Abstract
Background
Antimicrobial resistant Coagulase-negative Staphylococci (CoNS) have limited treatment options, rendered diseases untreatable and made hospitals to be reservoirs of the resistant strains. The aim of this study was to estimate the pooled prevalence and antimicrobial resistance of clinical isolates of CoNS from Ethiopia.
Results
The electronic database search yielded 6511 articles of which 21 met predefined inclusion criteria. The pooled prevalence of CoNS from Ethiopia was 12% (95% confidence interval (CI): 8, 16%). The analyses revealed high level of CoNS resistance to methicilin (37%[95% CI: 21, 55%]), vancomycin (911%[95% CI: 0, 35%]), penicillin (58%[95% CI: 42, 74%]), amoxicillin (42%[95% CI: 23, 61%]), amoxicillin-clavulanate (27%[95% CI: 2, 27%]), ampicillin (64%[95% CI: 46, 80%]), tetracycline (60% [95% CI: 49, 70%]), doxycycline (36%[95% CI:19,55%]), Sulfamethoxazole-trimethoprim (50%[95% CI: 36, 64%]), ceftriaxone (27% [95% CI: 18, 38%]), cephalothin (32% [95% CI: 7, 62%]), norfloxacin (39%[95% CI: 24, 56%]), chloramphenicol (40%[95% CI: 23, 58%]), clindamycin (11% [95% CI: 2, 27%]), ciprofloxacin (14%[95% CI: 6, 22%]), gentamicin (27%[95% CI:19,36%]) and erythromycin (30%[95% CI:20%,42%]). High heterogeneity, I2 ranging from 69.04 to 96.88%; p-values ≤0.01, was observed. Eggers’ test did not detect publication bias for the meta-analyses and low risk of bias was observed in included studies.
Conclusions
CoNS has gotten resistant to commonly used antimicrobials from Ethiopia. There is a need of launching national antimicrobial treatment, development and implementation of policy guidelines to contain the threat. Further research focusing on factors promoting resistance and the effect of resistance on treatment outcome studies are warranted.
Similar content being viewed by others
Background
The Coagulase-negative Staphylococci (CoNS) are normal flora which often cause infection associated with implanted appliances and devices, especially in the old, very young children and immune-compromised patients [1, 2]. On human body, there is a widespread distribution of CoNS from normal flora to those that cause severe diseases [3, 4]. Acquisition of genes result in the conversion of commensal staphylococci into invasive pathogens [5]. The vast majority of infections caused by CoNS results in hospitalization [6] and they are among the five most commonly reported pathogens in nosocomial infections [7]. Many of the CoNS species are commonly resistant to antimicrobials used currently against staphylococcal infections [8, 9]. The spread of multi-drug resistant CoNS strains has been promoted by the use of antibiotics in hospitals which has provided a reservoir of antimicrobial resistant strains. The genetic exchange between CoNS and Staphylococcus aureus (S. aureus) and the widespread prevalence of methicillin resistance among CoNS species is a great public health concern [5, 9, 10].
The rising trend of antimicrobial resistant staphylococci species including CoNS [11,12,13,14,15] worldwide requires national antimicrobial resistance prevention policy and updated treatment guidelines which are based on national antimicrobial resistance surveillance. However, developing countries like Ethiopia have financial constraints to conduct national antimicrobial resistance survey. Small scale studies funded by various organizations are conducted and these studies could provide ample evidence to fulfill the gap in national antimicrobial surveillance data if they are summarized and synthesized to draw national estimate of national antimicrobial resistance prevalence. To this end, we had previously conducted a meta-analysis of resistance of S. aureus to antimicrobial agents in Ethiopia [16]. The finding of the study revealed a very high level of S. aureus resistance to almost to all of antimicrobial agents commonly used in Ethiopia. Similarly, CoNS are important opportunistic pathogens showing a growing threat of antimicrobial resistance and becoming a difficult-to- treat pathogens. There is a need to have summarized evidence on AMR burden among CoNS to draw national estimate of CoNS antimicrobial resistance prevalence. The current study focuses on CoNS species while the previous study entirely focuses on S. aureus [16]. The two studies are separate and there is no overlapping data. However the two studies followed similar study design and objectives.
To the best of our knowledge, there is neither nationwide prevalence surveillance and resistance data nor meta-analysis or systematic review on prevalence of antimicrobial resistance of CoNS in Ethiopia. However, many small scale studies are conducted in many parts of the country. The studies had clear differences in setting, population, methodology, findings, and other characteristics. The purpose of this study was, therefore, to determine prevalence and antimicrobial resistance of CoNS using the best available literature from Ethiopia.
Methods
Study design
Meta-analysis of prevalence and antimicrobial resistance of CoNS was conducted using the best available evidence from Ethiopia.
Literature search strategy
To identify potentially eligible studies, databases of Pub Med, Google Scholar, Hinari, Scopus and directory of open Access Journals (DOAJ) were searched until December 2016. Two of the authors (SS and SF) independently searched for relevant studies to be included. Selection of the study was done by the two authors independently. Consensus was reached on discussion with the third author (SD) whenever disagreement arose. Endnote software was used to manage the references. Articles indexed in Pub Med were directly downloaded using Endnote while those not found in Pub Med are manually added to Endnote. The reference lists of the identified studies were manually searched to identify additional relevant studies for inclusion.
The search was done using various key words combined by Boolean search conjunctions ‘AND’, ‘OR’ and ‘NOT’. These combined key words are, ‘Staphylococci’ AND ‘antimicrobial resistance AND Ethiopia’, ‘Staphylococci’ AND ‘antimicrobial susceptibility’ AND ‘Ethiopia’, ‘Staphylococci’ AND ‘antibiotic resistance’ AND ‘Ethiopia’, ‘Staphylococci’ AND ‘antibiotic susceptibility’ AND ‘Ethiopia’, ‘Staphylococci’ AND ‘drug resistance’ AND ‘Ethiopia’, ‘Staphylococci’ AND ‘drug susceptibility’ AND ‘Ethiopia’, “Staphylococci’ AND ‘antibacterial resistance’ AND ‘Ethiopia’, ‘Staphylococci’ And ‘antibacterial susceptibility’ AND ‘Ethiopia’.
Study selection procedures and criteria
Two-stage selection of the articles was conducted independently by two of the authors (SS and SF). In the first stage, the titles and abstracts of all retrieved articles were reviewed and grouped as eligible when they address the study question, otherwise dropped from further review. In the second stage, eligible articles were reviewed in full detail for decision on inclusion.
Eligibility criteria
Articles were selected based on predefined inclusion criteria. Included articles in this study were those that had the following characteristics: Prospective or retrospective studies, original journal articles, with antimicrobial susceptibility test data according to the criteria of the Clinical Laboratory Standards Institute (CLSI) [17], studies which defined antimicrobial resistance range according to CLSI manual, and those that used only clinical isolates. Duplicate studies, studies with small number of isolates (1–9), studies conducted other than clinical subjects like on foods, food handlers’ belongings, health workers’ belongings, health workers’ carriage or animals and of non-infectious carriage were excluded.
Data extraction
Excel spreadsheets prepared by SD were used for data extraction. The characteristics of studies extracted included first author name, year of publication, place of study, study design, total number of CoNS, number of resistant CoNS isolates, and isolation source. The primary outcome of this study was prevalence and antimicrobial resistant CoNS. If the proportion of sensitive isolates (x) was reported, the number of resistant isolates was calculated by multiplying the number of total isolates (y) by one minus the proportion of sensitive isolates (1-x). The proportion of CoNS is obtained from the division of CoNS positive population by total population under the study.
Risk of bias assessment
The methodological quality of each included study was assessed using the quality assessment checklist for prevalence studies as used in the study [18]. Graphs of the summary of the risk of bias were developed using RevMan 5.3 (Cochrane Informatics and Knowledge Management Department, London, UK).
Statistical analysis
Statistical analyses were conducted using Stata version 13.0 (Statacorp, LP, college station, TX). The prevalence values were pooled using the metaprop command in Stata [19]. Heterogeneity of the studies was assessed using the I2 statistic. Because of significant heterogeneity amongst the studies, the random-effects model (REM) was used to estimate the pooled proportion and 95% CIs using the DerSimonian and Laird method [20]. The Freeman-Turkey double arcsine transformation was used to avoid the missing of proportions near or at 0 and 1 from meta-analysis [21]. Subgroup analysis was done by study design, isolation source and study area. The presence of publication bias was tested using Egger’s test [22]. This meta-analysis was conducted as per the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) guidelines [23].
Results
Included studies and characteristics
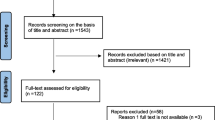
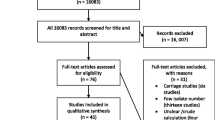
Pub Med database search yielded 591, and Google database yielded 5920. After removal of the duplications, 5329 articles remained for further examination. Title and abstract screening reduced the number of eligible articles to 42 for full text evaluation. Twenty-one articles [13,14,15, 24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41] satisfied the criteria for inclusion for meta-analysis and twenty-one studies were excluded with reasons. Five studies did not address the study question (proportion of resistant CoNS) and were excluded [42,43,44,45,46]. The other sixteen studies with small number of isolates (less than 10) [47,48,49,50,51,52,53,54,55,56,57,58,59] were also excluded (Fig. 1).
Among total of 8047 patients who tested for CoNS, 647 were positives and the isolates were from ear discharge, eye discharge, blood, wound infection, surgical site infection, mixed samples and urine sample. Eleven studies were primary data [13, 15, 25, 27, 28, 30, 36,37,38,39,40] while nine studies were secondary data (records from hospitals or regional laboratories) [14, 24, 26, 29, 31,32,33,34,35]. The characteristics of included studies were summarized in Table 1. Identification of CoNS was conducted using culture on blood agar, Gram-staining, catalase/coagulase test, and cocktail of biochemical assays for carbohydrate fermentation or utilization of specific substrate in all included studies. Kirby Bauer disk diffusion method was used for determination of antimicrobial susceptibility test.
Publication bias, heterogeneity and risk of bias assessment
High heterogeneity, I2 ranging from 67.65 to 96.76%; p-values ≤0.01 was observed. Eggers’ test didn’t detect publication bias. Overall, most of the prospective studies demonstrated a low risk of bias; however that of retrospective relatively a higher risk of bias. The methodological quality of the studies included is illustrated in (Fig. 2).
Prevalence of coagulase-negative staphylococci
The pooled prevalence of CoNS clinical isolates from Ethiopia was 12%(95% CI: 8, 16%), I2 = 96.88%. Subgroup analysis by study design showed significantly higher prevalence of CoNS in prospective studies compared to retrospective studies (16%(95% CI: 11, 21%) versus 6%(95% CI: 3, 10%)) (Fig. 3). Higher prevalence from surgical site infection (SSI) (19%) was observed compared to urinary tract infection (3%) (Fig. 4). A subgroup analysis by study region showed a higher value of CoNS in Addis Ababa 28%, significantly higher than Mekelle, Jimma, Dessie, and Gondar (Fig. 5).
Prevalence of antimicrobials resistant coagulase-negative staphylococci
High level of resistance by CoNS was observed to most commonly used antimicrobials from Ethiopia and the resistance level was summarized in Table 2. Methicilin resistance was 37% (95 %CI: 21, 55%), Additional file 1: Figure S1; to vancomycin 11% (95% CI: 0, 35%), Additional file 2: Figure S2; to penicillin 58%(95% CI: 42, 74%), Additional file 3: Figure S3; to amoxicillin 42% (95% CI: 23, 61%), Additional file 4: Figure S4; to amoxicillin-clavulanate 27%(95% CI: 2, 27%), Additional file 5: Figure S5; to ampicillin 64%(95% CI: 46, 80%), Additional file 6: Figure S6. Similarly, resistance to other antimicrobials was high; to tetracycline 60% (95% CI: 49, 70%), Additional file 7: Figure S7; to doxycycline 36% (95% CI:19,55, 81.18%, Additional file 8: Figure S8; to sulfametaxazole-trimethoprim 50% [95% CI: 36, 64%), Additional file 9: Figure S9; to ceftriaxone (27% [95% CI: 18, 38%]), Additional file 10: Figure S10; to cephalothin (32% [95% CI: 7, 62%], Additional file 11: Figure S11; to norfloxacin (39% [95% CI: 24, 56%], Additional file 12: Figure S12; to chloramphenicol (40% [95% CI: 23, 58%], Additional file 13: Figure S13; to clindamycin (11% [95% CI: 2, 27%]) Additional file 14: Figure S14; and to ciprofloxacin (14% [95% CI: 6, 22%]), Additional file 15: Figure S15; to gentamicin 27% (95%, CI:19, 36%), Additional file 16: Figure S16; and to erythromycin 30% (95%, CI:20, 42%) Additional file 17: Figure S17. Forest plots of the abovementioned antimicrobials resistance were placed as additional file respectively from methicilin to erythromycin (Additional file 1: Figure S1, Additional file 2: Figure S2, Additional file 3: Figure S3, Additional file 4: Figure S4, Additional file 5: Figure S5, Additional file 6: Figure S6, Additional file 7: Figure S7, Additional file 8: Figure S8, Additional file 9: Figure S9, Additional file 10: Figure S10, Additional file 11: Figure S11, Additional file 12: Figure S12, Additional file 13: Figure S13, Additional file 14: Figure S14, Additional file 15: Figure S15, Additional file 16: Figure S16 and Additional file 17: Figure S17) and the proportions were summarized in Table 2.
Comparisons of the prevalence of CoNS resistance to different antimicrobial agents were outlined in (Fig. 6). The magnitude of CoNS resistance to the different antimicrobials ranges from 11% to vancomycin and clindamycin to 64% to ampicillin.
Discussion
In this meta-analysis, we estimated the pooled prevalence and antimicrobial resistance of CoNS to 17 different antimicrobials commonly used in Ethiopia. Twenty-one studies were included in this meta-analysis and the number of studies included in each meta-analysis varies from 5 to 17. Overall, the 21 studies provided evidence concerning the prevalence and antimicrobial resistance of CoNS to different antimicrobials based on isolates identified from 8047 patients. The study revealed that CoNS resistances to commonly available antimicrobials were high, ranging from 11% (vancomycin) to 64% (clindamycin and amipicilin) in Ethiopia.
The emergence of antimicrobial resistant staphylococcus species is a global concern. Our previous studies showed that MRSA is unexpectedly high (97.0%) in Hawassa University comprehensive specialized hospital, Ethiopia [12] and meta-analysis on MRSA in Ethiopia demonstrated very high level of resistance (47% [95% CI: 33, 61%]) [16] and in other meta-analysis similar results were observed (32.5% [95% CI, 24.1 to 40.9%]) [60]. The current findings on MRCoNS showed similarly high resistance (34 95% CI [17, 53%]). These findings signified that methicillin resistance is increasing and becoming of public health concern in Ethiopia. The prevalence of MRCoNS was relatively lower than MRSA. This may be due to infrequent occurrence of infection with CoNS compared to S. aureus resulting in decreased antimicrobial exposure. However, CoNS is continuously evolving from commensal staphylococci into invasive pathogens and then to resistant strains and possibly gaining resistant genes from S. aureus [5].
There is no published meta-analysis focusing on antimicrobial resistance of CoNS for comparison, however the pooled prevalence of CoNS resistance in this study showed a higher level of resistance compared to other studies in different parts of the world [8, 61]. A higher prevalence of resistance was observed in a study conducted in India compared to the current study [62, 63]. A higher rate of resistance in this study may be due to a higher exposure, irrational use of antimicrobial agents and lack of infection prevention policies, especially in hospital settings.
Subgroup analysis by study design showed a higher pooled prevalence of CoNS in prospective studies (17%) compared to retrospective studies (6%). This could be attributed to missing data and poorly defined denominator in the case of retrospective design. A higher prevalence of CoNS in SSI may be due to the fact that CoNS are opportunistic pathogens causing infections in patients with implanted medical devices and surgical procedures [8, 64]. A higher prevalence of CoNS in Addis Ababa compared to other regions can be explained by lager exposure rate in Tikur Anbessa referral hospital where referral cases with severe diseases of recurrent infection treated from all over the country.
Aminoglycosides and fluoroquinolones showed relatively lower level of resistance in this study. A lower level of resistance observed could suggest the development of mutant resistant strains for commonly used first line agents made the microbes easily susceptible to less commonly used antimicrobials [65, 66]. The lower rate of resistance observed with clindamycin may be due to its infrequent use in Ethiopia resulting in lower exposure rate.
The cause of antimicrobial resistance is multifactorial, from lack of infection prevention to irrational use of antimicrobials by health professionals and patients [67]. It is a common practice that antimicrobials can be obtained over-the-counter in Ethiopia. This mishandling of antimicrobials is the main cause of emergence of resistance [68]. Absence of culture techniques and routine antimicrobial susceptibility testing and consequent empiric therapy is a reason for selection of resistant strains and spread. Therefore, in line to strategies for prevention and containment of resistance there is a need for innovative ways of halting resistance. Combination therapy and search for novel antimicrobials will provide a vital role to counter this global problem.
Significant risk of bias was observed with five retrospective studies. This risk of bias was due to unclear denominator in retrospective study during data retrieval and possibly loss of samples during retrieval resulting in non-response. In addition, all of the included studies were conducted in small particular localities and non-representative convenient samplings technique was used. Therefore, the limitations of this study arise from the characteristics of included studies. First, this is in vitro antimicrobial resistance testing and direct inference to clinical outcome calls for caution. Secondly, many studies were conducted in limited localities and mainly in teaching hospitals in bigger cities where patients with advanced and severe stages of diseases with recurrent infection are treated. For this reason, the resistance prevalence could have been overrated. Lastly, studies were not conducted on phenotypic characteristics of resistance, lacking details of molecular characteristic of resistant strains.
Conclusions
This meta-analysis revealed that CoNS has gotten resistant to many of common antibiotics used in Ethiopia. Launching national antimicrobial treatment and use policy guideline is essential for fighting the antimicrobial resistance. Further research focusing on factors promoting antimicrobial resistance, molecular genetics and outcome studies are warranted. Antimicrobial susceptibility should be determined prior to treatment of infections.
Abbreviations
- CI:
-
Confidence interval
- CLSI:
-
Clinical Laboratory Standards Institute
- CoNS:
-
Coagulase negative Staphylococci
- MRCoNS:
-
Methicillin-resistant coagulase-negative staphylococci
- MRSA:
-
Methicillin resistant Staphylococcus aureus
- S. aureus :
-
Staphylococcus aureus
- SSI:
-
Surgical sit infection
- VRSA:
-
Vancomycin resistant Staphylococcus aureus
References
Gill VJ, Selepak ST, Williams EC. Species identification and antibiotic susceptibilities of coagulase-negative staphylococci isolated from clinical specimens. J Clin Microbiol. 1983;18(6):1314–9.
Fleer A, Verhoef J. New aspects of staphylococcal infections: emergence of coagulase-negative staphylococci as pathogens. Antonie Van Leeuwenhoek. 1984;50(5–6):729–44.
Harris LF. Coagulase-negative staphylococci--emerging pathogens. Alaska Med. 1985;54(10):23–4. 26, 31
Gautam V, Sethuraman N, Kaur R, Sachdev S, Marwaha N, Ray P. Changing epidemiology of coagulase-negative staphylococci in normal flora of skin. Indian J Med Microbiol. 2017;35(2):277–8.
Yu W, Kim HK, Rauch S, Schneewind O, Missiakas D. Pathogenic conversion of coagulase-negative staphylococci. Microbes Infect. 2017;19(2):101–9.
Pinilla LC, Vasquez E, Moreno FA, Talavera D, Mendoza N, Donderis C, Toro J. Identification of coagulase-negative staphylococci in hospitalized patients. Rev Med Panama. 1985;10(2):111–5.
Kloos WE, Bannerman TL. Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev. 1994;7(1):117–40.
Szczuka E, Jablonska L, Kaznowski A. Coagulase-negative staphylococci: pathogenesis, occurrence of antibiotic resistance genes and in vitro effects of antimicrobial agents on biofilm-growing bacteria. J Med Microbiol. 2016;65(12):1405–13.
Gilani M, Usman J, Latif M, Munir T, Gill MM, Anjum R, Babar N. Methicillin resistant coagulase negative staphylococcus: from colonizer to a pathogen. Pak J Pharm Sci. 2016;29(4):1117–21.
Hanssen AM, Kjeldsen G, Sollid JU. Local variants of staphylococcal cassette chromosome mec in sporadic methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci: evidence of horizontal gene transfer? Antimicrob Agents Chemother. 2004;48(1):285–96.
Soumya KR, Philip S, Sugathan S, Mathew J, Radhakrishnan EK. Virulence factors associated with coagulase negative staphylococci isolated from human infections. 3 Biotech. 2017;7(2):140.
Deyno S, Toma A, Worku M, Bekele M. Antimicrobial resistance profile of staphylococcus aureus isolates isolated from ear discharges of patients at University of Hawassa comprehensive specialized hospital. BMC Pharmacol Toxicol. 2017;18(1):35.
Shiferaw B, Gelaw B, Assefa A, Assefa Y, Addis Z. Bacterial isolates and their antimicrobial susceptibility pattern among patients with external ocular infections at Borumeda hospital, Northeast Ethiopia. BMC Ophthalmol. 2015;15:103.
Muluye D, Wondimeneh Y, Moges F, Nega T, Ferede G. Types and drug susceptibility patterns of bacterial isolates from eye discharge samples at Gondar University hospital, Northwest Ethiopia. BMC Res Notes. 2014;7:292.
Mama M, Abdissa A, Sewunet T. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University specialized hospital, south-West Ethiopia. Ann Clin Microbiol Antimicrob. 2014;13:14.
Deyno S, Fekadu S, Astatkie A. Resistance of Staphylococcus aureus to antimicrobial agents in Ethiopia: a meta-analysis. Antimicrob Resist Infect Control. 2017;6:85.
Stephen J, Ronald J, Yvette S, José H, Ivonne D, Robert L, Susan E, Carol A. Manual of antimicrobial susceptibility testing. Washington: American Society for Microbiology; Marie, B. 2005. p. 39–40.
Ding C, Yang Z, Wang J, Liu X, Cao Y, Pan Y, Han L, Zhan S. Prevalence of Pseudomonas aeruginosa and antimicrobial-resistant Pseudomonas aeruginosa in patients with pneumonia in mainland China: a systematic review and meta-analysis. Int J Infect Dis. 2016;49(2016):119–28.
Nyaga VN, Arbyn M, Aerts M. Metaprop: a Stata command to perform meta-analysis of binomial data. Archives Public Health. 2014;72(39)
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88.
Miller JJ. The inverse of the freeman – Tukey double arcsine transformation. Am Stat. 1978;32(4):138.
Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–34.
Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. 2010;8(5):336–41.
Hailu D, Mekonnen D, Derbie A, Mulu W, Abera B. Pathogenic bacteria profile and antimicrobial susceptibility patterns of ear infection at Bahir Dar regional Health Research Laboratory center, Ethiopia. Springerplus. 2016;5:466.
Abera B, Alem A, Bezabih B. Methicillin-resistant strains of Staphylococcus aureus and coagulase-negative staphylococus from clinical isolates at Felege Hiwot Refferal hospital, North West Ethiopia. Ethiop Med J. 2008;46(2):149–54.
Biadglegne F, Abera B. Antimicrobial resistance of bacterial isolates from urinary tract infections at Felge Hiwot referral hospital, Ethiopia. Ethiop J Health Dev. 2009;23(3):236–8.
Guta M, Aragaw K, Merid Y. Bacteria from infected surgical wounds and their antimicrobial resistance in Hawassa university referral teaching hospital, southern Ethiopia. Afr J Microbiol Res. 2014;8(11):1118–24.
Mengesha RE, Kasa BG, Saravanan M, Berhe DF, Wasihun AG. Aerobic bacteria in post surgical wound infections and pattern of their antimicrobial susceptibility in Ayder teaching and referral hospital, Mekelle, Ethiopia. BMC Res Notes. 2014;7:575.
Dagnew M, Yismaw G, Gizachew M, Gadisa A, Abebe T, Tadesse T, Alemu A, Mathewos B. Bacterial profile and antimicrobial susceptibility pattern in septicemia suspected patients attending Gondar University hospital, Northwest Ethiopia. BMC Res Notes. 2013;6:283.
Godebo G, Kibru G, Tassew H. Multidrug-resistant bacterial isolates in infected wounds at Jimma University specialized hospital, Ethiopia. Ann Clin Microbiol Antimicrob. 2013;12:17.
Muluye D, Wondimeneh Y, Ferede G, Moges F, Nega T. Bacterial isolates and drug susceptibility patterns of ear discharge from patients with ear infection at Gondar University hospital, Northwest Ethiopia. BMC Ear Nose Throat Disorders. 2013;13(1):10.
Wasihun AG, Wlekidan LN, Gebremariam SA, Dejene TA, Welderufael AL, Haile TD, Muthupandian S. Bacteriological profile and antimicrobial susceptibility patterns of blood culture isolates among febrile patients in Mekelle hospital, northern Ethiopia. Springerplus. 2015;4:314.
Aweke T, Dibaba G, Ashenafi K, Kebede M. Bacterial pathogens of exterior ocular infections and their antibiotic vulnerability pattern in southern Ethiopia. African J Immunol Res. 2014;1(1):019–25.
Kibret M, Abera B. Antimicrobial resistance trend of Bacteria from clinical isolates: an 8-year retrospective study at Dessie regional laboratory, Northeast Ethiopia. Ethiopian Pharmac J. 2010;28:39–46.
Kibret M, Abera B. Prevalence and antibiogram of bacterial isolates from urinary tract infections at Dessie Health Research Laboratory, Ethiopia. Asian Pac J Trop Biomed. 2014;4(2):164–8.
Sewunet T, Demissie Y, Mihret A, Abebe T. Bacterial profile and antimicrobial susceptibility pattern of isolates among burn patients at Yekatit 12 hospital burn center, Addis Ababa, Ethiopia. Ethiop J Health Sci. 2013;23(3):209–16.
Tadesse E, Teshome M, Merid Y, Kibret B, Shimelis T. Asymptomatic urinary tract infection among pregnant women attending the antenatal clinic of Hawassa referral hospital, southern Ethiopia. BMC Res Notes. 2014;7:155.
Tenssay ZW. Multiple antimicrobial resistance in bacterial isolates from clinical and environmental sources of Jimma hospital, south West Ethiopia. Ethiopia J Sci. 2002;25(2):295–302.
Lema T, Woldeamanuel Y, Asrat D, Hunegnaw M, Baraki A, Kebede Y, Yamuah L, Aseffa A. The pattern of bacterial isolates and drug sensitivities of infected ulcers in patients with leprosy in ALERT, Kuyera and Gambo hospitals, Ethiopia. Lepr Rev. 2012;83(1):40–51.
Tesfaye T, Beyene T, Gelaw T, Bekele S, Saravanan M. Bacterial profile and antimicrobial susceptibility pattern of external ocular infections in Jimma University specialized hospital, Southwest Ethiopia. Amer J Infect Dis Microbiol. 2013;1(1):13–20.
Wasihun AG, Yilikal Zemene Y. Bacterial profile and antimicrobial susceptibility patterns of otitis media in AyderTeaching and referral hospital, Mekelle university, Northern Ethiopia. Springer Plus. 2015;4(701)
Argaw-Denboba A, Abejew AA, Mekonnen AG. Antibiotic-resistant Bacteria are major threats of otitis Media in Wollo Area, northeastern Ethiopia: a ten-year retrospective analysis. Int J Microbiol. 2016;2016:8724671.
Endris M, Takele Y, Woldeyohannes D, Tiruneh M, Mohammed R, Moges F, Lynen L, Jacobs J, van Griensven J, Diro E. Bacterial sepsis in patients with visceral leishmaniasis in Northwest Ethiopia. Biomed Res Int. 2014;2014:361058.
Gizachew M, Abdella H, Tiruneh M. Antimicrobial Susceptibility Patterns of Staphylococcus aureus at the University of Gondar Tertiary Hospital, Northwest Ethiopia: A Retrospective Cross Sectional Study. Bacteriol Parasitol. 2015;6:3.
Wondimeneh Y, Muluye D, Alemu A, Atinafu A, Yitayew G, Gebrecherkos T, Damtie D, Ferede G. Urinary tract infection among obstetric fistula patients at Gondar University hospital, Northwest Ethiopia. BMC Womens Health. 2014;14:12.
Worku M, Bekele M. Bacterial isolate and antibacterial resistance pattern of ear infection among patients attending at Hawassa university referral hospital, Hawassa, Ethiopia. Indian J Otol. 2014;20:155–9.
Abera B, Kibret M. Bacteriology and antimicrobial susceptibility of otitis Media at Dessie Regional Health Research Laboratory, Ethiopia. Ethiop J Health Dev. 2011;25(2):161–7.
Abrha A, Abdissa A, Beyene G, Getahun G, Girma T. Bacteraemia among severely malnourished children in Jimma university hospital, Ethiopia. Ethiop J Health Sci. 2011;21(3):175–82.
Alebachew G, Teka B, Endris M, Shiferaw Y, Tessema B. Etiologic agents of bacterial Sepsis and their antibiotic susceptibility patterns among patients living with human immunodeficiency virus at Gondar University teaching hospital, Northwest Ethiopia. Biomed Res Int. 2016;2016:5371875.
Assefa Y, Moges F, Endris M, Zereay B, Amare B, Bekele D, Tesfaye S, Mulu A, Belyhun Y. Bacteriological profile and drug susceptibility patterns in dacryocystitis patients attending Gondar University teaching hospital, Northwest Ethiopia. BMC Ophthalmol. 2015;15:34.
Beyene G, Tsegaye W. Bacterial uropathogens in urinary tract infection and antibiotic susceptibility pattern in Jimma university specialized hospital, southwest ethiopia. Ethiop J Health Sci. 2011;21(2):141–6.
Demilie T, Beyene G, Melaku S, Tsegaye W. Urinary bacterial profile and antibiotic susceptibility pattern among pregnant women in north West Ethiopia. Ethiop J Health Sci. 2012;22(2):121–8.
Derese B, Kedir H, Teklemariam Z, Weldegebreal F, Balakrishnan S. Bacterial profile of urinary tract infection and antimicrobial susceptibility pattern among pregnant women attending at antenatal Clinic in Dil Chora Referral Hospital, Dire Dawa, eastern Ethiopia. Ther Clin Risk Manag. 2016;12:251–60.
Dessalegn L, Shimelis T, Tadesse E, Gebre-selassie S. Aerobic bacterial isolates from post-surgical wound and their antimicrobial susceptibility pattern: a hospital based cross-sectional study. J Med Res. 2014;3(2):18–23.
Dessie W, Mulugeta G, Fentaw S, Mihret A, Hassen M, Abebe E. Pattern of bacterial pathogens and their susceptibility isolated from surgical site infections at selected referral hospitals, Addis Ababa, Ethiopia. Int J Microbiol. 2016;2016
Gebrehiwot A, Lakew W, Moges F, Moges B, Anagaw B, Yismaw G, Nega T, Unakal C, Kassu A. Bacterial profile and drug susceptibility pattern of neonatal sepsis in Gondar University hospital, Gondar Northwest Ethiopia. Pharm Lett. 2012;4(6):1811–6.
Gebremariam TT. Bacteriology and risk factors of bacterial keratitis in Jimma, Southwest Ethiopia. Ethiop Med J. 2015;53(4):191–7.
Ramos JM, Perez-Tanoira R, Garcia-Garcia C, Prieto-Perez L, Bellon MC, Mateos F, Tisisano G, Yohannes T, Reyes F, Gorgolas M. Leprosy ulcers in a rural hospital of Ethiopia: pattern of aerobic bacterial isolates and drug sensitivities. Ann Clin Microbiol Antimicrob. 2014;13:47.
Tesfahunegn Z, Asrat D, Woldeamanuel Y, Estifanos K. Bacteriology of surgical site and catheter related urinary tract infections among patients admitted in Mekelle hospital, Mekelle, Tigray, Ethiopia. Ethiop Med J. 2009;47(2):117–27.
Eshetie S, Tarekegn F, Moges F, Amsalu A, Birhan W, Huruy K. Methicillin resistant Staphylococcus aureus in Ethiopia: a meta-analysis. BMC Infect Dis. 2016;16(1):689.
Morgenstern M, Erichsen C, Hackl S, Mily J, Militz M, Friederichs J, Hungerer S, Buhren V, Moriarty TF, Post V, et al. Antibiotic resistance of commensal Staphylococcus aureus and coagulase-negative staphylococci in an international cohort of surgeons: a prospective point-prevalence study. PLoS One. 2016;11(2):e0148437.
Singh S, Dhawan B, Kapil A, Kabra SK, Suri A, Sreenivas V, Das BK. Coagulase-negative staphylococci causing blood stream infection at an Indian tertiary care hospital: prevalence, antimicrobial resistance and molecular characterisation. Indian J Med Microbiol. 2016;34(4):500–5.
Hashmi A, Abdullah FE, Abdullah NE, Kazmi SU. Species identification and antibiotic susceptibilities of coagulase- negative staphylococci isolated from urinary tract infection specimens. J Coll Physicians Surg Pak. 2016;26(7):581–4.
Fajardo M, Hidalgo R, Gaitan J, Sanchez-Silos R, Martin-Cordero P. About microbiological methods for detection of oxacillin resistance in coagulase-negative staphylococci. Rev Esp Quimioter. 2016;29(5):244–8.
Richardson JF, Marples RR. Changing resistance to antimicrobial drugs, and resistance typing in clinically significant strains of Staphylococcus epidermidis. J Med Microbiol. 1982;15(4):475–84.
Inouye Y, Hashimoto M, Sugiyama M, Takesue Y, Santo T, Yokoyama T. Susceptibility of methicillin-resistant Staphylococcus aureus clinical isolates to various antimicrobial agents. IV. Aminoglycoside-modifying enzyme AAC(6′)/APH(2″) is responsible for arbekacin-resistance enhanced by bleomycin. Hiroshima J Med Sci. 1994;43(3):87–92.
Yadesa TM, Gudina EK, Angamo MT. Antimicrobial use-related problems and predictors among hospitalized medical in-patients in Southwest Ethiopia: prospective observational study. PLoS One. 2015;10(12):e0138385.
Erku DA. Antimicrobial stewardship: a cross-sectional survey assessing the perceptions and practices of community pharmacists in Ethiopia. Interdiscipl Perspect Infect Dis. 2016;2016:5686752.
Acknowledgements
Serawit Deyno acknowledges PHARMBIOTRAC, Mbarara University of Science and Technology for providing stipend for the PhD study. We would like to appreciate the commitment from Andrew Glory Mtewa of the Malawi University of Science and Technology for assisting with language edition.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
SD and SF conceptualized the research idea. SS and SF conducted literature search, selection and data extraction. SD performed the statistical analyses and prepared the draft manuscript. All authors revised, edited and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S1. Forest plot of the proportion of CoNS resistance to methicillin. (DOCX 19 kb)
Additional file 2:
Figure S2. Forest plot of the proportion of CoNS resistance to vancomycin. (DOCX 16 kb)
Additional file 3:
Figure S3. Forest plot of the proportion of CoNS resistance to penicillin. (DOCX 19 kb)
Additional file 4:
Figure S4. Forest plot of the proportion of CoNS resistance to amoxicillin. (DOCX 18 kb)
Additional file 5:
Figure S5. Forest plot of the proportion of CoNS resistance to amoxicillin-clavulanate. (DOCX 17 kb)
Additional file 6:
Figure S6. Forest plot of the proportion of CoNS resistance to amipicillin. (DOCX 18 kb)
Additional file 7:
Figure S7. Forest plot of the proportion of CoNS resistance to tetracycline. (DOCX 20 kb)
Additional file 8:
Figure S8. Forest plot of the proportion of CoNS resistance to doxycycline. (DOCX 16 kb)
Additional file 9:
Figure S9. Forest plot of the proportion of CoNS resistance to sulfamethoxazole-trimethoprim. (DOCX 21 kb)
Additional file 10:
Figure S10. Forest plot of the proportion of CoNS resistance to ceftriaxone. (DOCX 20 kb)
Additional file 11:
Figure S11. Forest plot of the proportion of CoNS resistance to cephalothin. (DOCX 17 kb)
Additional file 12:
Figure S12. Forest plot of the proportion of CoNS resistance to norfloxacin. (DOCX 18 kb)
Additional file 13:
Figure S13. Forest plot of the proportion of CoNS resistance to chloramphenicol. (DOCX 18 kb)
Additional file 14:
Figure S14. Forest plot of the proportion of CoNS resistance to clinidamycin. (DOCX 17 kb)
Additional file 15:
Figure S15. Forest plot of the proportion of CoNS resistance to ciprofloxacin. (DOCX 19 kb)
Additional file 16:
Figure S16. Forest plot of the proportion of CoNS resistance to gentamicin. (DOCX 21 kb)
Additional file 17:
Figure S17. Forest plot of the proportion of CoNS resistance to erythromycin. (DOCX 21 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Deyno, S., Fekadu, S. & Seyfe, S. Prevalence and antimicrobial resistance of coagulase negative staphylococci clinical isolates from Ethiopia: a meta-analysis. BMC Microbiol 18, 43 (2018). https://doi.org/10.1186/s12866-018-1188-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-018-1188-6