Abstract
Background
The association between BIN1 rs744373 variant and Alzheimer’s disease (AD) had been identified by genome-wide association studies (GWASs) as well as candidate gene studies in Caucasian populations. But in East Asian populations, both positive and negative results had been identified by association studies. Considering the smaller sample sizes of the studies in East Asian, we believe that the results did not have enough statistical power.
Results
We conducted a meta-analysis with 71,168 samples (22,395 AD cases and 48,773 controls, from 37 studies of 19 articles). Based on the additive model, we observed significant genetic heterogeneities in pooled populations as well as Caucasians and East Asians. We identified a significant association between rs744373 polymorphism with AD in pooled populations (P = 5 × 10− 07, odds ratio (OR) = 1.12, and 95% confidence interval (CI) 1.07–1.17) and in Caucasian populations (P = 3.38 × 10− 08, OR = 1.16, 95% CI 1.10–1.22). But in the East Asian populations, the association was not identified (P = 0.393, OR = 1.057, and 95% CI 0.95–1.15). Besides, the regression analysis suggested no significant publication bias. The results for sensitivity analysis as well as meta-analysis under the dominant model and recessive model remained consistent, which demonstrated the reliability of our finding.
Conclusions
The large-scale meta-analysis highlighted the significant association between rs744373 polymorphism and AD risk in Caucasian populations but not in the East Asian populations.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is the preeminent cause for dementia in the elderly and causes 50–75% dementia types [1,2,3]. In the populations of Caucasian ancestry, large-scale genome-wide association studies (GWASs) have identified some common genetic risk factors for AD [4]. In addition to the APOE gene (encoding apolipoprotein E) as a major AD risk factor, recent GWASs of AD in Caucasian populations also identified several new genetic loci, including: bridging integrator 1 (BIN1), complement receptor 1 (CR1), clusterin (CLU), phosphatidylinositol binding clathrin assembly protein (PICALM), membrane-spanning 4-domains, subfamily A, member 4 (MS4A4)/membrane-spanning 4-domains, subfamily A, member 6E (MS4A6E), CD2-associated protein (CD2AP), CD33 molecule (CD33), EPH receptor A1 (EPHA1) and ATP-binding cassette transporter A7 (ABCA7) [4,5,6,7]. These genetic loci largely stimulate downstream analysis, for example the identification of relevant gene expression regulators [8,9,10,11,12,13,14,15], the construction gene expression regulatory networks [16,17,18,19,20], and so on.
Rs744373 is a single nucleotide polymorphism (SNP) that locates upstream of BIN1 gene. In populations of Caucasian ancestry, rs744373 polymorphism was consistently confirmed to be significantly associated with AD risk with P = 3.16 × 10 − 10 [21], P = 2.6 × 10 − 14 [6], P = 2.13 × 10 − 09 [22], P = 2.9 × 10 − 07 [23] and P = 1.1 × 10 − 04 [24]. Recently, the association has also been extensively investigated in East Asian populations. However, besides the positive associations, many studies have also identified negative results. Tan et al. did not report any significant association when analyzd 1224 Chinese individuals (612 cases and 612 controls) using allele test (P = 0.217) and genotype test (P = 0.547, 0.263 and 0.397 for dominant, recessive and additive logistic genetic models) [25]. The result from Li et al. was also negative [26]. Wang et al. identified a significant result in population from East China (P = 0.038), but not southwest China (P = 0.874). When combining the two parts of populations, they still did not identify any significant association (P = 0.187) [27]. In Brazilian Chinese population, Ramos et al. analyzed 241 individuals (82 cases and 159 controls) and didn’t find any significant results (P = 0.660 for dominant model and P = 0.547 for recessive model) [28]. Ohara et al. did not report significant association (P = 0.06 for additive model) when analyzed 825 AD cases and 2934 controls from Japan [29]. In 2013, we conducted a meta-analysis using all currently available samples (2022 AD cases and 4209 controls) and the results were significant (P = 1.19 × 10 − 02, 7.08 × 10 − 03 and 5.75 × 10 − 03 for the dominant model, recessive model and additive model) [30]. Another subsequent meta-analysis with more samples (11,832 AD cases and 18,133 controls) obtained a consistent result with us [3].
Given the inconsistent findings in East Asian populations, we believe that the relatively small sample sizes, as well as the genetic heterogeneity of AD susceptibility loci among different populations, may be important factors in the untrustworthiness of the results. In this study, we aimed to collect more studies and samples than before and obtain more statistically significant results by performing genetic heterogeneity test and meta-analysis of the rs744373 polymorphism in the Caucasians, East Asians, and pooled populations.
Materials and methods
Literature acquisition
In order to find all available association studies, we searched the PubMed database (https://www.ncbi.nlm.nih.gov/pubmed) and AlzGene database (http://www.alzgene.org/) with the Keywords “Alzheimer’s disease”, “Bridging Integrator 1” or “BIN1”. We also searched Google Scholar (http://scholar.google.com/) to acquire the articles citing the studies obtained in the PubMed and AlzGene databases. The literature acquisition was updated on December 12, 2017. In addition, we collected as much data as we could by directly contacting with authors. These datasets were not published due to not significant results, etc., and were not included in the previous meta-analysis of rs744373 polymorphism with AD.
Inclusion criteria
The studies inclusion criteria contained: (1) being a case-control study; (2) investigating the association between rs744373 polymorphism and AD; (3) being conducted in East Asian or Caucasian populations; (4) providing the numbers of rs744373 genotypes or sufficient data to calculate them or (5) providing an OR with 95% confidence interval (CI) and the P-value or sufficient data to calculate them.
Data extraction
The information was extracted from each study contained: (1) author names; (2) publication year; (3) the sample’s ethnicity; (4) the numbers of cases and controls; (5) the genotyping platform; (6) the frequencies of rs744373 genotypes or sufficient data to calculate them or (7) the OR with 95% CI or sufficient data to calculate them.
Genetic model
Since not all studies provided exact genotype numbers, we investigated the association between rs744373 polymorphism and AD risk in this meta-analysis primarily using the additive genetic model. We selected allele C as effect allele and T as reference allele, the additive model can be described as C allele versus T allele [31].
Comparison of MAF and OR in Caucasians and east Asians
We compared the minor allele frequency (MAF), which is the frequency of rs744373 allele C in this study, and the OR values between the Caucasian populations and East Asian populations. We used the t-test to investigate whether there were differences in the OR values and MAF values between these two populations. We used program R (http://www.r-project.org/) to perform the t-test and calculate the OR and MAF values that not available in the original articles.
Heterogeneity test
We used the Cochran’s Q test to investigate genetic heterogeneity among different studies. Cochran’s Q test approximately follows a chi-square distribution and its degree of freedom is k-1 (k represents the number of studies included in this studies). Statistics I2 can also use to measure the genetic heterogeneity, which is calculated as:
\( {\mathrm{I}}^2=\frac{\mathrm{Q}-\left(\mathrm{k}-1\right)}{\mathrm{Q}}\times 100\% \)
The statistics I2 is in the range of 0–100%, and we divided it into four parts: 0–25%, 26–50%, 51–75%, 76–100%, which respectively represent low, moderate, large and extreme heterogeneity [30]. We conducted Cochran’s Q test in East Asians, Caucasians, and pooled Populations respectively. All calculations of P-value and I2 value were completed using the program R (http://www.r-project.org/). We choose P < 0.05 or I2 > 50% as discriminant criterion for significant result of heterogeneity test.
Meta-analysis
In the meta-analysis, we used fixed effect model (Mantel–Haenszel) or random effect model (DerSimonian–Laird) to calculate the overall OR. And which model to choose depends on whether the genetic heterogeneity is significant or not. If the P-value of Cochran’s Q test was less than 0.05, and I2 value was greater than 50%, we selected the random effect model, otherwise we selected the fixed effect model. The signification of overall OR was measured by Z test.
Sensitivity analysis and publication Bias analysis
To further test the stability of our results, we conducted a sensitivity analysis by sequentially removing each study in the meta-analysis at a time. We used funnel plots to evaluate the potential publication bias. A symmetrical inverted funnel indicated the results were no bias, and an asymmetrical inverted funnel indicated bias results [4]. Begg’s test and Egger’s test was used to evaluate the asymmetry of the funnel plot [4]. The significant level was 0.01. All statistical tests above were also performed using the program R (http://www.r-project.org/).
Results
Literature search and data description
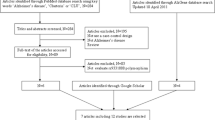
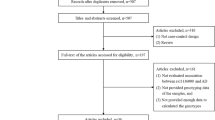
We obtained 126 articles by searching the PubMed database. Eighty-eight articles were excluded because they were (1) not Case-Control design, (2) not analyzed in East Asian or Caucasian populations, (3) not related with AD, (4) meta-analysis or (5) review articles. We further excluded 24 articles because they did not investigate the association between rs744373 polymorphism and AD or not provide sufficient data. The remaining 14 articles met the analysis requirements. According to the same criteria, we also obtained two articles from the AlzGene database. In addition, we had found one article by searching Google Scholar. We applied for two datasets of two articles (studies) by contacting the author directly. Finally, 37 studies in 19 articles, including 22,395 AD cases and 48,773 control samples, were included in this meta-analysis. More detailed information about selecting studies was described in Fig. 1. The main characteristics of included studies were described in Table 1.
Comparison of MAF and OR between Caucasian and east Asian
There were 11 studies belong to East Asian populations. The MAF values of rs744373, OR values and other information of these 11 studies listed in the top 11 rows in Table 1. The other studies listed in the last 26 rows in Table 1 belonged to Caucasian populations. By using the t-test to compare the MAF values between Caucasians and East Asians, we found a significant result with t = 5.89 and P = 1.53 × 10− 6. However, the result of comparison of OR values did not indicate a very significant distinction between the two populations (t = 1.75 and P = 0.11).
Heterogeneity test
We conducted heterogeneity test of rs744373 polymorphism in different populations, and identified significant genetic heterogeneity in Caucasians (P = 0.001, I2 = 52.3%), East Asians (P = 0.001, I2 = 65.1%) and pooled populations (P = 1.03 × 10− 5, I2 = 57.2%). Detailed results were described in Table 2 and Fig. 2.
Forest plot for the meta-analysis of the association between rs744373 and AD under the additive model. “OR” is the abbreviation of Odds Ratio. “Beta” indicates the ln (OR). “se” is the standard error of Beta. “Weight” represents the weight of each study when calculating the overall OR. The genetic heterogeneity test results (I2 and its P-value) and the meta-analysis results (overall OR and 95% CI) in pooled populations are listed at the bottom of the figure. The results for subgroup analysis are also listed by the grey font
Meta-analysis
Based on the results of Heterogeneity test, we used random effect model to calculate the overall OR values in East Asians, Caucasians and pooled populations, respectively. Meta-analysis results indicated significant correlation in Caucasians with P = 3.38 × 10− 08, OR = 1.16, 95% CI 1.10–1.22, and in pooled populations with P = 5 × 10− 07, OR = 1.12, and 95% CI 1.07–1.17 (Table 2). However, we did not find any association between rs744373 polymorphism and AD in East Asian populations with P = 0.39, OR = 1.06, and 95% CI 0.95–1.15. The detailed results and forest diagram were described in Table 2 and Fig. 2.
Sensitivity analysis and publication Bias analysis
Using sensitivity analysis, we identified that the results of meta-analysis remained largely unchanged by excluding any one study (Table 3). The symmetrical inverted funnel in the funnel plot suggested no publication bias of the results (Begg’s test, P = 0.471; Egger’s test, P = 0.428). Funnel diagram was described in Fig. 3.
Discussion
GWASs showed that SNPs located in upstream of BIN1, particular rs744373, are strongly associated with AD risk [41]. The expression quantitative trait loci (eQTL) analysis identified a pronounced association between rs744373 and the expression of BIN1 in brain tissue [3]. BIN1 gene have diverse functions, including endocytosis, trafficking, immune response, apoptosis, and tau metabolism, that are thought have potential roles in AD pathological mechanism [41, 42]. To some extent, investigating the association between rs744373 and AD risk is helpful for understanding the role of BIN1 in AD pathogenesis.
Based on the significant association between rs744373 polymorphism and AD risk identified by the GWASs in Caucasian populations, many recent studies had also explored this association in East Asian populations, as described in the introduction. However, the findings of the association studies in East Asian were always inconsistent. Considering a relatively small sample size may result in less statistical power, we collected 37 studies involving 22,395 AD cases and 48,773 controls for the meta-analysis. To the best of our knowledge, this was the largest sample size by far.
By meta-analysis of the 37 studies, we obtained significant association between rs744373 polymorphism and AD risk in pooled populations (P = 5 × 10− 07, OR = 1.12, and 95% CI 1.07–1.17) and also in Caucasian populations (P = 3.38 × 10− 08, OR = 1.16, 95% CI 1.10–1.22). The results were consistent with the previous studies. However, in East Asian populations, our results showed a significant genetic heterogeneity of rs744373 polymorphism (P = 0.001, I2 = 65.1%) and the meta-analysis did not show a significant association between rs744373 polymorphism with AD risk by using a random effect model (P = 0.393, OR = 1.057, and 95% CI 0.95–1.15). The insignificant publication bias results and the consistent sensitivity analysis results showed that our results were reliable.
To confirm the findings that were obtained by additive genetic model, we further used the dominant model (CC + CT versus TT) and recessive model (CC versus CT + TT) to investigate the association of rs744373 polymorphism with AD risk based on genotype data of 33,184 samples (12,717 AD cases and 20,467 controls). As same as the results of additive model, we obtained significant association between rs744373 and AD in pooled populations (P = 3.95 × 10− 11, OR = 1.17, 95% CI 1.12–1.23 for dominant model and P = 1.35 × 10− 05, OR = 1.19, 95% CI 1.10–1.29 for recessive model), as well as in Caucasian populations (P = 5.99 × 10− 11, OR = 1.20, 95% CI 1.14–1.27 for dominant model and P = 1.00 × 10− 05, OR = 1.26, 95% CI 1.14–1.39 for recessive model). We also obtained negative results in East Asian populations (P = 0.391, OR = 1.06, 95% CI 0.93–1.21 for dominant model and P = 0.806, OR = 1.03, 95% CI 0.81–1.31 for recessive model). The consistent results among the three kinds of genetic models demonstrated the reliability of our results. The data was described in Additional file 1 and the detailed results were described in Table 2, Table 4, Fig. 4, Fig. 5 and Additional file 1. The information about the samples and publication bias was described in Additional file. In summary, this large-scale meta-analysis highlighted the significant association between rs744373 polymorphism and AD in Caucasian populations but not in the East Asian populations.
Researchers have begun to focus on AD genetic heterogeneity between different races and ethnicities since the end of the last century [43]. They found that the frequency variations in ApoE subtypes existed among nine populations include Caucasians and East Asians [43]. Besides the most consistent genetic risk factor ApoE for Sporadic AD, some studies have also reported many genetic risk factors that appear distinct AD susceptibility between Caucasian and East Asian populations. For instance, following genes were proven to be only associated with AD risk in Caucasian populations but not in East Asian populations: Triggering Receptor Expressed On Myeloid Cells 2 (TREM2) [44, 45], Solute Carrier Family 24 Member 4 (SLC24A4) [46], NME/NM23 Family Member 8 (NME8) [47], GRB2 Associated Binding Protein 2 (GAB2) [48], Myocyte Enhancer Factor 2C (MEF2C) [49], Inositol Polyphosphate-5-Phosphatase D (INPP5D) [50], CLU [51], ABCA7, CD2AP, and EPHA1 [25], Fermitin Family Member 2 (FERMT2) [52]. Hence, the complex difference among different ethnicities and races probably cause the genetic heterogeneity of AD between Caucasians and East Asians.
Our samples of East Asian ancestry mainly came from Chinese, Japanese and Koreans populations. On the one hand, these samples may not be able to represent the East Asian populations completely. On the other hand, the specific differences in sample collection processes of different studies would lead to genetic heterogeneity among different populations. Considering these limitations, we believe that a large sample size GWAS in East Asian population is very necessary.
Conclusions
Until now, the genetic association between BIN1 rs744373 and AD risk in East Asian populations is still not deterministic. In the study, we conducted a meta-analysis with the largest sample size so far (22,395 AD cases and 48,773 controls). The meta-analysis results under the additive, dominant and recessive model indicated a significant association between rs744373 and AD risk in Caucasian populations but not in East Asian populations. The consistent results of sensitivity analysis, as well as the negative results of publication bias analysis, demonstrated the reliability of our findings. We believe that the greater statistically powerful results are helpful for the understanding of AD genetic risk factors in East Asian populations.
Availability of data and materials
Most of the summary statistics extracted from each study are included within the articles and its Additional files.
Abbreviations
- ABCA7:
-
ATP-binding cassette transporter A7
- AD:
-
Alzheimer’s disease
- APOE:
-
apolipoprotein E
- BIN1:
-
bridging integrator 1
- CD2AP:
-
CD2-associated protein
- CD33:
-
CD33 molecule
- CI:
-
confidence interval
- CLU:
-
clusterin
- CR1:
-
complement receptor 1
- EPHA1:
-
EPH receptor A1
- GTEx:
-
Genotype-tissue Expression
- GWAS:
-
Genome-wide association study
- MAF:
-
minor allele frequency
- MS4A4:
-
membrane-spanning 4-domains, subfamily A, member 4
- MS4A6E:
-
membrane-spanning 4-domains, subfamily A, member 6E
- OR:
-
odds ratio
- PICALM:
-
phosphatidylinositol binding clathrin assembly protein
- SE:
-
standard error
- SNP:
-
single nucleotide polymorphism
References
Liu GY, Jiang YS, Wang P, Feng RN, Jiang N, Chen XY, Song H, Chen ZG. Cell adhesion molecules contribute to Alzheimer's disease: multiple pathway analyses of two genome-wide association studies. J Neurochem. 2012;120:190–8.
Liu GY, Yao LF, Liu JF, Jiang YS, Ma GD, Chen ZG, Zhao B, Li KS, Alzheimer's GER: Cardiovascular disease contributes to Alzheimer's disease: evidence from large-scale genome-wide association studies. Neurobiol Aging 2014, 35:786–792.
Zhu RX, Liu X, He ZY. The bridging integrator 1 gene polymorphism rs744373 and the risk of Alzheimer's disease in Caucasian and Asian populations: an updated Meta-analysis. Mol Neurobiol. 2017;54:1419–28.
Liu GY, Wang HY, Liu JF, Li JB, Li HL, Ma GD, Jiang YS, Chen ZG, Zhao B, Li KS. The CLU gene rs11136000 variant is significantly associated with Alzheimer's disease in Caucasian and Asian populations. NeuroMolecular Med. 2014;16:52–60.
Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–U1061.
Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429.
Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–U1068.
Deng S-P, Cao S, Huang D-S, Wang Y-PJIAtocb, bioinformatics: Identifying stages of kidney renal cell carcinoma by combining gene expression and dna methylation data 2017, 14:1147–1153.
Guo W-L, Huang D-SJMB: An efficient method to transcription factor binding sites imputation via simultaneous completion of multiple matrices with positional consistency. 2017, 13:1827–1837.
Huang D-S, Zheng C-HJB: Independent component analysis-based penalized discriminant method for tumor classification using gene expression data. 2006, 22:1855–1862.
Peng C, Zou L, Huang D-SJIA: Discovery of Relationships Between Long Non-Coding RNAs and Genes in Human Diseases Based on Tensor Completion 2018, 6:59152–59162.
Shen Z, Bao W, Huang D-SJSr: Recurrent Neural Network for Predicting Transcription Factor Binding Sites. 2018, 8:15270.
Shen Z, Zhang Y-H, Han K, Nandi AK, Honig B, Huang D-SJC: miRNA-disease association prediction with collaborative matrix factorization 2017, 2017.
Yi H-C, You Z-H, Huang D-S, Li X, Jiang T-H, Li L-PJMT-NA: A deep learning framework for robust and accurate prediction of ncRNA-protein interactions using evolutionary information 2018, 11:337–344.
Yuan L, Zhu L, Guo W-L, Zhou X, Zhang Y, Huang Z, Huang D-SJIAToCB, Bioinformatics: Nonconvex penalty based low-rank representation and sparse regression for eQTL mapping 2017, 14:1154–1164.
Bao W, Jiang Z, Huang D-SJBb: Novel human microbe-disease association prediction using network consistency projection 2017, 18:543.
Deng S-P, Zhu L, Huang D-S. Mining the bladder cancer-associated genes by an integrated strategy for the construction and analysis of differential co-expression networks. BMC Genomics. 2015;S4.
Deng S-P, Zhu L, Huang D-SJIAToCB, Bioinformatics: Predicting hub genes associated with cervical cancer through gene co-expression networks 2016, 13:27–35.
Guo W, Zhu L, Deng S, Zhao X, Huang DJSCIS: Understanding tissue-specificity with human tissue-specific regulatory networks 2016, 59:070105.
Zhang H, Zhu L, Huang D-SJIAtocb, bioinformatics: DiscMLA: an efficient discriminative motif learning algorithm over high-throughput datasets. 2016.
Antunez C, Boada M, Gonzalez-Perez A, Gayan J, Ramirez-Lorca R, Marin J, Hernandez I, Moreno-Rey C, Moron FJ, Lopez-Arrieta J, et al. The membrane-spanning 4-domains, subfamily a (MS4A) gene cluster contains a common variant associated with Alzheimer's disease. Genome Med. 2011;3.
Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou FG, Crook JE, Pankratz VS, Sando SB, Aasly JO, et al. Replication of BIN1 association with Alzheimer's disease and evaluation of genetic interactions. J Alzheimers Dis. 2011;24:751–8.
Hu XL, Pickering E, Liu YC, Hall S, Fournier H, Katz E, Dechairo B, John S, Van Eerdewegh P, Soares H. Initi AsDN: Meta-analysis for genome-wide association study identifies multiple variants at the BIN1 locus associated with late-onset Alzheimer's disease. PLoS One. 2011;6.
Lambert JC, Zelenika D, Hiltunen M, Chouraki V, Combarros O, Bullido MJ, Tognoni G, Fievet N, Boland A, Arosio B, et al. Evidence of the association of BIN1 and PICALM with the AD risk in contrasting European populations. Neurobiol Aging. 2011;32.
Tan L, Yu JT, Zhang W, Wu ZC, Zhang Q, Liu QY, Wang W, Wang HF, Ma XY, Cui WZ. Association of GWAS-linked loci with late-onset Alzheimer's disease in a northern Han Chinese population. Alzheimers Dement. 2013;9:546–53.
Li HL, Yang P, Liu ZJ, Sun YM, Lu SJ, Tao QQ, Guo QH, Wu ZY. Common variants at Bin1 are associated with sporadic Alzheimer's disease in the Han Chinese population. Psychiatr Genet. 2015;25:21–5.
Wang HZ, Bi R, Hu QX, Xiang Q, Zhang C, Zhang DF, Zhang W, Ma XH, Guo WJ, Deng W, et al. Validating GWAS-identified risk loci for Alzheimer's disease in Han Chinese populations. Mol Neurobiol. 2016;53:379–90.
dos Santos LR, Belcavello L, Camporez D, de Magalhaes CIM, Zandonade E, Morelato RL, Errera FIV, Louro ID, Batitucci MDP, de Paula F. Association study of the BIN1 and IL-6 genes on Alzheimer's disease. Neurosci Lett. 2016;614:65–9.
Ohara T, Ninomiya T, Hirakawa Y, Ashikawa K, Monji A, Kiyohara Y, Kanba S, Kubo M. Association study of susceptibility genes for late-onset Alzheimer's disease in the Japanese population. Psychiatr Genet. 2012;22:290–3.
Liu GY, Zhang SY, Cai ZY, Li Y, Cui LL, Ma GD, Jiang YS, Zhang LC, Feng RN, Liao MZ, et al. BIN1 gene rs744373 polymorphism contributes to Alzheimer's disease in east Asian population. Neurosci Lett. 2013;544:47–51.
Li XW, Shen N, Zhang SY, Liu JF, Jiang QH, Liao MZ, Feng RN, Zhang LC, Wang GY, Ma GD, et al. CD33 rs3865444 polymorphism contributes to Alzheimer's disease susceptibility in Chinese, European, and north American populations. Mol Neurobiol. 2015;52:414–21.
Guo WL, Zhu L, Deng SP, Zhao XM, Huang DS. Understanding tissue-specificity with human tissue-specific regulatory networks. Sci China-Inf Sci. 2016;59.
Deng SP, Zhu L, Huang DS. Predicting hub genes associated with cervical Cancer through gene co-expression networks. IEEE-ACM Trans Comput Biol Bioinform. 2016;13:27–35.
Deng SP, Zhu L, Huang DS. Mining the bladder cancer-associated genes by an integrated strategy for the construction and analysis of differential co-expression networks. BMC Genomics. 2015;16.
Jiao B, Liu XY, Zhou L, Wang MH, Zhou YF, Xiao TT, Zhang WW, Sun R, Waye MMY, Tang BS, Shen L. Polygenic analysis of late-onset Alzheimer's disease from mainland China. PLoS One. 2015;10.
Gharesouran J, Rezazadeh M, Khorrami A, Ghojazadeh M, Talebi M. Genetic evidence for the involvement of variants at APOE, BIN1, CR1, and PICALM loci in risk of late-onset Alzheimer's disease and evaluation for interactions with APOE genotypes. J Mol Neurosci. 2014;54:780–6.
Carrasquillo MM, Khan QU, Murray ME, Krishnan S, Aakre J, Pankratz VS, Nguyen T, Ma L, Bisceglio G, Petersen RC, et al. Late-onset Alzheimer disease genetic variants in posterior cortical atrophy and posterior AD. Neurol. 2014;82:1455–62.
Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–40.
Nizamutdinov II, Andreeva TV, Stepanov VA, Marusin AV, Rogaev EI, Zasedatelev AS, Nasedkina TV. Biochip for determination of genetic markers of sporadic Alzheimer's disease risk in the Russian Slavic population. Mol Biol. 2013;47:827–35.
Moreno DJ, Ruiz S, Rios A, Lopera F, Ostos H, Via M, Bedoya G. Association of GWAS top genes with late-onset Alzheimer's disease in Colombian population. Am J Alzheimers Dis Other Demen. 2017;32:27–35.
Cuyvers E, Sleegers K. Genetic variations underlying Alzheimer's disease: evidence from genome-wide association studies and beyond. Lancet Neurol. 2016;15:857–68.
Chapuis J, Hansmannel F, Gistelinck M, Mounier A, Van Cauwenberghe C, Kolen KV, Geller F, Sottejeau Y, Harold D, Dourlen P, et al. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry. 2013;18:1225–34.
Hallman DM, Boerwinkle E, Saha N, Sandholzer C, Menzel HJ, Csazar A, Utermann G. The apolipoprotein E polymorphism: a comparison of allele frequencies and effects in nine populations. Am J Hum Genet. 1991;49:338–49.
Huang M, Wang D, Xu Z, Xu Y, Xu X, Ma Y, Xia Z. Lack of genetic association between TREM2 and Alzheimer's disease in east Asian population: a systematic review and meta-analysis. Am J Alzheimers Dis Other Dement. 2015;30:541–6.
Ma J, Zhou Y, Xu J, Liu X, Wang Y, Deng Y, Wang G, Xu W, Ren R, Liu X, et al. Association study of TREM2 polymorphism rs75932628 with late-onset Alzheimer's disease in Chinese Han population. Neurol Res. 2014;36:894–6.
Lu H, Zhu XC, Wang HF, Cao L, Tan MS, Tan CC, Jiang T, Yu JT, Tan L. Lack of association between SLC24A4 polymorphism and late-onset Alzheimer's disease in Han Chinese. Curr Neurovasc Res. 2016;13:239–43.
Liu SL, Wang XC, Tan MS, Wang HF, Zhang W, Wang ZX, Yu JT, Tan L. NME8 rs2718058 polymorphism with Alzheimer's disease risk: a replication and meta-analysis. Oncotarget. 2016;7:36014–20.
Hu Y, Zheng L, Cheng L, Zhang Y, Bai W, Zhou W, Wang T, Han Z, Zong J, Jin S, et al. GAB2 rs2373115 variant contributes to Alzheimer's disease risk specifically in European population. J Neurol Sci. 2017;375:18–22.
Tang SS, Wang HF, Zhang W, Kong LL, Zheng ZJ, Tan MS, Tan CC, Wang ZX, Tan L, Jiang T, et al. MEF2C rs190982 polymorphism with late-onset Alzheimer's disease in Han Chinese: a replication study and meta-analyses. Oncotarget. 2016;7:39136–42.
Jing H, Zhu JX, Wang HF, Zhang W, Zheng ZJ, Kong LL, Tan CC, Wang ZX, Tan L, Tan L. INPP5D rs35349669 polymorphism with late-onset Alzheimer's disease: a replication study and meta-analysis. Oncotarget. 2016;7:69225–30.
Lu SJ, Li HL, Sun YM, Liu ZJ, Yang P, Wu ZY. Clusterin variants are not associated with southern Chinese patients with Alzheimer's disease. Neurobiol Aging. 2014;35:2656 e2659–11.
Zhang QY, Wang HF, Zheng ZJ, Kong LL, Tan MS, Tan CC, Zhang W, Wang ZX, Tan L, Yu JT, Tan L. FERMT2 rs17125944 polymorphism with Alzheimer's disease risk: a replication and meta-analysis. Oncotarget. 2016;7:39044–50.
Acknowledgements
We thank Dr. Guiyou Liu and Dr. Lin Jiao of the School of Life Science and Technology for their technical and linguistic assistance.
About this supplement
This article has been published as part of BMC Bioinformatics Volume 20 Supplement 25, 2019: Proceedings of the 2018 International Conference on Intelligent Computing (ICIC 2018) and Intelligent Computing and Biomedical Informatics (ICBI) 2018 conference: bioinformatics. The full contents of the supplement are available online at https://bmcbioinformatics.biomedcentral.com/articles/supplements/volume-20-supplement-25.
Funding
This work was supported by the National Nature Science Foundation of China (61822108, 61571152), the Natural Science Foundation of Heilongjiang Province (F2015006) and the National Science and Technology Major Project (2017YFSF090117, 2017YFC1201200). The publication costs are funded by the National Nature Science Foundation of China (61571152).
Author information
Authors and Affiliations
Contributions
QHJ and SLJ proposed and designed the research. ZFH searched the database and processed the GWAS summary data with assistance from TW, RT, PPW, WYZ, PR, JZ and YH. ZFH preformed most of the statistical analyses. ZFH wrote the article with assistance from TW. QHJ supervised the writing and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
Meta-analysis under dominant and recessive model. Table S1. The selected studies investigating the association between rs744373 and AD using dominant model and recessive model Figure S1. Funnel plot of the publication bias analysis under dominant model. Figure S2. Funnel plot of the publication bias analysis under recessive model.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Han, Z., Wang, T., Tian, R. et al. BIN1 rs744373 variant shows different association with Alzheimer’s disease in Caucasian and Asian populations. BMC Bioinformatics 20 (Suppl 25), 691 (2019). https://doi.org/10.1186/s12859-019-3264-9
Published:
DOI: https://doi.org/10.1186/s12859-019-3264-9