Abstract
Background
Sorbicillinoids are a family of complex cyclic polyketides produced by only a small number of distantly related ascomycete fungi such as Trichoderma (Sordariomycetes) and Penicillium (Eurotiomycetes). In T. reesei, they are synthesized by a gene cluster consisting of eight genes including two polyketide synthases (PKS). To reconstruct the evolutionary origin of this gene cluster, we examined the occurrence of these eight genes in ascomycetes.
Results
A cluster comprising at least six of them was only found in Hypocreales (Acremonium chrysogenum, Ustilaginoidea virens, Trichoderma species from section Longibrachiatum) and in Penicillium rubens (Eurotiales). In addition, Colletotrichum graminicola contained the two pks (sor1 and sor2), but not the other sor genes. A. chrysogenum was the evolutionary eldest species in which sor1, sor2, sor3, sor4 and sor6 were present. Sor5 was gained by lateral gene transfer (LGT) from P. rubens. In the younger Hypocreales (U. virens, Trichoderma spp.), the cluster evolved by vertical transfer, but sor2 was lost and regained by LGT from C. graminicola. SorB (=sor2) and sorD (=sor4) were symplesiomorphic in P. rubens, whereas sorA, sorC and sorF were obtained by LGT from A. chrysogenum, and sorE by LGT from Pestalotiopsis fici (Xylariales). The sorbicillinoid gene cluster in Trichoderma section Longibrachiatum is under strong purifying selection. The T. reesei sor genes are expressed during fast vegetative growth, during antagonism of other fungi and regulated by the secondary metabolism regulator LAE1.
Conclusions
Our findings pinpoint the evolution of the fungal sorbicillinoid biosynthesis gene cluster. The core cluster arose in early Hypocreales, and was complemented by LGT. During further speciation in the Hypocreales, it became subject to birth and death evolution in selected lineages. In P. rubrens (Eurotiales), two cluster genes were symplesiomorphic, and the whole cluster formed by LGT from at least two different fungal donors.
Similar content being viewed by others
Background
Horizontal or lateral gene transfers (HGT and LGT) are important mechanisms of genome evolution that significantly contribute to the development of adaptive traits [1]. Although once considered a process of limited effect outside prokaryotes [2, 3], we now know that HGT and LGT have occurred in all major eukaryotic lineages (reviewed in [4]), including protozoans, plants, animals and fungi [5–7]. In fungi, HGT-driven gene innovation was shown to have resulted in refined repertoires of secreted and transporter proteins and increased metabolic capacities [8]. A survey of sixty fungal genomes detected hundreds of genes horizontally acquired from bacteria [9]. But the list of donors of fungal genetic material also includes plants [10], microbial eukaryotes [11, 12], and - most frequently - other fungi [13]. We will use the term LGT to describe the latter events.
Fungal secondary metabolites have a long history of positive (pharmaceuticals) and negative (toxins) impacts on mankind. Polyketides (PKS) make up a major group of them, most of which are formed by only a few, frequently not closely related species [14, 15]. The origin of PKS diversity has been explained as the result of gene duplication, HGT, LGT, recombination and domain shuffling [16]. However, most of these data have been obtained only for bacteria. Kroken et al. [17] postulated that the observed diversity in fungal PKS’s may not have resulted from HGT or LGT, but rather be due to birth-and-death evolution. However, increased sampling of genomic data from diverse taxonomic groups later provided evidence for the origin of several fungal PKS by HGT from bacteria, and also - in a few cases – by LGT from other fungi and plants [8, 18–22]. In almost all of these cases, translocation involved the whole secondary metabolite clusters (i.e. the PKS and the adjacently located genes encoding modifying enzymes, gene regulators and transporters) - rather than individual genes. To the best of our knowledge, the only exception is the demonstration of reacquisition of biotin prototrophy in Saccharomyces cerevisiae by stepwise HGT from bacterial donors [23].
Trichoderma is a genus of mycotrophic ascomycetes. Baker et al. [24] have recently compared the polyketide synthase (PKS) inventory of three Trichoderma species (T. reesei, T. virens and T. atroviride) and showed that two polyketide synthase encoding genes - pks10, pks11 - were unique to T. reesei. Pks10 and pks11 are located head-to-head in the center of chromosome 5 [25] and were shown to be responsible for the synthesis of sorbicillinoids [26]. These are complex cyclic polyketides, some of which have been shown to exhibit cytostatic and neuroprotective effects [27]. Sorbicillinoids are produced by T. reesei ([28, 29]; named T. longibrachiatum by the authors), but also some other fungal species belonging to the Sordariomycetes (e.g. Verticillium, Acremonium, Paecilomyces; for review see [27]) and the Eurotiomycete Penicillium notatum [30]. In support of this, a putative sorbicillinoid synthesizing cluster similar to the T. reesei cluster, is present in P. rubens [26, 31]. Moreover, the P. rubens orthologue of T. reesei pks11 (pks13) was shown to be essential for sorbicillinoid biosynthesis [32].
This limited, yet taxonomically widespread occurrence of sorbicillinoid biosynthesis in fungi led us to hypothesize that their evolution occurred by other mechanisms than vertical transfer. The goal of this study was to evaluate the evolutionary history of sorbicillin biosynthesis in T. reesei and other fungi. Here we show that this PKS cluster indeed originated from LGT, but in contrast to other reported cases [8, 18–22] it was not transferred as a whole cluster but formed by separate transfers of the individual genes from different donor species. The first almost complete cluster occurred in A. chrysogenum, from where it was transferred to P. rubens. In contrast, its further shaping in the Hypocreales occurred mainly via birth-and-death evolution and survived only in a few species including one of the most recent lineages of Trichoderma, the Longibrachiatum section.
Results
Identification of homologues of the sorbicillinoid biosynthetic clusters in Ascomycetes
To identify gene clusters potentially involved in sorbicillinoid biosynthesis in fungi, we first searched the National Center for Biotechnology Information (NCBI) protein database with the two PKS10- and PKS11- encoded proteins of T. reesei, which represent a non-reducing and a reducing PKS respectively, by bidirectional BLASTP (see Methods, (Additional file 1: Figure S1). Genes encoding proteins with highest similarity to both PKS10 and PKS11 were identified from the plant pathogenic fungus Colletotrichum graminicola (Sordariomycetes, Glomerellales), the opportunistic cephalosporin C-producer Acremonium chrysogenum (Sordariomycetes, Hypocreales), the “rice false smut” causing pathogen Ustilaginoidea virens (Sordariomycetes, Hypocreales) and Penicillium rubens (Eurotiomycetes, Eurotiales). Genes encoding proteins with still high similarity to PKS10 and PKS11 were also found in several other fungi (Eurotiomycetes and Sordariomycetes for PKS11, and – in addition – Dothidiomycetes for PKS10), but only the four species named above contained both of them.
Baker et al. [24] reported that PKS11 and PKS10 are unique to T. reesei, based on the absence in other Trichoderma species for which genome sequences were available at that date. Since the genomes of eight more Trichoderma spp. (i.e. T. harzianum, T. asperellum, T. hamatum, T. gamsii, T. longibrachiatum, T. citrinoviride and T. parareesei) are now available ([32–36]; http://genome.jgi.doe.gov/programs/fungi/index.jsf), we also screened them for the presence of pks10 and pks11 orthologs. The two genes were only found in T. longibrachiatum, T. citrinoviride and T. parareesei, which all are – as T. reesei – members of the Longibrachiatum Section of Trichoderma [37].
The PKS10 and PKS11 orthologs that were retrieved by BLASTP and by screening of the Trichoderma genomes shows that they form a significantly supported clade that contained all those species in which genomes both PKSs were present. C. graminicola occurred at a basal position in this clade (Fig. 1). To indicate that these two genes are part of the sorbicillin biosynthetic cluster, we will – in agreement with [26] - further name them sor1 (=pks11) and sor2 (=pks10) throughout the manuscript.
Phylogenetic analysis of SOR1/SorA (T. reesei PKS11) and SOR2/SorB (T. reesei PKS10) proteins by PhyML. Numbers at the nodes indicate the boostrap (1000 replica) support. Species printed in red belong to Hypocreales, brown to Glomerellales, black Pseudoeurotiaceae (all Sordariomycetes), blue Eurotiomycetes, green Dothidiomycetes. Accession numbers for proteins are given in Additional file 9: Table S1
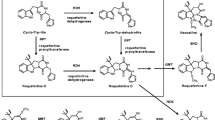
The sorbicillinoid biosynthetic gene clusters in T. reesei and P. rubens comprise 8 and 6 genes, located on chromosomes 5 and 1, respectively ([25, 26], http://www.ncbi.nlm.nih.gov/protein/CAP95405) (Fig. 2). sor3/sorC and sor4/sorD encode binuclear Zn2Cys6 transcription factors, of which sor4 is essential for the biosynthesis of sorbicillinoids in T. reesei [26]; sor5/sorE encodes a FAD-dependent monooxygenase responsible for the oxidative de-aromatisation of sorbicillin and dihydrosorbicillin to sorbicillinol and dihydrosorbicillinol, respectively [38]; and sor6/sorF encodes a transporter of the major facilitator superfamily (MFS).
Architecture of the sorbicillinoid biosynthesis gene cluster in those fungi that possess SOR1/SorA and SOR2/SorB orthologues. Red: PKS, blue/dark blue: Zn2Cys6 transcription factor; green, FAD monooxygenase; orange, MSF transporter; black, short chain reductase/dehydrogenase; white, FAD-dependent oxidase. The dotted cluster in A. chrysogenum specifies an unknown protein. Accession numbers for all proteins shown are given in Additional file 9: Table S1
We consequently analysed whether the other species that contain sor1 and sor2 would indeed also contain the other 4 or 6 genes that are present in the P. rubens and T. reesei cluster, respectively, and have them organized in a genomic cluster (Fig. 2): C. graminicola contained no homologues of any of them, but A. chrysogenum, U. virens and the other 3 Trichoderma spp. contained sor3, sor4, sor5 and sor6 (i.e. the genes encoding the two transcription factors, the MFS transporter the FAD-dependent monooxygenase, respectively). All of them were located in immediate vicinity of sor1 and sor2, although U. virens sor5 is located a few genes farther apart than in the other fungi.
The cluster in T. reesei contained two further genes – sor7 and sor8 - that were absent from most other fungi: sor7, encoding a short-chain dehydrogenase/reductase, for which a P. rubens ortholog (CAP92704.1) - is present in the genome but not located in the vicinity of the sorbicillinoid gene cluster In A. chrysogenum, another gene of unknown function is found at the position of sor7. sor8 encodes an FAD-dependent oxidase (Fig. 2), of which an ortholog is present in A. chrysogenum and U. virens, but not located in the vicinity of the sorbicillinoid cluster, and absent from the P. rubens genome (Fig. 2).
We also looked at possible synteny of the 5′ and 3′ flanking regions of the cluster: while there was considerable synteny between the four Trichoderma spp., no synteny was found between Trichoderma and the other fungi possessing the sorbicillinoid biosynthesis cluster.
We tested the null hypothesis that the phylogenetic history of SOR3 – SOR8 was consistent with a vertical transfer within fungi by implementing phylogenetic analyses (Additional file 2: Figure S2 A-F). This shows that only SOR5 forms a strongly supported clade containing – except for C. graminicola - all species that also contain the SOR1 and SOR2 proteins. SOR3, SOR4 and SOR6 are distributed in several clades, which – even after collapsing branches with poor bootstrap support (<75%) are not concordant with the established Ascomycota phylogeny (cf. [39]). On the other hand, SOR7 and SOR8 display a phylogeny that strongly resembles the Ascomycota phylogeny (see below; cf. Additional file 3: Figure S3). They are also present in Trichoderma spp. which lack the sorbicillinoid biosynthetic cluster.
Evolution of the sorbicillinoid gene cluster in filamentous fungi
The above described discordance between the phylogeny of SOR1-SOR6 homologues and the Ascomycota phylogeny suggested that they may have arisen by LGT from different ancestors. To test this hypothesis, we applied three complementary approaches: the bipartition dissimilarity test implemented in T-REX [40], which identifies HGT/LGT events by quantifying the proximity between two phylogenetic trees using a refinement of the Robinson and Foulds distance [41, 42]; the reconciliation of each gene tree to the fungal species phylogeny, thereby assigning costs to gene duplications, HGT/LGT, gene loss, and incomplete lineage sorting, as implemented in Notung [43]; and the Jane software tool that uses a polynomial time dynamic programming algorithm in conjunction with a genetic algorithm to find solutions pairs of trees [44]. We accept proof for HGT/LGT only for those cases where (i) at least two of these programs provided consistent results that were not rejected by the third, and (ii) where the protein tree topology was contradictory to the Ascomycota phylogeny and could not be more parsimoniously reconciled using a combination of differential gene duplications (GD) and gene loss.
The evolution displayed by the results from this analysis (Additional file 4: Figure S4, Additional file 5: Table S2) are summarized in Table 1: evidence for LGT was obtained for A. chrysogenum (SOR4), Trichoderma and U. virens (SOR2, SOR3), and P. rubens (SorA, SorC, SorE and SorF). Interestingly, three of the genes of P. rubens (SorA, SorC and SorF) were obtained from A. chrysogenum, whereas SOR4 of A. chrysogenum was obtained from P. rubens (SorD), indicating frequent LGTs between these two species. In Trichoderma and U. virens, only SOR2 appears to have been obtained by LGT from C. graminicola.
With the exception of SOR2, neither U. virens nor the four Trichoderma spp. appear to have received any of the other cluster genes by LGT. No LTG events could be inferred for sor7 and sor8 (neither by Notung, T-REX nor Jane) which is in agreement with the observation that these genes occupied positions concordant with Ascomycota phylogeny (vide supra).
Trichoderma SOR1 and SOR2 evolved by purifying selection
The Longibrachiatum Section of Trichoderma is one of the most recent branches in Trichoderma evolution [45]. The fact that we were unable to identify the sor genes in less evolutionary derived species of the genus but could not verify LGT as the mechanism of origin of the sorbicillioid gene cluster in the Longibrachiatum clade was thus unexpected. The alternative hypothesis to explain the absence of these genes in other species is that these genes have been lost. To test this hypothesis, we reconstructed the evolution of the eight SOR proteins by Count [46] and Gloome [47]. The results provided consistent evidence for loss of the respective genes in other infrageneric clades of Trichoderma and in those Hypocreaceae species that are close to Trichoderma but also lack them (Additional file 6: Figure S5). Interestingly, the ratio of the pairwise amino acid differences between SOR1/SorA and SOR2/SorB and the housekeeping genes used to construct the Ascomycota tree (see Methods), was significantly higher in the four Trichoderma spp. than in U. virens, P. rubens or A. chrysogenum (Table 2). This would be typical for LGT, as was found for SOR2. However, since sor1 has not been obtained by LGT, it may as well be due to a higher rate of evolution of these two genes in Trichoderma section Longibrachiatum. Determination of the Ka/Ks ratio for the Trichoderma sor1 and sor2 genes yielded values around 0.1, suggesting the operation of strong purifying selection.
Sorbicillinoid cluster gene expression in T. reesei
Many PKS synthesizing clusters in fungi are silenced [48]. We therefore used available oligonucleotide microarray data of T. reesei growing on glucose, glycerol, lactose or cellulose as carbon sources in submerged culture, or on glucose on agar plates to test whether the sor genes are indeed expressed. In fact the eight sor genes in the Trichoderma cluster are expressed at high levels under conditions of rapid growth (glucose, glycerol), whereas lower expression was detected on lactose which allows only slow vegetative growth (Fig. 3a). Most sor genes had only a low level of expression during asexual sporulation (Fig. 3a).
Heat plot of the expression of sor1 - sor8 in T. reesei during a growth on different carbon sources and onset of sporulation; Glc - glucose; Gly - glycerol, Lac - lactose, b during cultivation of a lae1 knock-out and a lae1 overexpressing strain on lactose, and c plotting of the ratio between the parent strain to the lae1 knock-out (white bars) and the lae1 overexpressing strain (black bars), respectively
The protein methyltransferase LaeA is a major regulator of secondary metabolism in Eurotiomycetes and some Sordariomycetes [48]. Its T. reesei orthologue LAE1 regulates some but not all PKS genes [49]. As shown in Fig. 3c, a lae1 knock-out mutant shows significantly decreased expression of sor1 and sor2, and interestingly also of sor7 and sor8. No increased expression was observed for these four genes in a strain overexpressing lae1 under a constitutive promoter. However, the genes encoding one of the two transcription factors (sor3), the MFS transporter (sor6) and the FAD monooxygenase (sor5) were significantly upregulated by lae1 overexpression.
Discussion
Although HGT and LGT occur in the majority of cases by transfer of single genes only (for review see [9, 50], the transfer of multiple genes or gene clusters has also been shown [11, 19, 50–52], particularly for genes encoding proteins for secondary metabolite synthesis [18, 20, 53–59]. In contrast, our data show that the fungal sorbicillin biosynthesis cluster evolved by complementing symplesiomorphous genes by LGT from other fungal donors, which in P. rubens occurred in at least two steps. Based on the species phylogeny and the LGT events found, A. chrysogenum is the most ancient known taxon that contains an almost complete cluster that misses only the transcription factor sor4, and which it obtained by LGT from P. rubens. The more recent Hypocreales (U. virens and the four Trichoderma spp. of section Longibrachiatum) regained one of the PKSs (sor2) from C. graminicola, implying that this gene was lost in one of the Hypocreales that are more recent than A. chrysogenum.
In contrast, the cluster is missing in the Eurotiales with the exception of P. rubens, at least with respect to species whose genome sequence is available. The Eurotiales only contain orthologs of the PKS SorB and the transcription factor SorD, which lends to speculate that these two genes are involved in the synthesis of another polyketide. It is interesting to note that three missing genes (sorA, sorC and sorF) were obtained from A. chrysogenum, to which P. rubens transferred its sorD, indicating a history of a frequent gene exchange between these two fungi. We cannot say, however, whether the LGT from A. chrysogenum to P. rubens occurred in one or several steps. It is also interesting that sorE (encoding the FAD monooxygenase crucial for sorbicillin formation [38]) had not been transferred to P. rubens but to an unknown Eurotiales ancestor from P. fici. Thus sorE must have been present in P. rubens before LGT of sorA, sorC and sorF.
The absence of sor3 – sor6 from C. graminicola could be due to gene loss. An alternative hypothesis, however, would be that SOR1 and SOR2 are synthesizing a different polyketide than sorbicillin in this fungus. Indeed, an annotation of the genes flanking the C. graminicola sor1/sor2 locus revealed an adjacent oxidoreductase gene whose encoded protein exhibited 83% amino acid similarity to an oxidoreductase CtnB involved in citrinin biosynthesis in Monascus aurantiacus (Eurotiales) [60], and a putative aldehyde dehydrogenase (Additional file 7: Figure S6). We therefore assume that the resulting polyketide synthesized by Colletotrichum is (or was) probably not a sorbicillinoid. Sorbicillinoid were thus in fact first produced in A. chrysogenum or closely related but as yet unknown ancestor.
Despite of the occurrence of the sorbicillinoid gene cluster in only U. virens and Trichoderma spp. from section Longibrachiatum, we found (with the exception of sor2) no evidence for their origin by further LGT events. Instead, our data show that the cluster evolved by vertical transfer, and has been lost by the operation of massive birth-and-death evolution [61] within the Hypocreales. In fact, a scenario of gene duplications followed by gene loss has earlier been suggested for the evolution of fungal non-reducing polyketide synthases [17], and claimed to be responsible for the todays patchy distribution of distantly related secondary metabolites.
Yet our finding that the sorbicillinoid cluster only survived in Trichoderma species belonging to section Longibrachiatum is interesting. It is consistent with the formation of the characteristic yellow pigment secreted by these species [62], because sorbicillinoids have a characteristic yellow-orange color [63]. Species from this section are known to have smaller genomes than other Trichoderma spp. and represent one of the youngest phylogenetic clades of the genus [38, 45]. The fact that the sorbicillinoid gene cluster has been maintained in these species but not in other Trichoderma spp. suggests that the respective products are of selective importance to fungi from this section. This is also supported by our findings of strong purifying selection acting on sor1 and sor2. Unfortunately, the function of sorbicillinoids is not known yet: although some sorbicillinoids were reported to inhibit the growth of tumour cells [27, 64], they usually display only low inhibitory activity against bacteria and fungi [27]. Their role as components of antagonism against other organisms is unlikely. Rather, their ecological importance may reside in their high antioxidant and radical scavenging activity [27, 65]. Our findings of high expression of the sor genes in T. reesei under conditions of fast growth, but not during sporulation, supports a role of sorbicillinoids in vegetative growth, which is corroborated by finding them in high concentrations during submerged growth of T. reesei (Additional file 8: Table S3). Interestingly, sor1 and sor2 are also strongly upregulated upon confrontation of T. reesei with plant pathogenic Thanatephorus spp./Rhizoctonia solani (Cantharellales, Basidiomycota) [66]. At a first glance, this contradicts the above conclusion that sorbicillinoids are not involved in antagonism. However, the protection against radicals formed by reactive oxygen species is an important defence reaction of fungi, plants and higher eukaryotes when confronted by other organisms [67–70]. It will be intriguing to find out whether the sorbicillinoids indeed play such a role and - if so – why their biosynthesis was just maintained in only a small group of fungal species.
Conclusions
Tracking the evolution of secondary metabolite synthesizing gene clusters by LGT or HGT have so far in most cases been restricted to the detection of transfer of the whole clusters between two fungi. Our findings show how a fungal secondary metabolite cluster was assembled by individual genes from different fungi by LGT before it became subject to birth-and-death evolution in selected lineages.
Methods
Identification of sorbicillinoid biosynthetic genes in fungi
The eight proteins of the sorbicillinoid biosynthetic cluster in T. reesei were used in a preliminary sequence similarity search by BLASTP of the NCBI database. One hundred best hits were collected. In addition, we searched the genomes of T. longibrachiatum, T. citrinoviride, T. asperellum, T. hamatum and T. parareesei for homologues to SOR1 - SOR8. Since the latter two are not available in a public database, we prepared a local BLAST databases for these two fungi. All these sequences were then aligned by CLUSTALW [71], and subjected to phylogenetic analysis with PhyML 3.0 using the Dayhoff model and 1000 boostrap replica [72]. The topology of the resulting tree was analysed, and all proteins that formed clades not related to that comprising the T. reesei proteins were removed. The resulting collection of sequences was re-aligned with MUSCLE [73] and CLUSTALW [71] and edited by GBLOCKS [74] to identify potential differences in the phylogenetic reconstruction due to the use of different methods. Individual trees were reconstructed with the individual edited protein alignments with PhyML 3.0 using 1000 bootstrap repetitions, and their topology concordance confirmed. The four alignments were then concatenated, and Bayesian analysis performed with TOPALI v2.5 [75], using the WAG model, gamma substitution and 100,000 generations.
Ascomycota tree reconstruction
To reconstruct the reference Ascomycota phylogeny containing all fungi putatively involved in the LGT events described in this paper, the amino acids inferred from four nuclear genes, which were shown to be good phylogenetic markers for fungal species trees reconstruction (i.e. histone acetyltransferase subunit of the RNA polymerase II holoenzyme, FG533; NAD-dependent glutamate dehydrogenase, FG570; translation initiation factor eIF-5, FG832; and Tsr1p, a protein required for processing of 20S pre-rRNA, MS277) were retrieved from FunyBase [76] (http://genome.jouy.inra.fr/funybase). Proteins from species not contained in FunyBase were retrieved by BLASTP search of the GenBank (http://www.ncbi.nlm.nih.gov/genbank/), the Joint Genome Institute (http://genome.jgi-psf.org/programs/fungi/index.jsf?projectList), EnsemblFungi (http://fungi.ensembl.org/index.html) and Broad Institute (http://www.broadinstitute.org/) databases (all databases accessed 28-12-2015). Their alignment, and analysis by PhyML 3.0 and Bayesian analysis were essentially performed as described above.
Inferring HGT/LGT events
To test for the occurrence of HGT, three approaches were used: first, the bipartition dissimilarity test implemented in T-REX [40], which quantifies the proximity between two phylogenetic trees using a refinement of the Robinson and Foulds (RF) distance, was used by applying midpoint rooting and HGT identification by iteration. Second, a gene tree-species phylogeny reconciliation was performed in Notung, using its duplication, transfer, loss and ILS aware parsimony-based algorithm [43]. To this end, gene tree nodes with less than 0.90 SH-like local support were collapsed, and the resulting tree rooted and its polytomies resolved against the bifurcating species phylogeny. This resolved gene tree was then reconciled to the multifurcating, consensus species phylogeny using a duplication cost of 1.5, loss cost of 1 and ILS cost of 0, and the option to prune taxa not present in the gene tree enabled. Third, Jane version 4, a software tool for cophylogeny reconstruction problems that attributes costs to cospeciation, duplication, host switch, and sorting was used [44]. For our analyses we employed default cost settings, and the population size was set 50-fold the number of generations.
Gene gain and loss analysis
Gene gain and loss was tested by two methods: (i) Count [46], which can perform ancestral genome reconstruction by posterior probabilities in a phylogenetic birth-and-death model. Rates were optimized using a gain–loss–duplication model, with default parameters and allowing different gain–loss and duplication–loss rates for different branches, and one hundred rounds of optimization. (ii) Gloome [47], which enables accurate inference of gain and loss events by a stochastic mapping approach, using a variable gain and loss ratio.
Analysis of selection pressure by Ka/Ks ratio
Tajima’s D statistic [77] was determined with DNASp 5.0 [78], using a sliding-window approach.
Transcriptome analysis
We used transcriptome data from our own earlier studies. These included: cultivation of T. reesei QM 9414 on D-glucose, glycerol, lactose, and wheat straw in batch cultures [79, 80], during induction of asexual sporulation [81], at the onset of confrontation with the basidiomycete Thanatephorus spp./Rhizoctonia solani [82], and during growth on lactose in lae1 knock-out and lae1-overexpressing strains [51]. All transcriptome data were obtained by oligonucleotide array hybridization, using a high-density oligonucleotide microarray (Roche-NimbleGen, Inc., Madison, WI) with 60-mer probes representing the 9123 genes of T. reesei. Values were normalized by quantile normalization [83] and the RMA algorithm [84]. After elimination of transcripts that exhibited an SD >20% of the mean value within replicates, false discovery rates [85] were used to assess the significance of values. All transcriptome data and the related protocols are available at the GEO web site (http://www.ncbi.nlm.nih.gov/geo) under the accession numbers given in the cited papers.
Abbreviations
- GD:
-
Gene duplications
- HGT:
-
Horizontal gene transfer
- LGT:
-
Lateral gene transfer
- MFS:
-
Major facilitator superfamily
- PKS:
-
Polyketide synthase
References
Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–87.
Dagan T, Artzy-Randrup Y, Martin W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci U S A. 2008;105:10039–44.
Kloesges T, Popa O, Martin W, Dagan T. Networks of gene sharing among 329 proteobacterial genomes reveal differences in lateral gene transfer frequency at different phylogenetic depths. Mol Biol Evol. 2011;28:1057–74.
Huang J. Horizontal gene transfer in eukaryotes: the weak-link model. Bioessays. 2013;35:868–75.
Yue J, Hu X, Sun H, Yang Y, Huang J. Widespread impact of horizontal gene transfer on plant colonization of land. Nat Commun. 2012;3:1152.
Li F-W, Villarreal JC, Kelly S, Rothfels CJ, Melkonian M, Frangedakis E, et al. Horizontal transfer of an adaptive chimeric photoreceptor from bryophytes to ferns. Proc Natl Acad Sci U S A. 2014;111:6672–7.
Szöllösi GJ, Davín AA, Tannier E, Daubin V, Boussau B. Genome-scale phylogenetic analysis finds extensive gene transfer among fungi. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140335.
Richards TA, Leonard G, Soanes DM, Talbot NJ. Gene transfer into the fungi. Fungal Biol Rev. 2011;25:98–110.
Marcet-Houben M, Gabaldon T. Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 2010;26:5–8.
Richards TA, et al. Phylogenomic analysis demonstrates a pattern of rare and ancient horizontal gene transfer between plants and fungi. Plant Cell. 2009;21:1897–911.
Slot JC, Hibbett DS. Horizontal transfer of a nitrate assimilation gene cluster and ecological transitions in fungi: a phylogenetic study. PLoS One. 2007;2:e1097.
Tiburcio RA, Costa GG, Carazzolle MF, Mondego JM, Schuster SC, Carlson JE, et al. Genes acquired by horizontal transfer are potentially involved in the evolution of phytopathogenicity in Moniliophthora perniciosa and Moniliophthora roreri, two of the major pathogens of cacao. J Mol Evol. 2010;70:85–97.
Wisecaver JH, Slot JC, Rokas A. The evolution of fungal metabolic pathways. PLoS Genet. 2014;10:e1004816.
Simpson TJ. Fungal polyketide biosynthesis - a personal perspective. Nat Prod Rep. 2014;31:1247–52.
Vederas JC. Explorations of fungal biosynthesis of reduced polyketides – a personal viewpoint. Nat Prod Rep. 2014;31:1253–9.
Wang H, Sivonen K, Fewer DP. Genomic insights into the distribution, genetic diversity and evolution of polyketide synthases and nonribosomal peptide synthetases. Curr Opin Genet Dev. 2015;35:79–85.
Kroken S, Glass NL, Taylor J, Yoder O, Turgeon B. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci U S A. 2003;100:15670–5.
Khaldi N, Collemare J, Lebrun MH, Wolfe KH. Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biol. 2008;9:R18.
Khaldi N, Wolfe KH. Evolutionary origins of the fumonisin secondary metabolite gene cluster in Fusarium verticillioides and Aspergillus niger. Int J Evol Biol. 2011;2011:423821–7.
Slot JC, Rokas A. Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between fungi. Curr Biol. 2011;21(2):134–9.
Soanes D, Richards TA. Horizontal gene transfer in eukaryotic plant pathogens. Annu Rev Phytopathol. 2014;52:583–614.
Richards TA, Talbot NJ. Horizontal gene transfer in osmotrophs: playing with public goods. Nat Rev Microbiol. 2013;11:720–7.
Hall C, Dietrich FS. The reacquisition of biotin prototrophy in Saccharomyces cerevisiae involved horizontal gene transfer, gene duplication and gene clustering. Genetics. 2007;177:2293–307.
Baker SE, Perrone G, Richardson NM, Gallo A, Kubicek CP. Phylogenetic analysis and evolution of polyketide synthase encoding genes in Trichoderma. Microbiology UK. 2012;158:147–54.
Druzhinina IS, Kopchinskiy A, Kubicek EM, Kubicek CP. A complete annotation of the chromosomes of the cellulase producer Trichoderma reesei provides new insights in gene clusters, their expression and reveals genes required for fitness. Biotechnol Biofuels. 2016;9:75.
Jørgensen MS, Larsen TO, Mortensen UH, Aubert D. Unraveling the secondary metabolism of the biotechnological important filamentous fungus Trichoderma reesei (Teleomorph Hypocrea jecorina). Kgs. Lyngby: Technical University of Denmark; 2013. p. 164.
Harned AM, Volp KA. The sorbicillinoid family of natural products: isolation, biosynthesis, and synthetic studies. Nat Prod Rep. 2011;28:1790–810.
Andrade R, Ayer WA, Mebe PP. The metabolites of Trichoderma longibrachiatum. Part 1. Isolation of the metabolites and the structure of trichodimerol. Can J Chem. 1992;70:2526–35.
Andrade R, Ayer WA, Trifonov LS. The metabolites of Trichoderma longibrachiatum. Part II The structures of trichodermolide and sorbiquinol. Can J Chem. 1996;74:371–9.
Maskey RP, Grun-Wollny RP, Laatsch H. Sorbicillin analogues and related dimeric compounds from Penicillium notatum. J Nat Prod. 2005;68:865–70.
Salo OV, Ries M, Medema MH, Lankhorst PP, Vreeken RJ, Bovenberg RA, et al. Genomic mutational analysis of the impact of the classical strain improvement program on ß-lactam producing Penicillium chrysogenum. BMC Genomics. 2015;16:937.
Salo OV, Guzmán-Chávez F, Ries MI, Lankhorst PP, Bovenberg RA, Vreeken RJ, et al. Identification of a polyketide synthase involved in sorbicillin biosynthesis by Penicillium chrysogenum. Appl Environ Microbiol. 2016; AEM.00350-16. [Epub ahead of print].
Yang D, Pomraning K, Kopchinskiy A, Karimi Aghcheh R, Atanasova L, Chenthamara K, et al. Genome Sequence and Annotation of Trichoderma parareesei, the Ancestor of the Cellulase Producer Trichoderma reesei. Genome Announc. 2015;3: e00885–15.
Studholme DJ, Harris B, Le Cocq K, Winsbury R, Perera V, Ryder L, et al. Investigating the beneficial traits of Trichoderma hamatum GD12 for sustainable agriculture-insights from genomics. Front Plant Sci. 2013;4:258.
Baroncelli R, Zapparata A, Piaggeschi G, Sarrocco S, Vannacci G. Draft whole-genome sequence of Trichoderma gamsii T6085, a promising biocontrol agent of Fusarium head blight on wheat. Genome Announc. 2016;4:e01747–15.
Xie BB, Qin QL, Shi M, Chen LL, Shu YL, Luo Y, et al. Comparative genomics provide insights into evolution of Trichoderma nutrition style. Genome Biol Evol. 2014;6:379–90.
Druzhinina IS, Komoń-Zelazowska M, Ismaiel A, Jaklitsch W, Mullaw T, Samuels GJ, et al. Molecular phylogeny and species delimitation in the section Longibrachiatum of Trichoderma. Fungal Genet Biol. 2012;49:358–68.
Fahad AA, Abood A, Fisch KM, Osipow A, Davison J, Avramović M, et al. Oxidative dearomatisation: the key step of sorbicillinoid biosynthesis. Chem Sci. 2014;5:523–7.
Wang H, Xu Z, Gao L, Hao B. A fungal phylogeny based on 82 complete genomes using the composition vector method. BMC Evol Biol. 2009;9:195.
Boc A, Diallo AB, Makarenkov V. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012;40(Web Server issue):W573–9.
Boc A, Philippe H, Makarenkov V. Inferring and validating horizontal gene transfer events using bipartition dissimilarity. Syst Biol. 2010;59:195–211.
Robinson DR, Foulds LR. Comparison of phylogenetic trees. Math Biosci. 1981;53:131–47.
Stolzer M, Lai H, Xu M, Sathaye D, Vernot B, Durand D. Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics. 2012;28:i409–15.
Conow C, Fielder D, Ovadia Y, Libeskind‐Hadas R. Jane: a new tool for the cophylogeny reconstruction problem. Algorithms Mol Biol. 2010;5:16.
Kubicek CP, Herrera-Estrella A, Seidl-Seiboth V, Martinez DA, Druzhinina IS, Thon M, Zeilinger S, et al. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 2011;12:R40.
Csurös M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics. 2010;26:1910–2.
Cohen O, Ashkenazy H, Belinky F, Huchon D, Pupko T. GLOOME: gain loss mapping engine. Bioinformatics. 2010;26:2914–5.
Bok JW, et al. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol. 2009;5:462–4.
Aghcheh RK, Kubicek CP. Epigenetics as an emerging tool for improvement of fungal strains used in biotechnology. Appl Microbiol Biotechnol. 2015;99:6167–81.
Wisecaver JH, Rokas A. Fungal metabolic gene clusters—caravans traveling across genomes and environments. Front Microbiol. 2015;6:161.
Karimi-Aghcheh R, et al. Functional analyses of Trichoderma reesei LAE1 reveal conserved and contrasting roles of this regulator. G3 (Bethesda). 2013;3:369–78.
Novo M, Bigey F, Beyne E, Galeote V, Gavory F, Mallet S, et al. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc Natl Acad Sci U S A. 2009;106:16333–8.
Slot JC, Rokas A. Multiple GAL pathway gene clusters evolved independently and by different mechanisms in fungi. Proc Natl Acad Sci U S A. 2010;107:10136–41.
Cheeseman K, Ropars J, Renault P, Dupont J, Gouzy J, Branca A, et al. Multiple recent horizontal transfers of a large genomic region in cheese making fungi. Nat Commun. 2014;5:2876.
Campbell MA, Rokas A, Slot JC. Horizontal transfer and death of a fungal secondary metabolic gene cluster. Genome Biol Evol. 2012;4:289–93.
Patron NJ, Waller RF, Cozijnsen AJ, Straney DC, Gardiner DM, Nierman WC, et al. Origin and distribution of epipolythiodioxopiperazine (ETP) gene clusters in filamentous ascomycetes. BMC Evol Biol. 2007;7:174.
Schmitt I, Lumbsch HT. Ancient horizontal gene transfer from bacteria enhances biosynthetic capabilities of fungi. PLoS One. 2009;4:e4437.
Proctor RH, Van Hove F, Susca A, Stea G, Busman M, van der Lee T, et al. Birth, death and horizontal transfer of the fumonisin biosynthetic gene cluster during the evolutionary diversification of Fusarium. Mol Microbiol. 2013;90:290–306.
Moore GG, Collemare J, Lebrun MH. Evolutionary mechanisms involved in development of fungal secondary metabolite gene clusters. In: Osbourn A, Goss RJ, Carter GT, editors. Natural products discourse, diversity, and design. Hoboken: Wiley; 2014. p. 343–56.
Li YP, Pan YF, Zou LH, Xu Y, Huang ZB, He QH. Lower citrinin production by gene disruption of ctnB involved in citrinin biosynthesis in Monascus aurantiacus Li AS3.4384. J Agric Food Chem. 2013;61:7397–402.
Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–52.
Samuels GJ, Ismaiel A, Mulaw TB, Szakacs G, Druzhinina IS, Kubicek CP, Jaklitsch WM. The Longibrachiatum Clade of Trichoderma: a revision with new species. Fungal Divers. 2012;55:77–108.
Trivonov LS, Hilpert H, Floersheim P, Dreiding AS. Bisvertinols: a new group of dimeric vertinoids from Verticillium intertextum. Tetrahedron. 1986;42:3157–79.
Mazzucco CE, Warr G. Trichodimerol (BMS-182123) inhibits lipopolysaccharide-induced eicosanoid secretion in THP-1 human monocytic cells. J Leukoc Biol. 1996;60:271–7.
Abe N, Murata T, Hirota A. Novel oxidized sorbicillin dimers with 1,1-Diphenyl-2-picrylhydrazyl-Radical scavenging activity from a fungus. Biosci Biotech Biochem. 1998;62:2120–6.
Atanasova L, Knox BP, Kubicek CP, Druzhinina IS, Baker SE. The polyketide synthase gene pks4 of Trichoderma reesei provides pigmentation and stress resistance. Eukaryotic Cells. 2013;12:1499–508.
Lambeth JD. Nox enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4:181–9.
Torres MA, Dangl JL. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol. 2005;8:397–403.
Silar P. Peroxide accumulation and cell death in filamentous fungi induced by contact with a contestant. Mycol Res. 2005;109:137–49.
Takemoto D, Tanaka A, Scott B. NADPH oxidases in fungi: diverse roles of reactive oxygen species in fungal cellular differentiation. Fungal Genet Biol. 2007;44:1065–76.
Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;9(22):4673–80.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21.
Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;9:113.
Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–77.
Milne I, Wright F, Rowe G, Marshall DF, Husmeier D, McGuire G. TOPALi: software for automatic identification of recombinant sequences within DNA multiple alignments. Bioinformatics. 2004;20(11):1806–7.
Marthey S, Aguileta G, Rodolphe F, Gendrault A, Giraud T, Fournier E, et al. FUNYBASE: a FUNgal phYlogenomic dataBASE. BMC Bioinformatics. 2008;9:456.
Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95.
Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2.
Bischof R, Fourtis L, Limbeck A, Gamauf C, Seiboth B, Kubicek CP. Comparative analysis of the Trichoderma reesei transcriptome during growth on the cellulase inducing substrates wheat straw and lactose. Biotechnol Biofuels. 2013;6:127.
Ivanova C, Bååth JA, Seiboth B, Kubicek CP. Systems analysis of lactose metabolism in Trichoderma reesei identifies a lactose permease that is essential for cellulase induction. PLoS One. 2013;8:e62631.
Metz B, Seidl-Seiboth V, Haarmann T, Kopchinskiy A, Lorenz P, Seiboth B, et al. Expression of biomass-degrading enzymes is a major event during conidium development in Trichoderma reesei. Eukaryot Cell. 2011;10:1527–35.
Atanasova L, Le Crom S, Gruber S, Coulpier F, Seidl-Seiboth V, Kubicek CP, et al. Comparative transcriptomics reveals different strategies of Trichoderma mycoparasitism. BMC Genomics. 2014;14:121.
Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93.
Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP, et al. Summaries of affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. JR Stat Soc Series B Stat Methodol. 1995;57:289–300.
Derntl C, Rassinger A, Srebotnik E, Mach RL, Mach-Aigner AR. Identification of the Main Regulator Responsible for Synthesis of the Typical Yellow Pigment Produced by Trichoderma reesei. Appl Environ Microbiol. 2016;82(20):6247-6257.
Acknowledgements
The authors acknowledge the permission by Igor V. Grigoriev to use sequence data from yet unpublished T. longibrachiatum and T. citrinoviride genomes sequenced by the Joint Genome Institute of the US Department of Energy. The authors are grateful to Michael Sulyok, University of Applied Life Sciences of Vienna, for performing the trichodermol analyses.
Funding
The work was supported by a grants from the Austrian Science Fund to CPK (I-1249) and ISD (P25613-B20).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article, its additional (supplementary) files and in the references specified in Materials and Methods.
Authors’ contributions
Planned and designed the study: CPK; analysed and interpreted data: ISD, EMK, CPK; wrote the paper: ISD, EMK and CPK. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
All authors have read the article and agreed on its publication.
Ethics approval and consent to participate
Not applicable.
Note added in Proof
After submission of this paper, Derntl et al. [86] reported the regulation of sorbicillin biosynthesis in T. reesei by the transcription factors SOR3 and SOR4.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Figure S1.
Architecture of Trire2: 73618 and Trire2:73621. The bar specifies the size of the proteins (in amino acid residues). Abbreviations: PKS, polyketide synthase; AT, acyltransferase; DH, dehydrogenase; AM, adenosyl-methionine transferase; ER, enoyl reductase; KR, keto reductase; TR, thioester reductase. (PDF 165 kb)
Additional file 2: Figure S2.
Phylogenetic analysis of SOR3/SorC (A), SOR4/SorD (B), SOR5/SorE (C), SOR6/SorF (D), SOR7/SorG (E) and SOR8 (F) proteins by PhyML. Numbers at the nodes indicate the boostrap (1000 replicas) support. Numbers at the nodes indicate the bootstrap (1000 replicas) support. Colour codes are used as in Fig. 1. In addition, bright brown specifies members of the Sordariales. Accession numbers for all proteins shown are given in Additional file 9: Table S1. (PDF 368 kb)
Additional file 3: Figure S3.
PhyML evolutionary tree of fungi, using protein sequences of the histone acetyltransferase subunit of RNA polymerase II, NAD-dependent glutamate dehydrogenase, translation initiation factor eIF-5, and Tsr1p, a protein required for processing of 20S pre-rRNA. For further details, see Methods. Species that contain a sorbicillinoid biosynthesis cluster are given in red. Donor species are printed in bold. Of Trichoderma, only T. reesei is shown for simplicity. (PDF 297 kb)
Additional file 4: Figure S4.
Output trees of the analysis of SOR1-SOR8 by Notung (a), T-Rex (b) and Jane (c). In (a), yellow arrows indicate LGT, red D indicate duplication events; in (b), species names are abbreviated due to constraints of the program, but can easily be identified by comparing them to the species shown in (a) and (c); also note that of Trichoderma sect. Longibrachiatum, only T. reesei was used in these analyses; in (c), black lines identify the species tree, whereas blue lines indicate the protein tree. Lines with arrows show LGT, accompanied by support values. (PDF 673 kb)
Additional file 5: Table S2.
Statistics of T-Rex, Notung and Jane analyses. (DOCX 15 kb)
Additional file 6: Figure S5.
Gain and loss of SOR1 – SOR6 in the Hypocreales. Open bars indicate gene loss, number at the nodes indicate the respective loss rates. (PDF 130 kb)
Additional file 7: Figure S6.
Gene structure of the 3′ end of supercontig 46 of the C. graminicola genome sequence (http://genome.jgi.doe.gov/Colgr1/Colgr1.home.html). No further genes are located 3′ of Colgr1:8017. (PDF 219 kb)
Additional file 8: Table S3.
Trichodermol concentration in the culture fluid of T. reesei. (DOCX 12 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Druzhinina, I.S., Kubicek, E.M. & Kubicek, C.P. Several steps of lateral gene transfer followed by events of ‘birth-and-death’ evolution shaped a fungal sorbicillinoid biosynthetic gene cluster. BMC Evol Biol 16, 269 (2016). https://doi.org/10.1186/s12862-016-0834-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12862-016-0834-6