Abstract
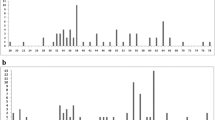
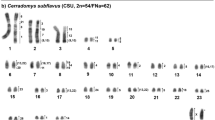
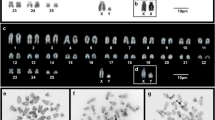
Oryzomyini is the most diverse tribe of subfamily Sigmodontinae, comprising 30 genera and 147 extant species. Cytogenetic studies on Oryzomyini reflect this diversity, revealing an exceptional range of karyotype variability, with chromosome numbers ranging from 2n = 16 to 2n = 88. In addition, some species exhibit autosomal and sex chromosomal polymorphisms, as well as the presence of B chromosomes. In the search for a more detailed understanding of the processes involved in karyotype diversification of Oryzomyini, we performed a genome-wide comparative study using chromosome probes of the entire chromosome set of Holochilus nanus and some autosomes of Oligoryzomys moojeni on metaphases of 15 species of Oryzomyini, in addition to one species of tribe Thomasomyini. We also performed a phylogenetic analysis of these species using chromosome painting data. The results show complete chromosome homology among the autosomal complement of species of Oryzomyini with the presence of pericentric inversions or centromeric shifts, paracentric inversion, Robertsonian rearrangements, tandem fusions/fissions, and translocations involved in the karyotype evolution of the tribe. In addition, sex chromosomes of these species present a homologous region between all analyzed species with amplification of the species-specific heterochromatin. The use of two different approaches to perform a phylogenetic analysis based on chromosome painting data revealed which one is more trustworthy and presents results similar to the findings obtained by molecular and morphological analyses. Finally, we propose that chromosomal diversity of the tribe Oryzomyini is promoted by genomic elements, such as repetitive DNA, that trigger the karyotype variability of these rodents.





Similar content being viewed by others
Data availability
Not applicable.
Code availability
Not applicable.
References
Andrades-Miranda J, Oliveira LF, Lima-Rosa CAV, Sana DA, Nunes AP, Mattevi MS (2002a) Genetic studies in representatives of genus Rhipidomys (Rodentia, Sigmodontinae) from Brazil. Acta Theriol 47(2):125–135. https://doi.org/10.1007/BF03192453
Andrades-Miranda J, Zanchin NIT, Oliveira LFB, Langguth AR, Mattevi MS (2002b) (T2AG3)n telomeric sequence hybridization indicating centric fusion rearrangements in the karyotype of the rodent Oryzomys subflavus. Genetica 114:11–16. https://doi.org/10.1023/A:1014645731798
Almeida EJC, Yonenaga-Yassuda Y (1991) Pericentric inversions and sex chromosome heteromorphisms in Oryzomys nigripes (Rodentia, Cricetidae). Caryologia 44:63–73. https://doi.org/10.1080/00087114.1991.10797020
Baker RJ, Koop BF, Haiduk MW (1983) Resolving systematic relationships with Gbands: a study of five genera of South American Cricetine rodents. Syst Zool 32(4):403–416. https://doi.org/10.1093/sysbio/32.4.403
Bakloushinskaya IYu, Matveevsky SN, Romanenko SA, Serdukova NA, Kolomiets OL, Spangenberg VE, Lyapunova EA, Graphodatsky AS (2012) A comparative analysis of the mole vole sibling species Ellobius tancrei and E. talpinus (Cricetidae, Rodentia) through chromosome painting and examination of synaptonemal complex structures in hybrids. Cytogen Genome Res 136:199–207. https://doi.org/10.1159/000336459
Barros MA, Reig OA, Perez-Zapata A (1992) Cytogenetics and karyosystematics of South American Oryzomyine rodents (Cricetidae: Sigmodontinae). Cytogenet Cell Genet 59:34–38. https://doi.org/10.1159/000133195
Bonvicino C, Almeida FC (2000) Karyotype, morphology and taxonomic status of Calomys expulsus (Rodentia: Sigmodontinae). Mammalia 64(3):339–352. https://doi.org/10.1515/mamm.2000.64.3.339
Burgin CJ, Colella JP, Kahn PL, Upham NS (2018) How many species of mammals are there? J Mammal 99(1):1–14. https://doi.org/10.1093/jmammal/gyx147
Camacho JPM, Sharbel TF, Beukeboom LW (2000) B chromosome evolution. Phil Trans R Soc London 355:163–178. https://doi.org/10.1098/rstb.2000.0556
Cantrell MA, Ederer MM, Erickson IK, Swier VJ, Baker RJ, Wichman HA (2005) MysTR: an endogenous retrovirus family in mammals that is undergoing recent amplifications to unprecedented copy numbers. J Virol 79(23):14698–14707. https://doi.org/10.1128/JVI.79.23.14698-14707.2005
Casavant NC, Scott L, Cantrell MA, Wiggins LE, Baker RJ, Wichman HA (2000) The end of the LINE?: lack of recent L1 activity in a group of South American rodents. Genetics 154(4):1809–1817. https://doi.org/10.1093/genetics/154.4.1809
D’Elía G, Pardiñas UFJ (2015) Subfamily Sigmodontinae Wagner, 1843. In: Patton J, Pardiñas UFJ, D’Elía G (eds) Mammals of South America, Volume 2: Rodents. University of Chicago Press, pp 229–249
D’Elía G, Fabre PH, Lessa EP (2019) Rodent systematics in an age of discovery: recent advances and prospects. J Mammal 100(3):852–871. https://doi.org/10.1093/jmammal/gyy179
D’Elía G, Pardiñas UFJ, Teta P, Patton JL (2007) Definition and diagnosis of a new tribe of Sigmodontine rodents (Cricetidae, Sigmodontinae), and a revised classification of the subfamily. Gayana 71(2):187–194
Di-Nizo CB, Banci KRS, Sato-Kuwabara Y, Silva MJJ (2017) Advances in cytogenetics of Brazilian rodents: cytotaxonomy, chromosome evolution and new karyotypic data. Comp Cytogenet 11(4):833–892. https://doi.org/10.3897/CompCytogen.v11i4.19925
Di-Nizo CB, Ferguson-Smith MA, Silva MJJ (2020) Extensive genomic reshuffling involved in the karyotype evolution of genus Cerradomys (Rodentia: Sigmodontinae: Oryzomyini). Genet Mol Biol. https://doi.org/10.1590/1678-4685-GMB-2020-0149
Di-Nizo CB, Ventura K, Ferguson-Smith MA, O’Brien PCM, Yonenaga-Yassuda Y, Silva MJJ (2015) Comparative chromosome painting in six species of Oligoryzomys (Rodentia, Sigmodontinae) and the karyotype evolution of the genus. PLoS ONE 10(2):e0117579. https://doi.org/10.1371/journal.pone.0117579
Dobigny G, Ducroz JF, Robinson TJ, Volobouev V (2004) Cytogenetics and cladistics. Syst Biol 53(3):470–484. https://doi.org/10.1080/10635150490445698
Erickson IK, Cantrell MA, Scott L, Wichman HA (2011) Retrofitting the genome: L1 extinction follows endogenous retroviral expansion in a group of muroid rodents. J Virol 85(23):12315–12323. https://doi.org/10.1128/JVI.05180-11
Ferguson-Smith MA, Trifonov V (2007) Mammalian karyotype evolution. Nat Rev Genet 8(12):950–962. https://doi.org/10.1038/nrg2199
Ford CF, Hamerton JL (1956) A colchicin hypotonic citrate squash sequence for mammalian chromosome. Stain Technol 31:247–251. https://doi.org/10.3109/10520295609113814
Freshney RI (1986) Animal cell culture - a practical approach. IRL Press, Oxford, p 247
Gardner AL, Patton JL (1976) Karyotypic variation in Oryzomyine rodents (Cricetinae) with comments on chromosomal evolution in the Neotropical Cricetine complex. Occas Pap Mus Zool 49:1–48
Gifalli-Iughetti C, Koiffmann CP (2009) The Y chromosome of the atelidae family (Platyrrhini): study by chromosome microdissection. Cytogenet Genome Res 125:46–53. https://doi.org/10.1159/000207518
Goloboff PA, Farris JS, Nixon KC (2008) TNT, a free program for phylogenetic analysis. Cladistics 24:774–786. https://doi.org/10.1111/j.1096-0031.2008.00217.x
Grahn RA, Rinehart TA, Cantrell MA, Wichman HA (2005) Extinction of LINE-1 activity coincident with a major mammalian radiation in rodents. Cytogenet Genome Res 110(1–4):407–415. https://doi.org/10.1159/000084973
Graphodatsky AS (1989) Conserved and variable elements of mammalian chromosomes. In: Halnan CRE (ed) Cytogenetics of animals. CAB International Press, UK, pp 95–123
Greenbaum IF, Gunn SJ, Smith SA, McAllister BF, Hale DW, Baker RJ, Engstrom MD, Hamilton MJ, Modi WS, Robbins LW, Rogers DS, Ward OG, Dawson WD, Elder FFB, Lee MR, Pathak S, Stangl JrFB (1994) Cytogenetic nomenclature of deer mice, Peromyscus (Rodentia): Revision and review of the standardized karyotype. Cytogenet Cell Genet 66:181–195. https://doi.org/10.1159/000133696
Grutzner F, Himmelbauer H, Paulsen M, Ropers HH, Haaf T (1999) Comparative mapping of mouse and rat chromosomes by fluorescence in situ hybridization. Genomics 55:306–313. https://doi.org/10.1006/geno.1998.5658
Guilly MN, Fouchet P, de Chamisso P, Schmitz A, Dutrillaux B (1999) Comparative karyotype of rat and mouse using bidirectional chromosome painting. Chromosome Res 7:213–221. https://doi.org/10.1023/A:1009251416856
Hass I, Sbalqueiro IJ, Müller S (2008) Chromosomal phylogeny of four Akodontini species (Rodentia, Cricetidae) from Southern Brazil established by Zoo-FISH using Mus musculus (Muridae) painting probes. Chromosome Res 16:75–88. https://doi.org/10.1007/s10577-007-1211-5
Hass I, Müller S, Artoni RF, Sbalqueiro IJ (2011) Comparative chromosome maps of neotropical rodents Necromys lasiurus and Thaptomys nigrita (Cricetidae) established by ZOO-FISH. Cytogenet Genome Res 135:42–50. https://doi.org/10.1159/000330259
Houck ML, Kumamoto AT, Gallagher DS Jr, Benirschke K (2001) Comparative cytogenetics of the African elephant (Loxodonta africana) and Asiatic elephant (Elephas maximus). Cytogenet Cell Genet 93:249–252. https://doi.org/10.1159/000056992
Jones N (2017) New species with B chromosomes discovered since 1980. Nucleus. https://doi.org/10.1007/s13237-017-0215-6
Kobayashi T, Yamada F, Hashimoto T, Abe S, Matsuda Y, Kuroiwa A (2008) Centromere repositioning in the X chromosome of X0/X0 mammals, Ryukyu spiny rat. Chromosome Res 16:587–593. https://doi.org/10.1007/s10577-008-1199-5
Leite RN, Kolokotronis SO, Almeida FC, Werneck FP, Rogers DS, Weksler M (2014) In the wake of invasion: tracing the historical biogeography of the South American cricetid radiation (Rodentia, Sigmodontinae). PLoS ONE 9(6):e100687. https://doi.org/10.1371/journal.pone.0100687
Malcher SM, Pieczarka JC, Geise L, Rossi RV, Pereira AL, O’Brien PCM, Asfora PH, Silva VF, Sampaio MI, Ferguson-Smith MA, Nagamachi CY (2017) Oecomys catherinae (Sigmodontinae, Cricetidae): evidence for chromosomal speciation? PLoS ONE 12(7):e0181434. https://doi.org/10.1371/journal.pone.0181434
Matsubara K, Nishida-Umehara C, Tsuchiya K, Nukaya D, Matsuda Y (2004) Karyotypic evolution of Apodemus (Muridae, Rodentia) inferred from comparative FISH analyses. Chromosome Res 12:383–395. https://doi.org/10.1023/B:CHRO.0000034103.05528.83
Matthey R (1972) Chromosomes and evolution. Triangle 11(3):107–112
Mlynarski EE, Obergfell CJ, O’Neill MJ, O’Neill RJ (2010) Divergent patterns of breakpoint reuse in muroid rodents. Mamm Genome 21:77–87. https://doi.org/10.1007/s00335-009-9242-1
Moreira CN, Di-Nizo CB, Silva MJJ, Yonenaga-Yassuda Y, Ventura K (2013) A remarkable autosomal heteromorphism in Pseudoryzomys simplex 2n = 56; FN = 54–55 (Rodentia, Sigmodontinae). Genet Mol Biol 36(2):201–206. https://doi.org/10.1590/S1415-47572013000200010
Moreira CN, Ventura K, Percequillo AR, Yonenaga-Yassuda Y (2020) A review on the cytogenetics of the tribe Oryzomyini (Rodentia: Cricetidae: Sigmodontinae), with the description of new karyotypes. Zootaxa 4876(1):1–111
Musser GG, Carleton MD, Brothers EM, Gardner AL (1998) Systematic studies of Oryzomyine rodents (Muridae, Sigmodontinae): diagnoses and distributions of species formerly assigned to Oryzomys “capito”. Bull Am Mus Nat Hist 236:1–376
Nagamachi CY, Pieczarka JC, O’Brien PCM, Pinto JA, Malcher SM, Pereira AL, Rissino JD, Mendes-Oliveira AC, Rossi RV, Ferguson-Smith MA (2013) FISH with whole chromosome and telomeric probes demonstrates huge karyotypic reorganization with ITS between two species of Oryzomyini (Sigmodontinae, Rodentia): Hylaeamys megacephalus probes on Cerradomys langguthi karyotype. Chromosome Res 21(2):107–119. https://doi.org/10.1007/s10577-013-9341-4
O’Brien SJ, Menotti-Raymond M, Murphy WJ, Nash WG, Wienberg J, Stanyon R, Copeland NG, Jenkins NA, Womack JE, Graves JAM (1999) The promise of comparative genomics in mammals. Science 286:458–481. https://doi.org/10.1126/science.286.5439.458
Pardiñas UFJ, Teta P, Salazar-Bravo J (2015) A new tribe of Sigmodontinae rodents (Cricetidae). Mastozoología Neotropical 22(1):171–186
Patton JL, Silva MN, Malcolm JR (2000) Mammals of the Rio Juruá and the evolutionary and ecological diversification of Amazonia. Bull Am Mus Nat Hist 244:1–306. https://doi.org/10.1206/0003-0090(2000)244%3C0001:MOTRJA%3E2.0.CO;2
Percequillo AR, Prado JR, Abreu EF, Dalapicolla J, Pavan AC, Chiquito EA, Brennand P, Steppan SJ, Lemmon AR, Lemmon EM, Wilkinson M (2021) Tempo and mode of evolution of Oryzomyine rodents (Rodentia, Cricetidae, Sigmodontinae): a phylogenomic approach. Mol Phylogenet Evol. https://doi.org/10.1016/j.ympev.2021.107120
Pereira AL, Malcher SM, Nagamachi CY, O’Brien PCM, Ferguson-Smith MA, Mendes-Oliveira AC, Pieczarka JC (2016) Extensive chromosomal reorganization in the evolution of New World Muroid Rodents (Cricetidae, Sigmodontinae): searching for ancestral phylogenetic traits. PLoS ONE 11(1):e0146179. https://doi.org/10.1371/journal.pone.0146179
Petit D, Couturier J, Viegas-Péquignot E, Lombard M, Dutrillaux B (1984) Great degree of homeology between the ancestral karyotype of squirrels (Rodents) and that of Primates and Carnivora. Ann Genet 27(4):201–212
Prado JR, Percequillo AR (2013) Geographic distribution of the genera of the tribe Oryzomyini (Rodentia: Cricetidae: Sigmodontinae) in South America: patterns of distribution and diversity. Arq Zool 44(1):1–120
Prado JR, Mendonça GC, Chiquito EA (2019) Topics in Oryzomyini rodents (Cricetidae, Sigmodontinae): natural history, diversity and the contribution of Brazilian researchers. Bol Soc Bras Mastozoolog 85:74–85
Prado JR, Knowles LL, Percequillo AR (2021) New species boundaries and the diversification history of marsh rat taxa clarify historical connections among ecologically and geographically distinct wetlands of South America. Mol Phylogenet Evol 155:106992. https://doi.org/10.1016/j.ympev.2020.106992
Reig OA, Aguilera M, Perez-Zapata A (1990) Cytogenetics and karyosystematics of South American oryzomyine rodents (Cricetidae: Sigmodontinae) II. High numbered karyotypes and chromosomal heterogeneity in Venezuelan Zygodontomys. Z Säugetierkunde 55(6):361–370
Rinehart TA, Grahn RA, Wichman HA (2005) SINE extinction preceded LINE extinction in sigmodontine rodents: implications for retrotranspositional dynamics and mechanisms. Cytogenet Genome Res 110:416–425. https://doi.org/10.1159/000084974
Romanenko SA, Volobouev V (2012) Non-sciuromorph rodent karyotypes in evolution. Cytogenet Genome Res 137:233–245. https://doi.org/10.1159/000339294
Romanenko SA, Lemskaya NA, Beklemisheva VP, Perelman PL, Serdukova NA, Graphodatsky AS (2010) Comparative cytogenetics of rodents. Russ J Genet 46(9):1138–1142. https://doi.org/10.1134/S1022795410090334
Romanenko SA, Perelman PL, Trifonov VA, Graphodatsky AS (2012) Chromosomal evolution in Rodentia. Heredity 108:4–16. https://doi.org/10.1038/hdy.2011.110
Romanenko SA, Serdyukova NA, Perelman PL, Pavlova SV, Bulatova NS, Golenishchev FN, Stanyon R, Graphodatsky AS (2017) Intrachromosomal rearrangements in rodents from the perspective of comparative region-specific painting. Genes 8(9):215. https://doi.org/10.3390/genes8090215
Salazar-Bravo J, Pardiñas UFJ, Zeballos H, Teta P (2016) Description of a New Tribe of Sigmodontine rodents (Cricetidae: Sigmodontinae) with an updated summary of valid tribes and their generic contents. Occas Pap Tex Tech Univ Mus 338:1–24
Seabright M (1971) A rapid being technique for human chromosomes. Lancet 2:971–972. https://doi.org/10.1016/S0140-6736(71)90287-X
Silva MJJ, Yonenaga-Yassuda Y (1998) Heterogeneity and meiotic behaviour of B and sex chromosomes, banding patterns and localization of (TTAGGG)n sequences by fluorescence in situ hybridization in the neotropical water rat Nectomys (Rodentia, Cricetidae). Chromosome Res 6:455–462. https://doi.org/10.1023/A:1009248311530
Silva MJJ, Yonenaga-Yassuda Y (2004) B chromosomes in brazilian rodents. Cytogenet Genome Res 106:257–263. https://doi.org/10.1159/000079296
Silva WO, Pieczarka JC, Ferguson-Smith MA, O’Brien PCM, Mendes-Oliveira AC, Sampaio I, Carneiro J, Nagamachi CY (2017) Chromosomal diversity and molecular divergence among three undescribed species of Neacomys (Rodentia, Sigmodontinae) separated by Amazonian rivers. PLoS ONE 12(8):e0182218. https://doi.org/10.1371/journal.pone.0182218
Silva WO, Pieczarka JC, Costa MJR, Ferguson-Smith MA, O’Brien PCM, Mendes-Oliveira AC, Rossi RV, Nagamachi CY (2019) Chromosomal phylogeny and comparative chromosome painting among Neacomys species (Rodentia, Sigmodontinae) from eastern Amazonia. BMC Evol Biol 19(1):184. https://doi.org/10.1186/s12862-019-1515-z
Silva WO, Rosa CC, Pieczarka JC, Ferguson-Smith MA, O’Brien PCM, Mendes-Oliveira AC, Rossi RV, Nagamachi CY (2020) Karyotypic divergence reveals that diversity in the Oecomys paricola complex (Rodentia, Sigmodontinae) from eastern Amazonia is higher than previously thought. PLoS ONE 15(10):e0241495. https://doi.org/10.1371/journal.pone.0241495
Steppan SJ, Adkins RM, Anderson J (2004) Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst Biol 53(4):533–553. https://doi.org/10.1080/10635150490468701
Steppan SJ, Schenk JJ (2017) Muroid rodent phylogenetics: 900-species tree reveals increasing diversification rates. PLoS ONE 12(8):e0183070. https://doi.org/10.1371/journal.pone.0183070
Suárez P, Nagamachi CY, Lanzone C, Malleret MM, O’Brien PCM, Ferguson-Smith MA, Pieczarka JC (2015) Clues on syntenic relationship among some species of Oryzomyini and Akodontini tribes (Rodentia: Sigmodontinae). PLoS ONE 10(12):e0143482. https://doi.org/10.1371/journal.pone.0143482
Svartman M, Almeida EJC (1992) The karyotype of Oryzomys capito laticeps (Cricetidae, Rodentia) from central Brazil. Rev Brasil Genet 15(4):963–972
Swier VJ, Bradley RD, Rens W, Elder FF, Baker RJ (2009) Patterns of chromosomal evolution in Sigmodon, evidence from whole chromosome paints. Cytogenet Genome Res 125:54–66. https://doi.org/10.1159/000218747
Trifonov VA, Kosyakova N, Romanenko SA, Stanyon R, Graphodatsky AS, Liehr T (2010) New insights into the karyotypic evolution in muroid rodents revealed by multicolor banding applying murine probes. Chromosome Res 18:265–275. https://doi.org/10.1007/s10577-010-9110-6
Ventura K, O’Brien PC, Yonenaga-Yassuda Y, Ferguson-Smith MA (2009) Chromosome homologies of the highly rearranged karyotypes of four Akodon species (Rodentia, Cricetidae) resolved by reciprocal chromosome painting: the evolution of the lowest diploid number in rodents. Chromosome Res 17:1063–1078. https://doi.org/10.1007/s10577-009-9083-5
Ventura K, Yonenaga-Yassuda Y, Ferguson-Smith MA (2012) Variable patterns of Y chromosome homology in Akodontini rodents (Sigmodontinae): a phylogenetic signal revealed by chromosome painting. Chromosome Res 20:427–433. https://doi.org/10.1007/s10577-012-9286-z
Ventura K, O’Brien PCM, Moreira CN, Yonenaga-Yassuda Y, Ferguson-Smith MA (2015) On the origin and evolution of the extant system of B chromosomes in Oryzomyini radiation (Rodentia, Sigmodontinae). PLoS ONE 10(8):e0136663. https://doi.org/10.1371/journal.pone.0136663
Veyrunes F, Dobigny G, Yang F, O’Brien PC, Catalan J, Robinson TJ, Britton-Davidian J (2006) Phylogenomics of the genus Mus (Rodentia; Muridae): extensive genome repatterning is not restricted to the house mouse. Proc Biol Sci 273:2925–2934. https://doi.org/10.1098/rspb.2006.3670
Viegas-Péquignot E, Petit D, Benazzou T, Prod’homme M, Lombard M, Hoffschir F, Descailleaux J, Dutrillaux B (1986) Phylogénie chromosomique chez les Sciuridae, Gerbillidae et Muridae, et étude d’espèces appartenant à d’autres familles de Rongeurs. Mammalia 50:164–202
Ward OG, Graphodatsky AS, Wurster-Hill DH, Eremina VR, Park JP, Yu Q (1991) Cytogenetics of beavers: a case of speciation by monobrachial central fusions. Genome 34:324–328. https://doi.org/10.1139/g91-053
Waters PD, Wallis MC, Graves JAM (2007) Mammalian sex—origin and evolution of the Y chromosome and SRY. Semin Cell Dev Biol 18:389–400. https://doi.org/10.1016/j.semcdb.2007.02.007
Weksler M (2006) Phylogenetic relationships of oryzomine rodents (Muroidea, Sigmodontinae): separate and combined analyses of morphological and molecular data. Bull Am Mus Nat Hist 296:1–149. https://doi.org/10.1206/0003-0090(2006)296[0001:PROORM]2.0.CO;2
Weksler M (2015) Tribe Oryzomyini Vorontsov, 1959. In: Patton J, Pardiñas UFJ, D’Elía G (eds) Mammals of South America, Volume 2: Rodents. University of Chicago Press, pp 918–926
Weksler M, Percequillo (2011) Key to the genera of the tribe Oryzomyini (Rodentia: Cricetidae: Sigmodontinae). Mastozool Neotrop 18(2):281–292
Weksler M, Percequillo AR, Voss RS (2006) Ten new genera of oryzomyine rodents (Cricetidae:Sigmodontinae). Bull Am Mus Nat Hist 3537:1–29. https://doi.org/10.1206/0003-0082(2006)3537[1:TNGOOR]2.0.CO;2
Yang F, Carter NP, Shi L, Ferguson-Smith MA (1995) A comparative study of karyotypes of muntjacs by chromosome painting. Chromosoma 103:642–652. https://doi.org/10.1007/BF00357691
Yonenaga-Yassuda Y, Prado RC, Mello DA (1987) Supernumerary chromosomes in Holochilus brasiliensis and comparative cytogenetic analysis with Nectomys squamipes (Cricetidae, Rodentia). Rev Brasil Genet 2:209–220
Yonenaga-Yassuda Y, Maia V, L’Abbate M (1988) Two tandem fusions and supernumerary chromosomes in Nectomys squamipes (Cricetidae, Rodentia). Caryologia 41(1):25–39. https://doi.org/10.1080/00087114.1988.10797845
Zanchin NIT, Langguth A, Mattev MS (1992) Karyotypes of brazilian species of Rhipidomys (rodentia, cricetidae). J Mammal 73:120–122. https://doi.org/10.2307/1381872
Zhao S, Shetty J, Hou L, Delcher A, Zhu B, Osoegawa K, Jong P, Nierman WC, Strausberg RL, Fraser CM (2004) Human, mouse, and rat genome large-scale rearrangements: stability versus speciation. Genome Res 14:1851–1860. https://doi.org/10.1101/gr.2663304
Acknowledgements
We are grateful to Dra. Angela Vianna Morgante for providing the facilities for cell culture, Camilla Bruno Di-Nizo, Fernanda Gianisela Pricoli, Maria Pinheiro, and Vítor Alves for technical support, and Renata Cecília Amaro for the suggestions to improve the manuscript. We thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico, and Fundação de Amparo à Pesquisa do Estado de São Paulo made this work possible.
Funding
This research was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 140027/2014-9 for CNM; CNPq 303477/2011-3 for YY) and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 18/09553-6 for CNM; FAPESP 09/16009-1 and 16/20055-2 for ARP; FAPESP 09/54300-00 for KV).
Author information
Authors and Affiliations
Contributions
CNM and KV conceived of or designed study. CNM performed research. CNM and KV analyzed data. MAF contributed new methods or models. CNM wrote the paper. CNM, ARP, YY, KV, MAF reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Handling editor: Svetlana Pavlova.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
do Nascimento Moreira, C., Percequillo, A.R., Ferguson-Smith, M.A. et al. Chromosomal evolution of tribe Oryzomyini (Rodentia: Cricetidae: Sigmodontinae). Mamm Biol 102, 441–464 (2022). https://doi.org/10.1007/s42991-022-00244-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42991-022-00244-4