Abstract
Background
The mitochondrial genomes of higher plants vary remarkably in size, structure and sequence content, as demonstrated by the accumulation and activity of repetitive DNA sequences. Incompatibility between mitochondrial genome and nuclear genome leads to non-functional male reproductive organs and results in cytoplasmic male sterility (CMS). CMS has been used to produce F1 hybrid seeds in a variety of plant species.
Results
Here we compared the mitochondrial genomes (mitogenomes) of Gossypium hirsutum sterile male lines CMS-2074A and CMS-2074S, as well as their restorer and maintainer lines. First, we noticed the mitogenome organization and sequences were conserved in these lines. Second, we discovered the mitogenomes of 2074A and 2074S underwent large-scale substitutions and rearrangements. Actually, there were five and six unique chimeric open reading frames (ORFs) in 2074A and 2074S, respectively, which were derived from the recombination between unique repetitive sequences and nearby functional genes. Third, we found out four chimeric ORFs that were differentially transcribed in sterile line (2074A) and fertile-restored line.
Conclusions
These four novel and recombinant ORFs are potential candidates that confer CMS character in 2074A. In addition, our observations suggest that CMS in cotton is associated with the accelerated rates of rearrangement, and that novel expression products are derived from recombinant ORFs.
Similar content being viewed by others
Background
Cytoplasmic male sterility (CMS), a phenomenon that the male reproductive structures fail to develop, is an important agronomical trait in higher plants. The CMS character is frequently used in crop breeding and commercial seed production to increase the yield of the crops such as rice, maize, rapeseed, and cotton [1,2,3,4,5]. CMS is maintained by the maintainer line which is similar to the CMS line in terms of the nuclear composition but is equipped with the male-fertile cytoplasm. Fertility is conferred by a third line which carries the nuclear restorer genes [6–8]. The CMS phenotype is closely associated with mutations in mitochondrial genomes (mitogenomes) [9, 10]. The different CMS phenotypes are the results of frequent recombination, gene shuffling and mutation [11, 12]. In CMS lines, mitogenomic sequences’ rearrangements produce chimeric genes which disrupt the normal physiological functions and cause male gametophyte abnormalities, such as pollen abortion [13]. Novel chimeric genes responsible for CMS were identified by evaluating the difference in mtDNA and transcriptional products among the following lines: CMS, maintainer, and restorer in maize [14], wheat [15], rice [16], pepper [17] and rapeseed [18,19,20]. Unfortunately, there are few reports on mitochondrial genomes rearrangement and the role of CMS in cotton.
Next-generation sequencing technology (NGS) has been applied to plant chloroplast (cp) genomes, with over 1200 species sequenced [21,22,23,24,25]. However, plant mtDNA has a large number of repeats sequences and rearrangements, thus limiting the use of NGS [26,27,28,29,30,31,32,32]. Likewise, RNA sequencing has been broadly used to study plant transcriptome and mtDNA [33,34,35,36,37]. However, the focuses of most studies were on mitochondrial global transcript levels. Comparative analysis of the mitochondrial transcriptomes of CMS, maintainer, F1 and restorer lines’ in the context of their nuclear genomes can provide insights into cytonuclear-related phenotypes, such as cytoplasmic male sterile [11, 38]. In this study, we performed a comparative analysis of the mtDNA of the CMS, restorer and maintainer lines from both Gossypium harknessii and G. hirsutum to determine candidate CMS factors. We also analyzed the expression patterns of uncharacterized ORFs, some of which are candidate genes for CMS. The results give some interesting clues about mitochondrial evolution and CMS generation, as well as generate a background for future studies on CMS molecular diversity and phenotypic variability in cotton.
Methods
Materials and mtDNA preparation
Line 2074A, an upland cotton cytoplasmic male sterile line with Gossypium harknessii Brandegee CMS-D2–2 cytoplasm, was from its original sterile line DES-HAMS277. Line 2074S, an upland cotton cytoplasmic male sterile line, was from G. hirsutum L. CMS-AD1. These two lines were genetically stable cotton sterile lines derived from 20 to 30 generations of backcross. Line 2074B, a cultivar of upland cotton ‘Sumian No. 20’, was the maintainer of these two cytoplasmic male sterile lines. The restorer E5903 is a nuclear restoring line with normal nuclear and normal male-fertile Gossypium harknessii Brandegee. 2074A, 2074S and E5903 cotton materials used in this study are developed in our own lab [39]. We breed these three cotton cultivars and the work started 20 years before. The mtDNA preparation was performed described previously [39].
Library construction and genome sequencing, assembly and sequence verification
The mitogenome Fosmid library was constructed according to the manufacturer’s protocols (MaxxPlaxTMLambda Packing Extract)/(CopyControlTM Fosmid Library Production Kits; Epicentre Technologies, Madison, WI). All these three mtDNAs Fosimd libraries have been constructed and screened with probes from sequences of conservative genes and scaffolds. From those libraries, 1000 clones were randomly selected and screened with 28 probes designed from sequences of mitochondrial genes. At last, 22, 26 and 21 positive clones were obtained from 2074A, E5903 and 2074S, from which 23 clones (seven for 2074A, nine for E5903, and seven for 2074S) were selected to cover larger repeats and sequenced the double-ends by shotgun strategy, with insert size of about 36.2 kb- 38.4 kb. Sequenced fragments were aligned using Blastn to determine the exactness of assembly [39].
The mtDNA samples were sequenced using Illumina strategy at BGI (Beijing Genomics Institute) and assembled primarily using SOAPdenovo [40]. The Illumina system produced 413–607 M usable reads in one run for genome assembly and about 700 × coverage with Solexa using paired-end (90 bp reads). Raw sequences were evaluated by two quality control tools, using the Trimmomatic [41] and FilterReads module in Kmernator (https://github.com/JGI-Bioinformatics/Kmernator) to remove potential undesirable artifacts, including adapters or low-quality or N bases or short sequences. The filtered reads Q30 > 85%. These filtered reads were a mixture of reads derived from chloroplast, mitochondrial and nuclear genomes; firstly, we removed the chloroplast and nuclear contaminant contigs through Blastn against nt/nr database (Additional file 1: Table S1). Through adjusting the software SOAPdenovo with the reasonable parameters (−s config_file -K 37 -R -D 1 [40]), we acquired 28–65 big contigs in 4 mitogenomes. Among them, the mitogenomic sequences of 2074B had been published and the sequence was deposited in GenBank database under the accession number: JX065074.1 [42]. In addition, known mitogenomic sequences from our previous studies, including G. hirsutum 2074B [42], G. barbadense [43], G. raimondii and G. arboreum [30], as well as eight other diploid and tetraploid species [31], were used to order/orient mitochondrial-type scaffolds.
Combined with the scaffolds’ information and one whole-genome backbone with positive clones, three procedures were adopted to finish the physics gaps. Firstly, we screened the library of the whole mitogenome according to the splicing sequence and the functional genes, constructed genome physical map and then sequenced the positive clones [39]. Secondly, according to the relationship of whole-genome physics map with the positive clones, we designed primers combination on the different scaffolds’ terminals, and used long-PCR to finish the gaps (Additional file 2: Table S2A). Thirdly, PCR amplification was performed based on primers pairs that consist of the terminal sequences of large repeats (Additional file 3: Table S2B). Finally, we assembled three circle mitogenomes (2074A, 2074S, E5903). To evaluate the quality and accuracy of these three mitogenomic sequences’ assemblies, pair-end reads were mapped onto their respective consensus sequences with BWA 0.7.10-r789 [44]. The resulting SAM files from BWA mapping were transformed into BAM files using samtools view program [45]. The BWA mapping results of these pair-end reads in BAM files were then used to calculate the depth of sequencing coverage using samtools depth program [45]. For three Gossypium species, the Illumina reads covered all parts of the genomes consistently, achieving an average sequencing depth of 214.3× in 2074A mitogenome (clean data, 413 M), 28.8× in 2074S mitogenome, 27.3× in E5903 mitogenome.
Analyses and annotations of mitogenomes and sequence data
Intersubspecific polymorphisms were firstly identified based on the MUMmer package (v3.06) [46]. The results were acquired using a custom-designed Perl script and were confirmed through careful visual inspection. We carried out analyses on repeat sequences using the Washington University (WU)-Blast, including forward, palindromic reverse, and complemented repeats with a minimal length of 20 bp. Cp-derived (chloroplast-derived) sequences were identified using BlastN search of mitogenomes against annotated cotton chloroplast genomes (Identity ≧90%, E-value ≦1e-5, and Length ≧30 bp). Nuclear-derived insertions were searched against the G. raimondii genome. The syntenic regions of mitogenomes between different cultivars were detected using Nucmer of the MUMmer package (v3.06) [47] with 50 bp exact minimal match. NCBI-BlastX and -BlastN searches of the genomes against databases of sequenced plant mitogenomes were performed to find protein-coding and structural RNA genes, respectively. tRNA genes were searched by tRNAscan-SE [48] and were identified by BlastN [47]. The annotated mitogenomes features, including gene coordinate and genome structures, in genomes were drawn by OGDRAW v1.1 [49] and R Project (https://www.R-project.org/).
We used YASS to analyze the genome complexity that was defined as the complete sequence information of a genome with only one copy of each duplicate (> 500 bp). We set parameters as follows: E-value < 1e-30 (with the score “+ 1” for one match and “-3” for one substitution); the rate of substitutions and insertions/deletions < 5% [50].
Analysis of candidate cytoplasmic male sterility genes
Based on the previous reports showing that CMS genes are chimeric [3, 7, 51, 52], a search for chimeric ORFs was conducted. Open reading frames (ORFs) were identified by ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder/) and EMBOSS (6.3.1: getorf) [53]. All ORFs at least 300 bp in length were compared to the mitogenomes of the maintainer line 2074B and the restorer line E5903 using BlastN with an identity of 99% and an E-value cut off of 1 × 10− 5. ORFs containing at least 30 bp of an identified mitochondrial gene were characterized as chimeric, excluding any ORFs that overlap the genomic position of an identified gene. Transmembrane domains in each candidate ORF were predicted using TMHMM Server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
Sequencing of the cotton mitochondrial transcriptome
The extracted mitochondrial RNA from the flower buds (3–5 mm size) in CMS line 2074A, its maintainer 2074B and fertile material F1 (2074A × E5903) were sequenced on an Illumina HiSeq2000 at BGI (Beijing Genomics Institute). Ribosomal RNAs were removed from the extracted mitochondrial RNA using Ribo-Zero (Epicentre, Madison, WI) and the mitochondrial RNA libraries were prepared using Illumina’s TruSeq RNAseq Sample Prep kit. Libraries were sequenced on one lane with 4 Gb clean reads/samples of an average length of 90 nt for paired-end. RNA sequence data quality was checked using FastQC to remove the adapters, low-quality, containing N bases and short sequences with reads Q30 > 85%. The reads were mapped to the assembled mitogenome of CMS line 2074A using bowtie2 [54] with the following parameters: -D 5 -R 1 -N 0 -L 25 -i S, 1, 2.00. Then, the resulting SAM files from bowtie2 mapping were transformed into BAM files using samtools view program [45]. The bowtie2 mapping results of these pair-end reads in BAM files were then used to calculate read count for each gene through HTSeq-count program [55]. Differentially expressed genes that showed up and down regulation between samples were defined based on the standards of cutoff: two-fold change and a p-value of less than 0.05.
Results and discussion
Structures and contents of CMS, maintainer, and restorer mitochondrial genomes
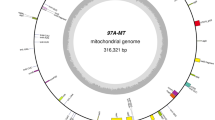
Cotton is the first species that the mitogenome is sequenced among the large numbers of malvales. We performed de novo sequencing of three mitogenomes lines: a) CMS-2074A, b) CMS-2074S, and c) E5903 (a restorer line). Lines 2074A and E5903 were derived from integrating the cytoplasm of diploid species G. harknessii (CMS-D2–2) into tetraploid G. hirsutum; while, 2074S was a result of alloplasmic G. hirsutum with G. hirsutum L. CMS-AD1-derived cytoplasm [39]. The mitogenomes of the three lines were highly conserved with the sequence identity more than 96%, indicating the preservation of the mitochondrial genome during cross-breeding (Table 1; Fig. 1). The mitogenomes of the three lines were 666,081 bp (E5903), 668,584 bp (2074A) and 668,464 bp (2074S), and there was about 3 kb difference detected (Table 1). These observations were close to the previous estimations based on restriction digestion patterns (690 kb – 710 kb) [56, 57]. Compared to the maintainer line 2074B, the above three lines (E5903, 2074A and 2074S) had more repeats. In four lines, the mitogenomic sequences belonging to the coding genes (including duplicated genes) and the plastid-derived sequences varied by less than 1% (Table 1, Additional file 4: Table S3). Both the proportions of nuclear-derived intergenic sequences and large repeats varied by 1–2%. Notably, the two CMS lines, 2074A and 2074S, contained two large inverse and direct repeats. Overall, the three mitogenomes, 2074A, 2074S, and E5903, had similar syntenic arrangements and were 87% identical in sequences’ similarity with the maintainer line, indicating general conservation among the varieties within species.
Linear maps of the four cotton mitogenomes. Known protein-coding genes, tRNA and rRNA genes, and gene fragments are shown on the line. Genes on the right side and left side of the line are transcribed direct and inverted, respectively. Colors indicate genes by function: Complex I (nad; yellow), Complex II (sdh; green), Complex III (cob; yellowish green), Complex IV (cox; light pink), Complex V (atp; olive-green), ribosomal proteins (brown), maturase (matR; orange), other genes (ccm and tRNA; purple), intron (white)
The main cycle (MC) molecules of two male sterile lines (2074A and 2074S) are 47 kb larger than that of the maintainer line (2074B). The reason is that they contain three large repeats, for examples, one is 10 kb larger than that of 2074B, which are similar as observed in the sterile line Ks3 that contains four repeats larger than 20 kb [41], and the sterile line TK18-MS that contains a pair of repeats of 86 kb in its MC molecule [58]. The intergenic regions of plant mtDNAs often contain retrotransposons from nuclear [25], chloroplast [59, 60] and other plant mitogenome [28]. 2074A and 2074S have more retrotransposons than 2074B and E5903, and they contain six unique regions with a total length of 31,694–36,741 bp. Furthermore, these sequences are novel and most are located in the intergenic regions, showing that they have a faster rate of evolution as similarly reported by Palmer et al., 2000 [61].
Gene evolution among CMS, restorer, and maintainer lines
Cotton mitogenomes typically contain 36 genes (Table 2; Fig. 1) whose content is largely conserved among species and across angiosperms, with few differences (for example, contrary to other sequenced plants, rpl2 in cotton lacks an intron [42, 43, 58]). As the case in the mitogenomes of other angiosperms, most of the genes encode proteins involved in ATP-generating electron transport: nine subunits of complex I (nad 1, nad 2, nad 3, nad 4, nad 4 L, nad 5, nad 6, nad 7 and nad 9), two subunits of complex II (sdh 3 and sdh 4), one subunit of complex III (cob), three subunits of complex IV (cox 1, cox 2 and cox 3), and five subunits of complex V (atp 1, atp 4, atp 6, atp 8 and atp 9); four genes involved in biogenesis of cytochrome c (ccm C, ccm B, ccm FC and ccm FN); 10 genes encode for ribosomal proteins (rpl 2, rpl 5, rpl 10, rpl 16, rps 3, rps 4, rps 7, rps 10, rps 12 and rps 14), while the numbers of the ribosomal proteins are usually variable among different species [58]. In addition, one gene (mttB) is involved in independent membrane targeting and translocation system, and one maturase gene (matR) is mapped within the 4th intron of nad1. 9 protein-encoding genes (ccmFC, cox2, rps10, rps3, nad1, nad2, nad4, nad5 and nad7) contain a total of 23 group II introns, among of which three are trans-spliced (nad 1, nad 2 and nad 5). As previously noted, the cotton mitogenomes presented here lack rps1, rps2, rps11, rps13, rps19 and sdh2, and only partial sequences of these genes were detected in cotton mitogenomes.
As reported previously, several mitochondrial genes exist in repeat regions and in multiple copies (Additional file 5: Table S4). However, unlike the mitogenome of G. hirsutum maintainer line, those of 2074A, 2074S and E5903 contain duplicated trnM (CAU) and ccmFC in large repeats. In addition, congruent with prior results, rps3 is located at a repeat’s boundary and varied in structure among these four Gossypium mitogenomes. In cotton, rps3 contains a central domain (pfam00013) which has been lost in the incomplete duplicates (pseudogene rps3–2) of 2074A, 2074S, and 2074B.
A total of 47 SNPs exists within 21 protein-coding genes in four mitogenomes analyzed, and only 11 SNPs were synonymous mutations (1 in 2074A mitogenome and 10 in 2074B mitogenome, Table 3). Remarkably, the numbers of nonsynonymous mutations (36 SNPs) are over three times as that of synonymous mutations, and nonsynonymous mutations were nearly evenly distributed among the mitochondrial genomes (10, 10, 9, and 7 unique nonsynonymous SNPs in 2074A, E5903, 2074S and 2074B, respectively). Most of these SNPs represent transversions rather than transitions (29 versus 7), and many of them were found in ribosomal protein-coding genes, (i.e., rpl2, rpl5, rpl10, rpl16, rps3, rps4 and rps10; Table 3). As protein-coding genes are extraordinarily conserved and exhibit slow evolutionary rates, the abundance of non-synonymous changes reported here may represent CMS-related candidate genes, although this needs to be functionally verified in each case. Mitochondrial genes, rps3 and rpl2, separately containing 3 and 2 nonsynonymous changes between the mitogenomes of 2074A (and 2074S)/E5903 and 2074B, might represent suitable candidates.
In the four mitogenomes analyzed, we found many gene editing events, for example, ACG was edited into AUG as start codon in three genes (rps10, nad1 and nad4L), and AUU was modified into AUG in one gene (mttB). There were five cases where gene editing generated stop codon, thereinto, four cases were the conversion of TAG into CGA in rps10, ccmC, atp9 and ccmFC genes; however, TAG was converted to CAA in atp6 gene. of Evolutionary rates analysis (ka/ks or ω) revealed that the ratios of ka to ks of nine genes (rps12, matR, atp1, mttB, rps4, rrn18 and nad1) were greater than 1, which implied a positive selection. In addition, that of two genes (rpl5 and cox3) were less than 1, which implied a purifying selection. By contrast, the non-coding regions appeared to be rapidly diverged (Additional file 6: Table S5).
Repeated sequences and unique sequences
The plant mitogenomes harbor massive repeated sequences, and the genome sizes tend to increase the genomic coverage by large repeats [62,63,64,65]. Our analysis revealed duplications were the main reason for the difference in size among the four lines. The duplicate lengths varied from 504 bp to 29 kb, which constituted 9.4–12.0% of the total genome lengths (Table 4). Two duplicated fragments 11,191 bp and 10,632 bp were present in the mitogenomes of all four lines. There was a common duplicate in three mitogenomes of 2074A, 2074S and E5903, but it was absent in 2074B. The mitogenomes of 2074A and 2074S were mostly identical, with one exception that a repeat sequence was present in 2074A but absent in 2074S. The mitogenome of 2074A is made up of a repeat sequence (29,277 bp), whereas that of 2074S consists of two repeat sequences (24,378 and 4621 bp) that are separated by a gap (Table 4). Total backbone DNA sequences represented a concatenation containing all basic fragments among all mitogenomes. When considering only one copy of each duplicated sequence, we found the genomic variations became small, especially from the same origin. The sizes of the backbone mtDNA sequences of 2074A and E5903 are same, and have a minimal difference with that of 2074S. Other repeats are smaller in size, and distribute distinctly and vary in copy number (Fig. 2).
The size distribution of repetitive content by the number of repeat pairs and total repeat length. The X position is repeat size category, which contains more than 10 kb, 1–10 kb, 0.5–1 kb, 101–500 bp, 41–100 bp, 31–40 bp, 21–30 bp. The Y positions are number of repeats pairs (primary axis) and proportion of total repeat length (secondary axis). The (a), (b), (c), (d) present 2074A, 2074S, 2074B, E5903 mitogenome, respectively
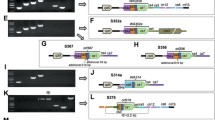
Intra-genomic recombination is an active phenomenon in the mitogenomes of plants [25]. Recombination frequency depends on the size of repeats, for example, large and direct repeats (> 1 kb) are associated with homologous recombination that will lead to the formation of sub-genomic molecules [66]. These four cotton mitogenomes exist as 4–7 larger repeats that produced subcircles. In 2074A, two pairs of subcircles are mediated by direct-repeat AR1, whereas one MC genomic circle may be produced by inverted-repeat AR4. However, 8–12 positive Fosmid clones covered all these large repeats and nine positive Fosmid clones covered all these repeats in E5903, which suggests that these four big repeats didn’t formed subcircles in 2074A and E5903 mitogenomes (Additional file 7: Figure S1). Importantly, every nodal point of large repeats is verified by long-PCR with special primers designed from their sequences (the PCR products were overlapped with repeats and non-repeats regions more than 300 bp, Additional file 2: Table S2A, Additional file 3: Table S2B).
Cytoplasmic male sterility is frequently associated with novel, chimeric, and often disruptive ORFs [3, 64]. In this study, we evaluated the mitogenomes of two CMS lines in the context of their maintainer and restorer lines for unique sequences that contain novel coding regions responsible for sterility in G. hirsutum. Six unique sequences (U1 to U6) were similar in two CMS lines, but were absent in the maintainer and restorer lines (Table 5). The total lengths of these regions were 31,694 bp in 2074A and 36,741 bp in 2074S, respectively (Table 5). Overall, these regions were unique with little similarities to known mitochondrial and nuclear sequences of other plants. These unique regions were frequently associated with repeats’ boundaries, which might indicate an origin from new sequence migration and recombination.
Mitochondrial genome syntenic evolution and organization
In addition, compared with 2074A, we analyzed the syntenic evolution of 4 cotton mitogenomes. We found 22 syntenic regions (named as S1 - S22), ranging from 2824 to 147,683 bp, which possessed at least 98% identity (Additional file 8: Figure S2). 2074B has lost syntenic segments S1 (U1), S6 (U3), S14 (U5), and S22 (U6). However, some segments are conserved in four mitogenomes, such as S5 - S6, S8 - S9, S10 - S13 and S15 - S20; the terminal sequences of S3, S4, S7, S13 and S21 are four large repeats (AR4, AR3, AR1, AR2, AR1, respectively), and the former sequence S20 was AR1 (as the difference of these larger repeats). The syntenic regions are broken, which suggests the repeat sequences are more dynamic and have undergone recombination in breeding process. S10 and S2 are broken by unique sequences U4 and U2, while other syntenic regions are more or less interrupted by insertion or deletion. These two cytoplasmic male sterile lines are more complex in nucleotide sequence composition, which suggests that male sterility may have been favored by faster rates of rearrangement and evolution, or CMS itself might have caused faster rearrangement and evolution.
CMS in two cytoplasmic male sterile lines of Gossypium hirsutum
CMS is a widespread phenomenon in plants and is associated with abnormal mitochondrial ORFs [7]. The occurrence of male sterility is an important feature in cotton breeding system. CMS is expected to be affected by mitochondrial gene(s), ORF content(s) and diversity during the emergence and selection of CMS specific mitochondrial genes. In other plants, several CMS-associated aberrant genes are located upstream or downstream of certain known genes and co-transcribed together [7, 67]. Since novel ORFs may be relevant to CMS, we analyzed all the predicted ORFs about their origin, conservation, function and expression. We compared all ORFs of 2074A and 2074S with that of the maintainer line 2074B, we observed 28 and 30 novel ORFs in 2074A and 2074S, respectively (Tables 6 and 7). The ORFs of 2074A were named as Aorf1 to Aorf28, and Aorf4 was duplicated in 2074A; while, those of 2074S were named as Sorf1 to Sorf30, and Sorf4 also was duplicated in 2074S. 11 of the ORFs are common in 2074A and 2074S. The length of polymorphisms in ORFs was frequently caused by frame shift mutations with several nucleotides’ insertions/deletions.
We categorized the specific ORFs into three basic groups: I) ORFs near the functional genes, which is transcribed in the same direction with adjacent positioned genes either up or down stream, and could be co-transcript relevant to CMS (Aorf4, Aorf25, Aorf27, Aorf28, Sorf4, Sorf8, Sorf14, Sorf27 and Sorf28); 2) Special ORFs in unique regions of sterile lines, which always have short-sequences homology to chloroplast or mitochondrial sequences of other plants; such as Aorf2, Aorf18, Sorf15, Sorf16 and Sorf2 that were found in unique sequences of two sterile lines. Mostly, they are similar to chloroplast or mitochondrial sequences of other plants, or have no homology sequences in NCBI-NR database. In the third group, the ORFs are comprised of homologous sequences of 2074B and unique sequences such as Aorf14, Sorf13 and Sorf14.
To further verify whether these ORFs were functionally associated with CMS, we profiled the expression of mitochondrial genes and ORF’s based on RNA-seq data of flower buds (3–5 mm in size) in CMS 2074A, maintainer 2074B, and the fertility-restored F1 (2074A × E5903). Among all the three lines, the expression of mitochondrial genes was highest in F1 and lowest in 2074B (Fig. 3, P < 0.05). The expression levels of shd3 and rpl10 genes were higher in 207A than in 2074B (Fig. 3, P < 0.05). Taking the sequences of 28 predicted ORFs in 2074A as a pool; we used Blastn to match all three-transcript data (2074A, 2074B and F1). As a result, 10 ORFs were expressed at high levels (10 fold) as compared to the similar sequence (with 1–3 gap) in 2074B; five ORFs were expressed at high levels as compared to the similar sequence in F1; the five ORFs were not expressed in 2074B (Additional file 9: Figure S3). Based on the first group principle, the ORFs near to functional genes, we found that four pairs of ORFs and their nearby genes (Aorf4 and atp8, Aorf9 and rrn26, Aorf4–2 and rpl2, Aorf28 and cox1/cox3) have same expression trend both in 2074B/2074A and F1/2074A, therefore, these four ORFs might be co-transcribed with functional genes and relevant to CMS.
Furthermore, we analyzed 16 reported CMS-associated ORFs. We found that these ORFs (78–488 bp) are near to co-transcribed genes and form a bicistronic complex with many functional genes. In this context, six ORFs in CMS2074S (Sorf25, Sorf4, Sorf4–2, Sorf29, Sorf8 and Sorf27) and five ORFs in CMS2074A (Aorf12, Aorf4, Aorf4–2, Aorf28 and Aorf9) were close to functional genes within 565 bp, and six (Sorf4, Sorf29, Sorf8, Aorf4, Aorf28 and Aorf9) of them are the products of rearrangements by large repeats. Additionally, these ORFs have transmembrane domain (except Aorf28, Table 5, Fig. 4) and same expression trend with their nearby genes. More important, four ORFs (Aorf4, Aorf28, Aorf9 and Aorf4–2) and their functional genes (atp8, cox1, cox3, rrn26 and rpl2) might have higher expression in CMS-2074A compared to F1. Aorf4 (561 bp) is found at the downstream 565 bp of atp8. Besides, the first 45 bp of Aorf4 are derived from rps3, while other partial sequences are identical to sdh3 (the 5′-end of orfH79 has 84 bp homology to cox1) and have same expression trends with atp8 in 2074B/2074A (− 0.3) and F1-A/2074A (2.6~ 2.9). Aorf4–2 (561 bp) is found in the downstream 444 bp of rpl2 and have same expression trends with rpl2 in 2074B/2074A (− 0.2~ 0.5) and F1-A/2074A (2.9~ 3.2). Aorf28 (867 bp), located at the downstream 241 bp of cox1 and the upstream 311 bp of cox3 (331 bp in 2074S), shows 66% identity with Arabidopsis mitogenome and is close to AR1. In addition, the expression trend of Aorf28, cox1 and cox3 were same. Aorf9 (357 bp), located at the downstream 19 bp of rrn26, keeps same expression trends with rrn26 in 2074B/2074A (− 0.7) and F1-A/2074A (2.4); as well, Aorf9 also has 76 bp identity with nad7 and 89% identity with Ricinus mitogenome. These four ORFs show the characters of CMS-associated genes and are similar to other ORFs, such as T-urf13 of maize [14], S-orf355/orf77 [66], orf224 of rape [8, 68,69,70], orf256 of wheat [15, 71], orf125 of radish [72], etc. All above chimeric ORFs from other plants are always near and co-transcribed with functional genes, which makes functional genes transcribe improperly and causes abortion [73,74,75,76]. As to now, these results were only based on the genome and RNA-seq data, more experiments, including functional validation of overexpression or CRISPR/Cas9 these orfs, are needed to confirm the real CMS gene of upland cotton.
Conclusions
The two almost identical male sterile lines, 2074A and 2074S, share high identity with the restore line E5903 but are different from their maintainer line 2074B, especially in non-coding regions. The cotton mtDNAs are 621,884–668,584 bp in length, and harbor 36 known protein-coding genes, three rRNAs (18S, 26S, and 5S rRNAs) as well as 18 different tRNAs. The rates of the coding genes (including duplicated genes) accounting for the total genomes’ length are almost similar, but the repeat sequences show a few differences. In addition, five genes (rps1, rps2, rps13, rps19 and sdh2) have been lost and 38 nonsynonymous mutations occurred in 21 protein-coding genes, though they are functionally irrelevant. Out of 28 ORFs in CMS 2074A, four ORFs (Aorf4, Aorf9, Aorf4–2 and Aorf28) are close to the functional genes and show similar characters to CMS-associated genes in other plants. These four ORFs may be the potential candidates conferring CMS in cotton.
Abbreviations
- CMS:
-
Cytoplasmic male sterility
- cp:
-
Chloroplast
- G.:
-
Gossypium
- MC:
-
Main cycle
- mitogenome:
-
Mitochondrial genome
- mtDNA:
-
Mitochondrial DNA
- NGS:
-
Next-generation sequencing technology
- ORFs:
-
Open reading frames
- rRNAs:
-
Ribosomal RNAs
- tRNAs:
-
Transfer RNAs
References
Pruitt KD, Hanson MR. Transcription of the Petunia mitochondrial CMS-associated Pcf locus in male sterile and fertility-restored lines. Mol Gen Genet. 1991;227:348–55.
Budar F, Pelletier G. Male sterility in plants: occurrence, determinism, significance and use. C R Acad Sci III. 2001;324:543–50.
Chase CD. Cytoplasmic male sterility: a window to the world of plant mitochondrial–nuclear interactions. Trends Genet. 2007;23:81–90.
Li SQ, Yang DC, Zhu YG. Characterization and use of male sterility in hybrid rice breeding. J Integr Plant Biol. 2007;49:791–804.
Tester M, Langridge P. Breeding technologies to increase crop production in a changing world. Science. 2010;327:818–22.
Fujii S, Kazama T, Yamada M, Toriyama K. Discovery of global genomic re-organization based on comparison of two newly sequenced rice mitochondrial genomes with cytoplasmic male sterility-related genes. BMC Genomics. 2010;11:209–23.
Luo DP, Xu H, Liu ZL, Guo JX, Li HY, Chen LT, Fang C, Zhang QY, Bai M, Yao N, Wu H, Wu H, Ji CH, Zheng HQ, Chen YL, Ye S, Li XY, Zhao XC, Li RQ. Liu YG. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat Genet. 2013;45:573–7.
An H, Yang ZH, Yi B, Wen J, Shen JX, Tu JX, Ma CZ. Fu TD. Comparative transcript profiling of the fertile and sterile flower buds of pol CMS in B. napus. BMC Genomics. 2014;15:258–67.
Horn R, Gupta KJ, Colombo N. Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion. 2014;19(3):198–205.
Hu J, Huang W, Huang Q, Qin X, Yu C, Wang L, et al. Mitochondria and cytoplasmic male sterility in plants. Mitochondrion. 2014;19:282–8.
Shearman JR, Sangsrakru D, Ruang-Areerate P, Sonthirod C, Uthaipaisanwong P, Yoocha T, Poopear S, Theerawattanasuk K, Tragoonrung S, Tangphatsornruang S. Assembly and analysis of a male sterile rubber tree mitochondrial genome reveals DNA rearrangement events and a novel transcript. BMC Plant Biol. 2014;14:45–55.
Szklarczyk M, Szymanski M, Wojcik-Jagla M, Simon PW, Weihe A, Borner T. Mitochondrial atp9 genes from petaloid male-sterile and male-fertile carrots differ in their status of heteroplasmy, recombination involvement, post-transcriptional processing as well as accumulation of RNA and protein product. Theor Appl Genet. 2014;127:1689–701.
Matera JT, Monroe J, Smelser W, Gabay-Laughnan S, Newton KJ. Unique changes in mitochondrial genomes associated with reversions of S-type cytoplasmic male sterility in maizemar. PLoS One. 2011;6:e23405.
Kennell JC, Pring DR. Initiation and processing of atp6, T-urf13 and ORF221 transcripts from mitochondria of T cytoplasm maize. Mol Gen Genet. 1989;216:16–24.
Song J, Hedgcoth C. A chimeric gene (orf256) is expressed as protein only in cytoplasmic male-sterile lines of wheat. Plant Mol Biol. 1994;26:535–9.
Kim DH, Kang JG, Kim BD. Isolation and characterization of the cytoplasmic male sterility-associated orf456 gene of chili pepper (Capsicum annuum L.). Plant Mol Biol. 2007;63:519–32.
Ji JJ, Huang W, Li Z, Chai WG, Yin YX, Li DW, Gong ZH. Tapetum-specific expression of a cytoplasmic orf507 gene causes semi-male sterility in transgenic peppers. Front Plant Sci. 2015;6:272–85.
Iwabuchi M, Koizuka N, Fujimoto H, Sakai T, Imamura J. Identification and expression of the kosena radish (Raphanus sativus cv. Kosena) homologue of the ogura radish CMS-associated gene, orf138. Plant Mol Biol. 1999;39:183–8.
Yang J, Liu X, Yang X, Zhang M. Mitochondrial-targeted expression of a cytoplasmic male sterility-associated orf220 gene causes male sterility in Brassica juncea. BMC Plant Biol. 2010;10:231–40.
Kumar P, Vasupalli N, Srinivasan R, Bhat SR. An evolutionarily conserved mitochondrial orf108 is associated with cytoplasmic male sterility in different alloplasmic lines of Brassica juncea and induces male sterility in transgenic Arabidopsis thaliana. J Exp Botany. 2012;63:2921–32.
Iorizzo M, Senalik D, Szklarczyk M, Grzebelus D, Spooner D, Simon P. De novo assembly of the carrot mitochondrial genome using next generation sequencing of whole genomic DNA provides first evidence of DNA transfer into an angiosperm plastid genome. BMC Plant Biol. 2012;12:61–77.
Xu Q, Xiong GJ, Li PB, He F, Huang Y, Wang KB, Li ZH, Hua JP. Analysis of complete nucleotide sequences of 12 Gossypium chloroplast genomes: origin and evolution of allotetraploids. PLoS One. 2012;7:e37128.
Zhang TW, Fang YJ, Wang XM, Deng X, Zhang XW, Hu SN, Yu J. The complete chloroplast and mitochondrial genome sequences of Boea hygrometrica: insights into the evolution of plant organellar genomes. PLoS One. 2012;7:e30531.
Chen ZW, Feng K, Grover CE, Li PB, Liu F, Wang YM, Xu Q, Shang MZ, Zhou ZL, Cai XY, Wang XX, Wendel JF, Wang KB, Hua JP. Chloroplast DNA structural variation, phylogeny, and age of divergence among diploid cotton species. PLoS One. 2016;11:e0157183.
Chen ZW, Grover CE, Li PB, Wang YM, Nie HS, Zhao YP, Wang MY, Liu F, Zhou ZL, Wang XX, Cai XY, Wang KB, Wendel JF, Hua JP. Molecular evolution of the plastid genome during diversification of the cotton genus. Mol Phylogenet Evol. 2017;112:268–76.
Backert S, Nielsen BL, Borner T. The mystery of the rings: structure and replication of mitochondrial genomes from higher plants. Trends Plant Sci. 1997;2(12):477–83.
Alverson AJ, Rice DW, Dickinson S, Barry K, Palmer JD. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell. 2011;23:2499–513.
Rice DW, Alverson AJ, Richardson AO, Young GJ, Sanchez-Puerta MV, Munzinger J, Barry K, Boore JL, Zhang Y, dePamphilis CW, Knox EB, Palmer JD. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science. 2013;342:1468–73.
Skippington E, Barkman TJ, Rice DW, Palmer JD. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. P Natl Acad Sci USA. 2015;112(27):E3515–24.
Chen Z, Nie H, Grover CE, Wang Y, Li P, Wang M, et al. Entire nucleotide sequences of Gossypium raimondii and G. arboreum mitochondrial genomes revealed A-genome species as cytoplasmic donor of the allotetraploid species. Plant Biol. 2017;19(3):484–93.
Chen ZW, Nie HS, Wang YM, Pei HL, Li SS, Zhang LD, Hua JP. Rapid evolutionary divergence of diploid and allotetraploid Gossypium mitochondrial genomes. BMC Genomics. 2017;18:876.
Chen ZW, Zhao N, Li SS, Grover CE, Nie HS, Wendel JF, Hua JP. Plant mitochondrial genome evolution and cytoplasmic male sterility. Crit Rev Plant Sci. 2017;36:55–69.
Picardi E, Horner DS, Chiara M, Schiavon R, Valle G, Pesole G. Large-scale detection and analysis of RNA editing in grape mtDNA by RNA deep-sequencing. Nucleic Acids Res. 2010;38:4755–67.
Fang YJ, Wu H, Zhang TW, Yang M, Yin YX, Pan LL, Yu XG, Zhang XW, Hu SN, Al-Mssallem IS, Yu J. A complete sequence and transcriptomic analyses of date palm (Phoenix dactylifera L.) mitochondrial genome. PLoS One. 2012;7:e37164.
Islam MS, Studer B, Byrne SL, Farrell JD, Panitz F, Bendixen C, Møller IM, Asp T. The genome and transcriptome of perennial ryegrass mitochondria. BMC Genomics. 2013;14:202.
Grimes BT, Sisay AK, Carroll HD, Cahoon AB. Deep sequencing of the tobacco mitochondrial transcriptome reveals expressed ORFs and numerous editing sites outside coding regions. BMC Genomics. 2014;15:31.
Tomohiko K, Kinya T. Whole mitochondrial genome sequencing and re-examination of a cytoplasmic male sterility-associated gene in Boro-taichung-type cytoplasmic male sterile rice. PLoS One. 2016. https://doi.org/10.1371/journal.pone.0159379.
Grewe F, Edger PP, Keren I, Sultan L, Pires JC, Ostersetzer-Biran O, et al. Comparative analysis of 11 Brassicales mitochondrial genomes and the mitochondrial transcriptome of Brassica oleracea. Mitochondrion. 2014;19:135–43.
Li SS, Liu GZ, Chen ZW, Wang YM, Li PB, Hua JP. Construction and initial analysis of five Fosmid libraries of mitochondrial genomes of cotton (Gossypium). Chin Sci Bull. 2013;58:4608–15.
Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18.
Liu GZ, Cao D, Li SS, Grover CE, Nie HS, Hua HP. The complete mitochondrial genome of Gossypium hirsutum and evolutionary analysis of higher plant mitochondrial genomes. PLoS One. 2013;8:e69476.
Liu GZ, Cao D, Li SS, Grover CE, et al. The complete mitochondrial genome of Gossypium hirsutum and evolutionary analysis of higher plant mitochondrial genomes. PLoS One. 2013;8(8):e69476.
Tang MY, Chen ZW, Grover CE, Wang MY, Li SS, Ma ZY, Wendel JF, Hua JP. Rapid evolutionary divergence of Gossypium barbadense and G. hirsutum mitochondrial genomes. BMC Genomics. 2015;16:770.
Li H, Durbin R. Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics. 2010;26(5):589–95.
Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics. 2003;10:1–18.
Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics Chapter. 2003:10.3.1–10.3.18.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alinment search tool. J Mol Biol. 1990;215:403–10.
Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64.
Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 2007;52:267–74.
Noé L, Kucherov G. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res 2005;33:W540–W543.
Schnable PS, Wise RP. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 1998;3:175–80.
Hanson MR, Bentolila S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell. 2004;16:S154–69.
Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–7.
Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9:357–4.
Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9.
Darracq A, Varre JS, Touzet P. A scenario of mitochondrial genome evolution in maize based on rearrangement events. BMC Genomics. 2010;11:233–48.
Hsu CL, Mullin BC. Physical characterization of mitochondrial DNA from cotton. Plant Mol Biol. 1989;13:467–8.
Lei B, Li S, Liu G, Chen Z, Su A, Li P, Li Z, Hua J. Evolution of mitochondrial gene content: loss of genes, tRNAs and introns between Gossypium harknessii and other plants. Plant Syst Evol. 2013;299:1889–97.
Goremykin VV, Salamini F, Velasco R, Viola R. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol Biol Evol. 2009;26:99–110.
Sandra S, Mandy K, Stephan G, Ralph B. Horizontal transfer of chloroplast genomes between plant species. Proc Natl Acad Sci U S A. 2012;109(7):2434–8.
Palmer JD, Adams KL, Cho Y, Parkinson CL, Qiu YL, Song K. Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates. Proc Natl Acad Sci U S A. 2000;97:6960–6.
Knoop V. Genomics of chloroplasts and mitochondria. Springer Netherlands. 2012;35:175–200.
Medha M, Karyn SG, Daqing S, Fayaz K, Stephanie LM, Brent LN. DNA recombination activity in soybean mitochondria. J Mol Biol. 2006;356:288–99.
Yoshiyuki T, Mizue T, Keita Y, Hiroshi Y, Toru T. A complete mitochondrial genome sequence of Ogura-type male-sterile cytoplasm and its comparative analysis with that of normal cytoplasm in radish (Raphanus sativus L.). BMC Genomics. 2012;13:352.
Bentolila S, Stefanov S. A reevaluation of rice mitochondrial evolution based on the complete sequence of male-fertile and male-sterile mitochondrial genomes. Plant Physiol. 2012;158:996–1017.
Sloan D, Alverson A, Storchova H, Palmer J, Taylor D. Extensive loss of translational genes in the structurally dynamic mitochondrial genome of the angiosperm Silene latifolia. BMC Evol Biol. 2010;10:274–88.
Heng S, Wei C, Jing B, Wan Z, Wen J, Yi B, Ma C, Tu J, Fu T, Shen J. Comparative analysis of mitochondrial genomes between the hau cytoplasmic male sterility (CMS) line and its iso-nuclear maintainer line in Brassica juncea to reveal the origin of the CMS-associated gene orf288. BMC Genomics. 2014;15:322.
Gallagher LJ, Betz SK, Chase CD. Mitochondrial RNA editing truncates a chimeric open reading frame associated with S male-sterility in maize. Curr Genet. 2002;42:179–84.
L’Homme Y, Stahl RJ, Li XQ, Hameed A, Brown GG. Brassica nap cytoplasmic male sterility is associated with expression of a mtDNA region containing a chimeric gene similar to the pol CMS-associated orf224 gene. Curr Genet. 1997;31:325–35.
Menassa R, L’Homme Y, Brown GG. Post-transcriptional and developmental regulation of a CMS-associated mitochondrial gene region by a nuclear restorer gene. Plant J. 2002;17:491–9.
Liu H, Cui P, Zhan K, Lin Q, Zhuo G, Guo X, Ding F, Yang W, Liu D, Hu S. Comparative analysis of mitochondrial genomes between a wheat K-type cytoplasmic male sterility (CMS) line and its maintainer line. BMC Genomics. 2011;163-176(67):12.
Koizuka N, Imai R, Iwabuchi M, Sakai T, Imamura J. Genetic analysis of fertility restoration and accumulation of ORF125 mitochondrial protein in the kosena radish (Raphanus sativus cv. Kosena) and a Brassica napus restorer line. Theor Appl Genet. 2000;100:949–55.
Kubo T, Newton KJ. Angiosperm mitochondrial genomes and mutations. Mitochondrion. 2008;8:5–14.
Yang JH, Huai Y, Zhang MF. Mitochondrial atpA gene is altered in a new orf220-type cytoplasmic male-sterile line of stem mustard (Brassica juncea). Mol Biol Rep. 2009;36:273–80.
Yang JH, Zhang MF, Yu JQ. Mitochondrial nad2 gene is co-transcripted with CMS-associated orfB gene in cytoplasmic male-sterile stem mustard (Brassica juncea). Mol Biol Rep. 2009;36:345–51.
Adams KL, Qiu YL, Stoutemyer M, Palmer JD. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci U S A. 2002;99:9905–12.
Acknowledgments
We deeply thank Dr. Yi Huang (Institute of Oil Crops, Chinese Academy of Agricultural Sciences) and Corrinne E. Grover (Iowa State University) for their valuable comments and discussion on previous versions of the manuscript. We are also grateful to anonymous reviewers for their helpful suggestions and comments.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31671741) and National Key R & D Program for Crop Breeding (2016YFD0100203). The funders did not have any role in the design, collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
Complete mtDNA sequences were deposited in GenBank database (JX944505.1 for CMS-2074S, JX536494.1 for CMS-2074A, and JX944506.1 for E5903). Other data sets supporting the results of this article are included within the article and its additional files.
Author information
Authors and Affiliations
Contributions
SSL assembled the mitochondrial genome, annotated the mitochondrial genomes, performed the data analysis and prepared the original manuscript. ZWC performed the transcriptomes data analyses, attended discussion and revised MS. NZ critically revised MS and attended data re-interpreting discussion. YMW maintained the experimental platform and participated in the bench work. HSN attended the transcriptomes data analyses and discussion. JPH designed the experiments, provided research platform, and revised the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Summary of the four mitogenomes sequencing and assembly. (DOCX 14 kb)
Additional file 2:
Table S2A. The verification about breaking point of scaffolds between 2074A and 2074B. (DOCX 15 kb)
Additional file 3:
Table S2B. The verification about breaking point of large repeats between 2074A and 2074B. (DOCX 15 kb)
Additional file 4:
Table S3. The chloroplast-derived sequences (> 70 bp) found in four mitogenomes. (DOCX 19 kb)
Additional file 5:
Table S4. List of multi-copy genes in cotton mtDNA. (DOCX 16 kb)
Additional file 6:
Table S5. Nucleotide differences relative to the 2074B mitogenome. (DOCX 14 kb)
Additional file 7:
Figure S1. The end sequencing positive clones in E5903 and 2074A. (DOCX 63 kb)
Additional file 8:
Figure S2. The syntenic regions in four mitochondrial genomes. (DOCX 80 kb)
Additional file 9:
Figure S3. Differential expression of CMS candidate ORFs in 2074A, 2074B and F1-A. Log2 transformations of the expression fold changes (2074B/2074A and F1-A/2074A) are represented by bars. Y axis denotes the levels of transformed expression fold changes. (DOCX 19 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Li, S., Chen, Z., Zhao, N. et al. The comparison of four mitochondrial genomes reveals cytoplasmic male sterility candidate genes in cotton. BMC Genomics 19, 775 (2018). https://doi.org/10.1186/s12864-018-5122-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-018-5122-y