Abstract
Background
Chromosomal painting in manatees has clarified questions about the rapid evolution of sirenians within the Paenungulata clade. Further cytogenetic studies in Afrotherian species may provide information about their evolutionary dynamics, revealing important insights into the ancestral karyotype in the clade representatives. The karyotype of Trichechus inunguis (TIN, Amazonian manatee) was investigated by chromosome painting, using probes from Trichechus manatus latirostris (TML, Florida manatee) to analyze the homeologies between these sirenians.
Results
A high similarity was found between these species, with 31 homologous segments in TIN, nineteen of which are whole autosomes, besides the X and Y sex chromosomes. Four chromosomes from TML (4, 6, 8, and 9) resulted in two hybridization signals, totaling eight acrocentrics in the TIN karyotype. This study confirmed in TIN the chromosomal associations of Homo sapiens (HSA) shared in Afrotheria, such as the 5/21 synteny, and in the Paenungulata clade with the syntenies HSA 2/3, 8/22, and 18/19, in addition to the absence of HSA 4/8 common in eutherian ancestral karyotype (EAK).
Conclusions
TIN shares more conserved chromosomal signals with the Paenungulata Ancestral Karyotype (APK, 2n = 58) than Procavia capensis (Hyracoidea), Loxodonta africana (Proboscidea) and TML (Sirenia), where TML presents less conserved signals with APK, demonstrating that its karyotype is the most derived among the representatives of Paenungulata. The chromosomal changes that evolved from APK to the T. manatus and T. inunguis karyotypes (7 and 4 changes, respectively) are more substantial within the Trichechus genus compared to other paenungulates. Among these species, T. inunguis presents conserved traits of APK in the American manatee genus. Consequently, the karyotype of T. manatus is more derived than that of T. inunguis.
Similar content being viewed by others
Background
Paenungulata (Afrotheria) includes the orders Proboscidea ILLIGER 1811, Hyracoidea HUXLEY 1869, and Sirenia ILLIGER 1811, established by morphological, genomic and cytogenetic evidence, despite the controversial phylogenetic position between these orders [1,2,3,4,5].
The order Sirenia are exclusively aquatic herbivorous mammals, composed of two families, Dugongidae (dugongs) and Trichechidae (manatees), that probably diverged in the early Eocene, 56 million years ago (myr) [6,7,8,9,10]. The Trichechidae family is divided into Miosireninae (extinct) and Trichechinae (current manatees) subfamilies. Three species of the Trichechus genus represent the current manatees, Trichechus manatus LINNAEUS 1758 (West Indian manatee), Trichechus senegalensis LINK 1795 (African manatee) and Trichechus inunguis NATTERER 1883 (Amazonian manatee). The taxon is distributed in the tropical and subtropical regions of the Atlantic Ocean: T. manatus lives in the Atlantic coastal region of the Americas, T. senegalensis in the rivers and coastal areas of western Africa and T. inunguis is endemic to Amazonian rivers [11].
Morphological data established the first phylogenetic relationships of trichequid representatives, suggesting that the first manatees have ancestry from estuarine regions and freshwater environments in South America [7, 12, 13]. Fossil analysis, through studies of tooth morphology, inferred that Ribodon limbatus AMEGHINO 1883 is an ancestor of the genus Trichechus [7, 12, 14]. Domning [7, 12] proposed that T. inunguis is the most recent species among the representatives of Trichechus based on morphology and paleogeographic history.
The mitochondrial gene data described by Vianna et al. [15] strengthened the phylogenetic relationship between T. manatus and T. senegalensis, corroborating the morphological phylogenetic interpretations [7, 12]. However, Cyt b genes in T. inunguis showed a lower degree of sequence changes concerning T. manatus and T. senegalensis, indicating the sequence in T. inunguis as the most conserved among Trichechus, although the study concluded that T. inunguis would be the most recent species. De Souza et al. [16] analyzed the mitochondrial genomes of Trichechus representatives and proposed the time of evolutionary divergence between the species at 6.5 myr. In addition, the study presented T. senegalensis as the oldest species among the Trichechus. It established a closer relationship between T. manatus and T. inunguis, mainly considering the divergence time at 1.34 myr between the two species. These divergence times are very short, considering the significant phenotypic differences between these species [11, 16]. From a morphological perspective, it is possible to confirm the proximity between T. manatus and T. senegalensis due to the similarity in habitat and niches of these species, which contribute to the preservation of typical phenotypes in marine manatees. However, despite the genomic data by Vianna et al. [15] reinforcing this proximity of T. manatus and T. senegalensis, the findings in T. inunguis were controversial in relation to the phylogenetic interpretations already described for the species. The similarity of mitogenomes between T. manatus and T. inunguis described by De Souza et al. [16] proposes, for the first time, a different phylogenetic interpretation for the group.
Chromosome painting has been effective in clarifying information about evolutionary aspects of mammals and assessing karyotypic and phylogenetic ancestry, as well as evolutionary divergence between taxonomic groups [17, 18]. Cytogenetic analyzes available in the literature for Trichechus showed the established diploid number (2n) and autosomal fundamental number (FN) for T. inunguis as 2n = 56/FN = 82 [19,20,21,22] and 2n = 48/FN = 92 for T. manatus [22,23,24]. This variation in karyotypes is remarkable, with a difference of four Robertsonian rearrangements [19] between T. manatus and T. inunguis, considering the short divergence time (1.34 myr) between these species. More recent data from Noronha et al. [22] and De Oliveira et al. [20], based on karyotypic analysis, demonstrated chromosomes rearrangements and the natural occurrence of hybrids from reproduction between T. inunguis and T. manatus or different generations (F1, F2). Cytogenetic data for T. senegalensis have not yet been described.
Cytogenetic analyzes of the African elephant (Loxodonta africana, 2n = 56), Florida manatee (Trichechus manatus latirostris, 2n = 48), and hyrax (Procavia capensis, 2n = 54), by chromosome painting and comparative analysis with Homo sapiens (HSA), show chromosomal signatures that validate the ancestral karyotype of Eutheria (EAK), with HSA 3/21, 7/16, 12/22, 14/15, and 16/19 syntenies, in addition to consolidating the Paenungulata clade with HSA 2/3, 8/22, and 18/19 syntenies [2, 18, 25]. Furthermore, Pardini et al. [2], using chromosome painting in T. m. latirostris (Sirenia), L. africana (Proboscidea), and P. capensis (Hyracoidea), established the karyotypic differences between these species and confirmed 11 synapomorphies that characterize the Paenungulata clade, in addition to establishing the ancestral karyotype (APK, 2n = 58).
Therefore, the verification and number of chromosomal changes that have occurred during the divergence of T. manatus and T. inunguis could help to elucidate the phylogenetic interpretations described for the genus Trichechus. Here, data on chromosome painting in Trichechus inunguis, and the evolutionary aspects that differentiate the manatees T. manatus and T. inunguis and their phylogenetic relationships, are shown for the first time on a comparative chromosomal analysis with other representatives of the Paenungulata clade available from the published data.
Results
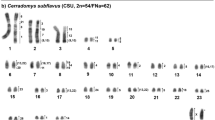
The karyotype of Trichechus inunguis (TIN) presents 2n = 56, FN = 92, and an XX/XY sex chromosome system. Of the autosome chromosomes, 19 pairs are bi-armed and 8 one-armed; the X is submetacentric, and the Y is acrocentric.
Hybridization of T. m. latirostris (TML) probes in TIN demonstrates 31 homeologous segments. Of these, we identified nineteen (TML 1, 2, 3, 5, 7, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, and 23) that hybridized to a single autosomal chromosomes of TIN (TIN 1, 3, 5, 2, 4, 6, 17, 7, 8, 13, 9, 14, 12, 10, 11, 20, 18, 21, and 23, respectively), in addition to the TML X and Y in TIN X and Y, respectively; four TML chromosomes showed two hybridization signals: TML 4 (TIN 16 and 26), TML 6 (TIN 15 and 27), TML 8 (TIN 19 and 22), and TML 9 (TIN 24 and 25) (Fig. 1 and Fig. 2; Table 1).
G-banded karyotype of Trichechus inunguis (2n = 56, FN = 92) [22], with chromosomal mapping plotted from hybridizations with whole chromosome probes from Trichechus manatus latirostris (2n = 48, FN = 92)
Additionally, when comparing by G band and chromosome painting the TML, TIN, Loxodonta africana (LAF) and Procavia capensis (PCA) species, we observed that TIN 1 underwent a pericentric inversion when compared to TML 1; and, TIN 2 (TML 5) and TIN 4 (TML 7) underwent centromere inversion/repositioning when compared to LAF (LAF 5 and LAF 17; LAF 4) and PCA (PCA 4; PCA 3), respectively [2, 22].
Discussion
Comparative analysis between TIN and TML
The comparative analysis between TIN and TML was proposed based on the results of Kellogg et al. [25], with hybridizations of Homo sapiens (HSA) probes in TML and the effects of hybridizations with TML probes in TIN of the present study. Therefore, the data found in TML were used as an intermediary to infer the chromosomal associations of HSA in TIN due to the high degree of genome similarity observed in the hybridizations between these species.
Common associations were observed in the ancestral Eutheria karyotype (AEK) with the HSA syntenies 3/21 (TIN 9), 7/16 (TIN 25), 12/22 in two blocks (TIN 4 and TIN 14), 14/15 (TIN 8), and 16/19 (TIN 13); and the association HSA 5/21 (TIN 1) for the Afrotheria clade, despite the HSA 5/21 gap in the karyotype of Procavia capensis [2]. Paenungulata ancestral karyotype (APK) associations were also found in T. inunguis, with HSA 2/3 syntenies in two blocks (TIN 9 and TIN 12), 18/19 (TIN 7), 8/22 (TIN 14) (see Fig. 3 and Table 3). HSA 4/8 synteny is common in AEK and has been detected in Afroinsectiphilia (African insectivores) [26,27,28,29]. However, it was not observed in T. inunguis, as well as in L. africana, T. m. latirostris, and P. capensis [2, 25, 30], reinforcing that this association was lost in the representatives of Paenungulata.
Comparative analysis by chromosome painting between T. m. latirostris (TML; red bar) and T. inunguis (TIN) (present study) and Homo sapiens (HSA; blue bar) with TML [25]. (*) represent centromeric regions
Comparative analyzes of the Paenungulata Ancestral Karyotype (APK) in Amazonian manatee
Cytogenetic studies on sirenians are still restricted to manatees T. manatus and T. inunguis [2, 19, 21, 23,24,25]. The two species have strikingly different karyotypes (T. inunguis 2n = 56; T. manatus 2n = 48), with a difference of four Robertsonian translocations and one pericentric inversion [22].
Comparative analysis by chromosome painting with TML probes between the TIN karyotype and the paenungulate representatives (Trichechus manatus latirostris – TML, Loxodonta Africana – LAF and Procavia capensis – PCA) corroborate the data found by Pardini et al. [2] who described the Ancestral Karyotype of Paenungulata (APK) (Table 3 and Fig. 4a and b). Comparative analysis by chromosome painting showed that the TIN (2n = 56) and TML (2n = 48) karyotypes differ by 4 fusion/fission events between 8 acrocentric pairs in TIN and 4 submetacentric pairs in TML (Fig. 1). The alterations detected in the TIN karyotype involving the TML chromosomes 4, 6, 8, and 9 also occurred in PCA and LAF, which are fragmented into two to three blocks in these karyotypes, respectively (Fig. 4b; Table 2) [2]. Considering the four Robertsonian rearrangements in TIN (Based on TML chromosomes 4, 6, 8 and 9 hybridization) we suggest that the TIN karyotype is more ancestral than the TML karyotype, since the latter is more similar to the Ancestral Paenungulate Karyotype (APK).
a) Representative idiograms of chromosome painting in Trichechus inunguis (TIN, 2n = 56), Loxodonta africana (LAF, 2n = 56), and Procavia capensis (PCA, 2n = 54) with T. m. latirostris (TML, 2n = 48); b) Chromosomal changes involving TML chromosomes 4, 6, 8, and 9 and the possible corresponding chromosomes of APK, 2n = 58 in TIN, PCA, and LAF
Our data corroborate those of Pardini et al. [2] and confirms that the TIN karyotype maintained the 11 synapomorphies proposed in the paenungulate representatives TML, LAF, and PCA, validated by the karyotype of the outgroup, aardvark (Orycteropus afer, 2n = 20). Furthermore, the study showed that the Ancestral Paenungulata Karyotype (APK) would consist of 2n = 58 chromosomes, validated by the karyotype of the outgroup, aardvark (2n = 20). Comparative analyzes from the APK indicate that L. africana (2n = 56) underwent 5 fusions, 4 fissions, and 1 inversion/centromere repositioning on chromosome 3 (LAF 3) to constitute the current karyotype; P. capensis (2n = 54) underwent 4 fusions and 2 fissions; T. m. latirostris underwent 5 fusions and 2 inversion/centromere repositioning (TML 5 and 7) [2]. From the same perspective of analysis by Pardini et al. [2], the analysis from this present study showed that T. inunguis showed a karyotype modification of 1 fusion (in TIN 9), 1 pericentric inversion (TIN 1) (by Noronha et al. [22]) and 2 inversion/centromere repositioning (TIN 2 and 4), indicating a more conserved karyotype with APK than other paenungulates (Table 3).
The rapid dissemination of the Trichechus genus
The paleoenvironmental dynamics that occurred in South America during the Cenozoic were responsible for the diversification and distribution of the first representatives of the genus Trichechus [12]. During the formation of the Amazon basin, the Andean elevation generated different landscapes that benefited the diversity of the South American biota [31,32,33]. The discovery of the Potamosiren fossil links the first manatees to the estuarine and freshwater environments of South America [7, 12]. The constant marine transgressions that occurred on the continent in the Neogene (Miocene and Pliocene) may have caused the reintroduction of sirenians into fresh waters, as the broad community of sirenians of the Tertiary was marine in origin [6, 9, 13, 32,33,34].
The first Trichechus diverged by allopatry in marine and freshwater environments. Within the Amazon basin, the Trichechus genus modified its diet; the high production of macrophytes and other abrasive grasses selected the first isolated Trichechus; outside the Amazon basin, marine Trichechus took different routes and diversified; Trichechus senegalensis, in coastal regions and rivers of tropical West Africa; and Trichechus manatus, in the coastal area of the American continents [12]. Fossil data for these manatees are still too scarce to suggest past distribution. However, the diversity of Trichechus manatus in the lineage-subspecies T. manatus bakerorum (extinct), T. manatus latirostris (Florida manatee), T. manatus manatus (Antillean manatee), and T. manatus manatus (Brazilian T. manatus) along the American Atlantic coast support a state of rapid diversification within the genus Trichechus, validated by morphological, genomic and cytogenetic characteristics [14, 15, 35,36,37,38].
Although phylogenetic positions are still controversial among extant Trichechus [12, 15, 16, 39], genomic data have estimated the time of evolutionary divergence between these species. The analysis by Cantanhede et al. [36] with D-loop between T. manatus and T. inunguis estimated the time of evolutionary divergence from 3.1 to 0.65 myr, while the complete mitochondrial genomes analyzed by De Souza et al. [16] showed an evolutionary divergence between T. manatus and T. inunguis of 1.34 myr. The short time of divergence between these species can be seen in our data due to the high chromosomic similarity found in the present study, which can also support the existence of natural hybridization between T. manatus and T. inunguis in the Amazon estuary [20, 22]. The estimated rate of chromosomal changes in Paenungulata is considered slow to moderate (0.09 – 0.16 changes per 1 million years – changes/myr) compared to other mammalian groups [2]. The chromosomal changes for the paenungulate of the orders Hyracoidea (P. capensis – 2n = 54) and Proboscidea (L. africana – 2n = 56) show a difference of 6 to 9 changes in APK, respectively, given that the evolutionary divergence of these taxa has been approximately 56 myr [1, 40]. In addition, other known representatives of Hyracoidea (Dendrohyrax arboreus, 2n = 54; Heterohyrax hrucei: 2n = 54) and Proboscidea (Elephas maximus, 2n = 56) still maintain a conserved diploid number [41, 42]. However, the difference of four Robertsonian translocations and a pericentric inversion between T. inunguis (2n = 56) and T. manatus (2n = 48) reveals a high rate of chromosomal changes within the genus Trichechus, between 1 to 5 changes/myr.
The analysis of the Cyt b gene by Vianna et al. [15] suggested that T. inunguis might belong to an older lineage of manatees adapted to freshwater. Therefore, the species may have a more conserved gene sequence than T. manatus and T. senegalensis. The new insights of De Souza et al. [16] on the phylogenetic relationship of T. manatus and T. inunguis provide more specific answers about the differences between these species, which were also reinforced in the present study. The chromosomal changes in APK that led to the karyotype of T. manatus and T. inunguis range from 7 to 4 changes, respectively; this indicates that T. inunguis shares a more conserved karyotype with APK, while T. manatus presents apomorphies that show a condition that is more derived from APK. Notably, the chromosomal evolution of the Trichechus genus will be elucidated only after the application of TML probes to T. senegalensis.
Conclusion
Here, we evaluated by chromosome painting important data on the karyotypic differences between the species Trichechus manatus and Trichechus inunguis and the phylogenetic relationships of these species to other representatives of Paenungulata. The high rate of chromosomal changes in manatees shows them as outliers of the Afrotheria clade. Despite this, the homeologies between the paenungulate karyotypes are still very conserved, with evidence even in the G-banding pattern. The shared HSA syntenies in T. inunguis reveal it as a representative of the placental mammalian taxons Afrotheria and Paenungulata. The phylogenetic signals found in T. inunguis show that the species shares more conserved chromosomal signals with the ancestral karyotype of Paenungulata (APK) compared to hyrax (Procavia capensis), the African elephant (Loxodonta africana), and Florida manatee (Trichechus manatus latirostris). From a phylogenetic perspective, the karyotype of T. m. latirostris is the most derived among the representatives of Paenungulata. Furthermore, the data from this study also point to the phylogenetic position between T. manatus and T. inunguis, showing that T. manatus presents a more recent condition than T. inunguis among the American Trichechus. However, complete understanding of the chromosomal evolution of the genus will be possible only after chromosomal painting of T. senegalensis.
Methods
Blood samples were collected from a male and a female of Trichechus inunguis under the SISBIO license number (Number: 44915–1). Chromosomal preparations were obtained from temporary lymphocyte cultures. Cultivation was performed in RPMI 1640 medium (Vitrocell) with fetal bovine serum (FBS) and phytohemagglutinin and incubated at 37ºC in 5% CO2 for 96 h. Metaphases were analyzed according to chromosome morphology and organized karyotype according to Assis et al. [19]. The G-banding pattern was performed using Seabright’s protocols [43], the best G banded karyotype was published for us in Noronha et al. [22]. The whole chromosome probes used in this study were described by Pardini et al. [2], where 23 peaks were generated from a male of Trichechus manatus latirostris (TML; 2n = 48) by flow-sorted, with 17 peaks of a single chromosome (TML 1, 5, 8, 9, 10, 11, 12, 14, 15, 16, 17, 18, 20, 20, 22, 23, Y) and 3 peaks composed of two chromosomes (2 + 4, 3 + 7 and 6 + X). The TML 20 chromosome is present in 2 separate peaks, possibly due to the heterochromatin difference between homologs carrying the nucleolus organizer region (NOR) and presenting nonspecific markings on the chromosomes. TML chromosomes 11, 14, and 17 have both peaks in their pure form and also mixed peaks with other chromosomes, such as 11 + 13, 14 + 19, and 17 + 21, making it possible to characterize the TML chromosome 19 in hybridizations (Table 1).
In situ hybridizations were performed according to Yang and Graphodatsky [44], photographed with a Zeiss Axiocam camera, coupled to a Zeiss microscope, and analyzed with AxioVision Rel software. 4.6. The analyzes followed the interpretation of the presence/absence of signals in the chromosomes; comparative idiograms were set up in Photoshop CS6 software for cytogenetic analysis between the investigated species.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- 2n:
-
Diploid number
- APK:
-
Ancestral Paenungulata Karyotype
- CO2 :
-
Carbon Dioxide
- Cyt b:
-
Cytochrome B
- D-loop:
-
Displacement loop
- AEK:
-
Ancestral Eutherian Karyotype
- FBS:
-
Fetal bovine serum
- FISH:
-
Fluorescence In-Situ Hybridization
- FN:
-
Fundamental number
- HSA:
-
Homo sapiens
- LAF:
-
Loxodonta africana
- NOR:
-
Nucleolar organizer regions
- OAF:
-
Orycteropus afer
- PCA:
-
Procavia capensis
- SISBIO:
-
Sistema de Autorização e Informação em Biodiversidade
- TIN:
-
Trichechus inunguis
- TMA:
-
Trichechus manatus
- TML:
-
Trichechus manatus latirostris
References
Meredith RW, Janečka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simão TLL, Stadler T, Rabosky DL, Honeycutt RL, Flynn JJ, Ingram CM, Steiner C, Williams TL, Robinson TJ, Burk-Herrick A, Westerman M, Ayoub NA, Springer MS, Murphy WJ. Impacts of the Cretaceous Terrestrial Revolution and KPg Extinction on Mammal Diversification. Science. 2011;334:521–4.
Pardini AT, O’brienFuBondeElder PCMBRKFFB, Ferguson-Smith MA, Yang F, Robinson TJ. Chromosome painting among Proboscidea, Hyracoidea and Sirenia: support for Paenungulata (Afrotheria, Mammalia) but not Tethytheria. Proc Royal Soc B. 2007;274:1333–40.
Poulakakis N, Stamatakis A. Recapitulating the evolution of Afrotheria: 57 genes and rare genomic changes (RGCs) consolidate their history. Syst Biodivers. 2010;8(3):395–408.
Seiffert ER. A new estimate of afrotherian phylogeny based on simultaneous analysis of genomic, morphological, and fossil evidence. BMC Evol Biol. 2007;7(1):224.
Springer MS, Burk-Herrick A, Meredith R, Eizirik E, Teeling E, O’brien SJ, Murphy WJ. The Adequacy of Morphology for Reconstructing the Early History of Placental Mammals. Syst Biol. 2007;56(4):673–84.
Benoit J, Adnet S, Mabrouk EE, Khayati H, Mustapha BHA, Marivaux L, Maezeraud G, Merigeaud S, Vianey-Liaud M, Tabuce R. Cranial Remain from Tunisia Provides New Clues for the Origin and Evolution of Sirenia (Mammalia, Afrotheria) in Africa. PLoS ONE. 2013;8(1):54307.
Domning PD. A phylogenetic analysis of the Sirenia. In Contributions in Marine Mammal Paleontology Honoring Frank C Whitmore Jr. Edited by Berta A. and Deméré TA. Proc San Diego Soc Nat Hist; 1994, 29:177–189.
Domning DP. Sirenian Evolution. In: Würsig B, Thewissen JGM, Perrin W, editors. Encyclopedia of Marine Mammals. 2nd ed. San Diego: Academic Press; 2008. p. 1016–9.
Velez-Juarbe J, Domning DP, Pyenson ND. Iterative Evolution of Sympatric Seacow (Dugongidae, Sirenia) Assemblages during the Past ∼26 Million Years. PLoS ONE. 2012;7(2):31294.
Voss M. On the invalidity of Halitherium schinzii Kaup, 1838 (Mammalia, Sirenia), with comments on systematic consequences. Zoosystematics and Evolution. 2014;90(1):87–93.
Reynolds JE, Powell JA, Keith Diagne LW, Barton SL, Scolardi KM. Manatees: Trichechus manatus, T. senegalensis and T. inunguis. In Encyclopedia of marine mammals 3rd edition. Edited by Wursig JGM and Thewissen JGM. Academic Press; 2018:558–566.
Domning PD. Evolution of manatees: a speculative history. J Paleontol. 1982;56(3):599–619.
Suarez C, Gelfo JN, Moreno-Bernal JW, Velez-Juarbe J. (2021) An early Miocene manatee from Colombia and the initial Sirenian invasion of freshwater ecosystems. J S Am Earth Sci. 2021;109:103277.
Domning DP, Hayek LC. Interspecific and intraspecific morphological variation in manatees (Sirenia: Trichechus). Mar Mamm Sci. 1986;2(2):87–144.
Vianna JA, Bonde RK, Caballero S, Giraldo JP, Lima RP, Clarl A, Marmontel M, Morales-Vela B, Souza MJ, Parr L, Rodriguez-Lopez MA, Mignucci-Giannoni AA, Powell JA, Santos FR. Phylogeography, phylogeny and hybridization in trichechid sirenians: implications for manatee conservation. Mol Ecol. 2006;15(2):433–47.
De Souza ÉMS, Freitas L, Da Silva Ramos EK, Selleghin-Veiga G, Richard-Ribeiro C, Silva FA, Marmontel M, Dos Santos FR, Laudisoit A, Verheyen E, Domning PD, Nery MF. The evolutionary history of manatees told by their mitogenomes. Sci Rep. 2021;11:3564.
Ferguson-Smith M, Trifonov V. Mammalian karyotype evolution. Nat Rev Genet. 2007;8:950–62.
Graphodatsky AS, Trifonov VA, Stanyo R. The genome diversity and karyotype evolution of mammals. Mol Cytogenet. 2011;4(1):22.
Assis MFL, Best RC, Barros RMS, Yonenaga-Yassuda Y. Cytogenetic study of Trichechus inunguis (Amazonian manatee). Rev Bras Genetics. 1989;11:41–50.
De Oliveira EHC, Gomes AJB, Costa AF, Emin-Lima R, Bonvicino CR, Viana MC, Reis LMA, Vidal MD, Cavalcanti MVG, Attademo FLN, Luna FO, Siciliano S. Karyotypical confirmation of natural hybridization between two manatee species, Trichechus manatus and Trichechus inunguis. Life. 2022;12(5):616.
Loughman W, Frye F, Herald E. The chromosomes of a male manatee. International Zoo Yearbook. 1970;10(1):151–2.
Noronha RCR, Almeida BRR, Chagas MCS, Tavares FS, Cardoso AL, Bastos EMC, Silva NKN, Klautal AGCM, Luna FO, Attademo FLN, Lima DS, Sabioni LA, Sampaio MIC, Martins C, Vicari MR, Oliveira JM, Nascimento LAS, Nagamachi CY, Pieczarka JC. Karyotypes of Manatees: new insights into hybrid formation (Trichechus inunguis x Trichechus m. manatus) in the Amazon estuary. Genes. 2022;13(7):1263.
Gray BA, Zori RT, Mcguire PM, Bonde RK. A first-generation cytogenetic ideogram for the Florida manatee (Trichechus manatus latirostris) based on multiple chromosome banding techniques. Hereditas. 2002;137(3):215–23.
White JR, Harkness DR, Isaacks RE, Duffield DA. Some studies on blood of the florida manatee, Trichechus manatus latirostris. Comp Biochem Physiol A Physiol. 1976;55(4):413–7.
Kellogg ME, Burkett S, Dennis TR, Stone G, Gray BA, Mcguire OM, Zori RT, Stanyon R. Chromosome painting in the manatee supports Afrotheria and Paenungulata. BMC Evol Biol. 2007;7(1):6.
Richard F, Lombard M, Dutrillaux B. Reconstruction of the ancestral karyotype of eutherian mammals. Chromosome Res. 2003;11(6):605–18.
Robinson TJ, Ferguson-Smith MA, Yang F. Cross-species chromosome painting in the golden mole and elephant-shrew: support for the mammalian clades Afrotheria and Afroinsectiphillia but not Afroinsectivora. Proc R Soc B. 2004;271:1477–84.
Svartman M, Stone G, Page JE, Stanyon R. A chromosome painting test of the basal eutherian karyotype. Chromosomal Res. 2004;12(1):45–53.
Yang F, Alkalaeva EZ, Perelman PL, Pardini AT, Harrison WR, O’brien PCM, Fu B, Graphodatsky AS, Ferguson-Smith MA, Robinson TJ. Reciprocal chromosome painting among human, aardvark, and elephant (superorder Afrotheria) reveals the likely eutherian ancestral Karyotype. Proc Natl Acad Sci. 2003;100(3):1062–6.
Fröenicke L, Wienberg J, Stone G, Adams L, Stanyon R. Towards the delineation of the ancestral eutherian genome organization: comparative genome maps of human and the African elephant (Loxodonta africana) generated by chromosome painting. Proc Royal Soc B: Biol Sci. 2003;270(1522):1331–40.
Albert JS, Carvalho TP, Petry P, Holder MA, Maxime EL, Espino J, Corahua I, Quispe R, Rengifo B, Ortega H, Reis RE. Aquatic biodiversity in the Amazon: habitat specialization and geographic isolation promote species richness. Animals. 2011;1(2):205–41.
Hoorn C, Wesselingh FP, Ter Steege H, Bermudez MA, Mora A, Sevink J, Sanmartín I, Sanchez-Meseguer A, Anderson CL, Figueireido JP, Jaramillo C, Riff D, Negrí FR, Hooghiemstra H, Lundberg T, Sadler T, Särkinen T, Antonelli A. Amazonia Through Time: Andean Uplift, Climate Change, Landscape Evolution, and Biodiversity. Science. 2010;330:927–31.
Hoorn C, Bogotá-A GR, Romero-Baez M, Lammertsma EL, Flantua SGA, Dantas EL, Dino R, Do Carmo DA, Chemale F. The Amazon at sea: Onset and stages of the Amazon River from a marine record, with special reference to Neogene plant turnover in the drainage basin. Global Planet Change. 2017;153:51–65.
Domning DP. The readaptation of Eocene sirenians to life in water. Hist Biol. 2000;14(1–2):115–9.
Barros HMDR, Meirelles ACO, Luna FO, Marmontel M, Cordeiro-Estrela P, Santos N, Astua D. Cranial and chromosomal geographic variation in manatees (Mammalia: Sirenia: Trichechidae) with the description of the Antillean manatee karyotype in Brazil. J Zoological Syst Evol Res. 2016;55(1):73–87.
Cantanhede AM, Da Silva VMF, Farias IP, Hrbek T, Lazzarini SM, Alves-Gomes J. Phylogeography and population genetics of the endangered Amazonian manatee, Trichechus inunguis Natterer, 1883 (Mammalia, Sirenia). Mol Ecol. 2005;14(2):401–13.
Domning DP. Fossil Sirenia of the West Atlantic and Caribbean Region, VII, Pleistocene Trichechus manatus Linnaeus, 1758. J Vertebr Paleontol. 2005;25(3):685–701.
Hunter ME, Mignucci-Giannoni AA, Tucker KP, King TL, Bonde RK, Gray BA, McGuire PM. Conserv Genet. 2012;13(6):1623–35.
Perini FA, Nascimento ER, Cozzuol MA. A new species of Trichechus Linnaeus, 1758 (Sirenia, Trichechidae), from the upper Pleistocene of southwestern Amazonia, and the evolution of Amazonian manatees. J Vertebr Paleontol. 2020;39:5.
Tabuce R, Asher RJ, Lehmann T. Afrotherian mammals: a review of current data. Mammalia. 2008;72(1):2–14.
Houck ML, Kumamoto AT, Gallagher DS Jr, Benirschke K. Comparative cytogenetics of the African elephant (Loxodonta africana) and Asiatic elephant (Elephas maximus). Cytogenet Genome Res. 2001;93(3–4):249–52.
Prinsloo P, Robinson TJ. Comparative cytogenetics of the Hyracoidea: chromosomes of two Hyrax species from South Africa. Z Sa ̈ugetierkunde. 1991;56:331–8.
Seabright M. A rapid banding technique for human chromosomes. Lancet. 1971;298:971–2.
Yang F, Graphodatsky AS. Animal probes and Zoo-FISH. In: Liehr T, editor. Fluorescence In Situ Hybridization (FISH) — Application Guide. Berlin: Springer protocols handbooks; 2016. p. 323–46.
Acknowledgements
The authors are grateful to members of Laboratory of Cytogenetics UFPA in the field work and chromosomal preparations. We also thankfull to the Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) for granting license to collect biological samples (Registration: 44915-10). We thankfully acknowledge the support by Zoológico da Universidade da Amazônia (ZOOUNAMA) and Institute of Biology and Conservation of Aquatic Mammals of the Amazon (BioMA) responsible for the management of animals and collection of blood samples. The authors also thank to Pró-reitoria de Pesquisa e Pos-graduação (PROPESP/UFPA), Fundo Brasileiro para a Biodiversidade (FUNBIO) and Dr. Leonardo Sena for the support in the collection of biological samples.
Funding
FST, MSc scholarship and Researcher scholarships (WOdS, RCRN, CYN and JCP) were funded by CNPQ, FST MSc scholarship was funded by CNPQ (PPGEAP/UFPA). This research was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) through the Edital Universal (proc. 475013/2012–3) and Coordenacão de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) through the Edital 047/2012 PRÓAMAZÔNIA: Biodiversidade e Sustentabilidade on a Project coordinated by CYN; by Banco Nacional de Desenvolvimento Econômico e Social–BNDES (Operação 2.318.698.0001) on a project coordinated by JCP and CYN (308428/2013–7), and JCP (308401/2013–1) are grateful to CNPq for Productivity Grants. RCRN received financial support from ULTRACARGO (5700009722).
Author information
Authors and Affiliations
Contributions
Conceptualization, FST, WOdS, MAFS, JCP, CYN and RCRN; cytogenetic analysis, FST and WOdS; samples collect, AGCMK, JMO, ALFR, GMS; investigation, FST, WOdS; resources, FST, RCRN, JCP and CYN; writing original draft preparation, FST, WOdS and RCRN; writing review and editing, FST, WOdS, RCRN, MAFS, JCP and CYN; visualization, MAFS; supervision, RCRN, CYN. and JCP; project administration, RCRN, JCP and CYN; funding acquisition, FST, RCRN, MAFS, JCP and CYN. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The animal study protocol was approved by the Ethics Committee in Research with experimental animals from the Universidade Federal do Pará, reference number 68–2015 and Instituto Chico Mendes de Conservação da Biodiversidade (ICMBio) for the collection permit (Registration: 44915–10). All methods were performed in accordance with the relevant guidelines and regulations by including a statement in the Declarations section to this effect. And the study is reported in accordance with ARRIVE guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare there is no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tavares, F.d.S., Oliveira da Silva, W., Ferguson-Smith, M.A. et al. Ancestral chromosomal signatures of Paenungulata (Afroteria) reveal the karyotype of Amazonian manatee (Trichechus inunguis, Sirenia: Trichechidae) as the oldest among American manatees. BMC Genomics 24, 38 (2023). https://doi.org/10.1186/s12864-023-09129-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-023-09129-3