015;6:170.
Romero I, Vazquez-Hernandez M, Escribano MI, Merodio C, Sanchez-Ballesta MT. Expression Profiles and DNA-Binding Affinity of Five ERF Genes in Bunches of Vitis vinifera cv. Cardinal Treated with High Levels of CO2 at Low Temperature. Front Plant Sci. 2016;7:370.
Van Loon LC, Rep M, Pieterse CMJ. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol. 2006;44:135–62.
Alexander D, Goodman RM, Gut-Rella M, Glascock C, Weymann K, Friedrich L, et al. Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. P Natl A Sci. 1993;90(15):7327–31.
Li X, Xia B, Jiang Y, Wu Q, Wang C, He L, et al. A new pathogenesis-related protein, LrPR4, from Lycoris radiata, and its antifungal activity against Magnaporthe grisea. Mol Biol Rep. 2010;37(2):995–1001.
El-Kereamy A, El-Sharkawy I, Ramamoorthy R, Taheri A, Errampalli D, Kumar P, et al. Prunus domestica pathogenesis-related protein-5 activates the defense response pathway and enhances the resistance to fungal infection. PLoS One. 2011; doi: 10.1371/journal.pone.0017973.
Park CJ, Kim KJ, Shin R, Park JM, Shin YC, Paek KH. Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J. 2004;37(2):186–98.
Müller M, Munné-Bosch S. Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 2015;169(1):32–41.
Gutterson N, Reuber TL. Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr Opin Plant Biol. 2004;7(4):465–71.
Nakano T, Fujisawa M, Shima Y, Ito Y. The AP2/ERF transcription factor SlERF52 functions in flower pedicel abscission in tomato. J Exp Bot. 2014;65(12):3111–9.
Zhang G, Chen M, Li L, Xu Z, Chen X, Guo J, et al. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot. 2009;60(13):3781–96.
Liu D, Chen X, Liu J, Ye J, Guo Z. The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J Exp Bot. 2012;63(10):3899–911.
Zhu Q, Zhang J, Gao X, Tong J, Xiao L, Li W, et al. The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses. Gene. 2010;457(1):1–12.
Chen XJ, Guo ZJ. Tobacco OPBP1 enhances salt tolerance and disease resistance of transgenic rice. Int J Mol Sci. 2008;9(12):2601–13.
Hu L, Liu S. Genome-wide identification and phylogenetic analysis of the ERF gene family in cucumbers. Genet Mol Biol. 2011;34(4):624–33.
Zhang HM, Zhang L, Ma YS, Shang T, Wang S, Yang Q, et al. Studies of an ap2/erf transcriptional factor regulating bi gene of Cucumis sativus. Acta Hortic Sin. 2014;41(4):672–80.
Zhang Y, Wang X, Yang S, Chi J, Zhang G, Ma Z. Cloning and characterization of a Verticillium wilt resistance gene from Gossypium barbadense and functional analysis in Arabidopsis thaliana. Plant Cell Rep. 2011;30:2085–96.
Zhang Z, Zhang H, Quan R, Wang XC, Huang R. Transcriptional regulation of the ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco. Plant Physiol. 2009;150(1):365–77.
Zhang Y, Wang X, Li Y, Wu L, Zhou H, Zhang G, et al. Ectopic expression of a novel ser/Thr protein kinase from cotton (Gossypium barbadense), enhances resistance to Verticillium dahliae infection and oxidative stress in Arabidopsis. Plant Cell Rep. 2013;32:1703–13.
Lai Y, Dang F, Lin J, Yu L, Shi Y, Xiao Y, et al. Overexpression of a Chinese cabbage BrERF11 transcription factor enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Physiol Biochem. 2013;62:70–8.
Lu X, Jiang W, Zhang L, Zhang F, Zhang F, Shen Q, et al. AaERF1 positively regulates the resistance to Botrytis cinerea in Artemisia annua. PLoS One.2013; doi: 10.1371/journal.pone.0057657.
McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, et al. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 2005;139(2):949–59.
Oñate-Sánchez L, Anderson JP, Young J, Singh KB. AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol. 2006;143(1):400–9.
Zhang H, Zhang D, Chen J, Yang Y, Huang Z, Huang D, et al. Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Mol Biol. 2004;55(6):825–34.
Wang S, Yang S, Yin Y, Xi J, Li S, Hao D. Molecular dynamics simulations reveal the disparity in specific recognition of GCC-box by AtERFs transcription factors super family in Arabidopsis. J Mol Recognit. 2009;22(6):474–9.
Zuo KJ, Qin J, Zhao JY, Ling H, Zhang LD, Cao YF, et al. Over-expression GbERF2 transcription factor in tobacco enhances brown spots disease resistance by activating expression of downstream genes. Gene. 2007;391(1–2):80–90.
Ponstein AS, Bres-Vloemans SA, Sela-Buurlage MB, van den Elzen PJ, Melchers LS, Cornelissen BJ. A novel pathogen-and wound-inducible tobacco (Nicotiana tabacum) protein with antifungal activity. Plant Physiol. 1994;104:109–18.
Lebeda A, Cohen Y. Cucurbit downy mildew (Pseudoperonospora cubensis)—biology, ecology, epidemiology, host-pathogen interaction and control. Eur J Plant Pathol. 2011;129:157–92.
Déon M, Bourré Y, Gimenez S, Berger A, Bieyssee D, Lamotted FD, et al. Characterization of a cassiicolin-encoding gene from Corynespora cassiicola, pathogen of rubber tree (Hevea brasiliensis). Plant Sci. 2012;185–186:227–37.
Zhang G, Chen M, Chen X, Xu Z, Guan S, Li LC, et al. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J Exp Bot. 2008;59(15):4095–107.
Palti J, Cohen Y. Downy mildew of cucurbits (Pseudoperonospora cubensis): the fungus and its hosts, distribution, epidemiology and control. Phytoparasitica. 1980;8(2):109–47.
Liu D, Qin Z, Zhang Y, Zhou X, Xin M. Histological observation of cucumber infected with Corynespora cassiicola. Eur J Plant Pathol. 2017; doi: 10.1007/s10658-017-1195-8.
Liu D, Xin M, Zhou X, Wang C, Qin Z. Resistance evaluation of the main cucumber germplasms. J Northeast Agr Univ. 2017;48(3):10–6.
Hong JK, Hwang BK. Influence of inoculum density, wetness duration, plant age, inoculation method, and cultivar resistance on infection of pepper plants by Colletotrichum coccodes. Plant Dis. 1998;82(10):1079–83.
Cohen Y, Rubin AE. Mating type and sexual reproduction of Pseudoperonospora cubensis, the downy mildew agent of cucurbits. Eur Plant Pathol. 2012;132(4):577–92.
Liang YC, Sun WC, Si J, Römheld V. Effects of foliar- and root-applied silicon on the enhancement of induced resistance to powdery mildew in Cucumis sativus. Plant Pathol. 2005;54(5):678–85.
Oh SK, Jang HA, Lee SS, Cho HS, Lee DH, Choi D, et al. Cucumber Pti1-L is a cytoplasmic protein kinase involved in defense responses and salt tolerance. J Plant Physiol. 2014;171(10):817–22.
Huang S, Li R, Zhang Z, Li L, Gu XF, Fan W, et al. 2009. The genome of the cucumber, Cucumis sativus L. Nat Genet. 2009;41:1275–81.
Wu T, Qin Z, Zhou X, Feng Z, Du Y. Transcriptome profile analysis of floral sex determination in cucumber. J Plant Physiol. 2010;167(11):905–13.
Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem. 2010;399(2):257–61.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–8.
Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2(7):1565–72.
Chen S, Songkumarn P, Liu J, Wang GL. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 2009;150(3):1111–21.
Jyothishwaran G, Kotresha D, Selvaraj T, Srideshikan SH, Rajvanshi PK, Jayabaskaran C. A modified freeze–thaw method for efficient transformation of Agrobacterium tumefaciens. Current Sci India. 2007;93(6):770–2.
Zhang Y, Zhang X, Liu B, Wang W, Liu X, Chen C, et al. A GAMYB homologue CsGAMYB1 regulates sex expression of cucumber via an ethylene-independent pathway. J Exp Bot. 2014;65(12):3201–13.
Miao M, Yang X, Han X, Wang K. Sugar signalling is involved in the sex expression response of monoecious cucumber to low temperature. J Exp Bot. 2011;62(2):797–804.
Palva TK, Hurtig M, Saindrenan P, Palva ET. Salicylic acid induced resistance to Erwinia carotovora subsp. carotovora in tobacco. MPMI-Mol Plant Microbe Interact. 1994;7(3):356–63.
Acknowledgements
We acknowledge Associate Professor Yongguang Li (the Key Laboratory of Northeastern Soybean Biology and Genetic Breeding of the Ministry of Agriculture, China) for providing the PCXSN-1250 vector. We also thank Huizhe Wang (Tianjin Kerun Cucumber Research Institute, China), who provided the C. cassiicola isolate.
Funding
This study was funded by the National Key Research and Development Program for Breeding Seven Key Crops (2016YFD0101700) and the Key Project of the Heilongjiang Province Natural Science Foundation (ZD2016003).
Availability of data and materials
The data sets supporting the results of this article are included within the article and Additional file 1: Table S1. Materials are available by contacting the corresponding author. The raw transcriptome sequencing data have been uploaded to the National Center for Biotechnology Information (NCBI) (GenBank accession no. SRX2468535). Data regarding the identification of disease-related genes are in Table 1 and Fig. 1. Data on bioinformatics are in Fig. 2. Data on the expression pattern under pathogen stress are in Fig. 3. Data on response pattern in response to hormone induction are in Fig. 4. Data on subcellular localization are in Fig. 5. Data on the CsERF004-over-expressing plants inoculated with P. cubensis and C. cassiicola are in Table 2 and Fig. 6. Data on the expression of PR genes in CsERF004-over-expressing plants are in Fig. 7. Data on the content of SA and ET in CsERF004-over-expressing plants are in Fig. 8. Data on the cis-elements in the CsPR1 and CsPR4 promoters are in Additional file 1: Table S1. All sequences data in Fig. 2 are in Additional file 2: Table S2.
Authors’ contributions
DL wrote the manuscript and performed the following experiments: artificial inoculation, RNA extraction, high-throughput sequencing data analysis, expression pattern analysis, transformation of cucumber, determination of ET and SA, and subcellular localization. ZQ conceived of this study, organized the entire project and helped to prepare the manuscript. XZ, MX and CW prepared the plant materials, analyzed the data and revised the manuscript. YZ provided the P. cubensis isolate, performed the experiment of artificial inoculation and revised the manuscript. All authors have read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Table S1.
Locations and sequences of cis-elements in the promoter regions of the CsPR1 and CsPR4 genes. (docx 19.4 KB)
Additional file 2: Table S2.
All sequences data in Fig. 2. (docx 16.7 KB)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liu, D., Xin, M., Zhou, X. et al. Expression and functional analysis of the transcription factor-encoding Gene CsERF004 in cucumber during Pseudoperonospora cubensis and Corynespora cassiicola infection. BMC Plant Biol 17, 96 (2017). https://doi.org/10.1186/s12870-017-1049-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-017-1049-8