Abstract
Proteomics has emerged as a highly promising bioanalytical technique in various aspects of applied biological research. In Indian academia, proteomics research has grown remarkably over the last decade. It is being extensively used for both basic as well as translation research in the areas of infectious and immune disorders, reproductive disorders, cardiovascular diseases, diabetes, eye disorders, human cancers and hematological disorders. Recently, some seminal works on clinical proteomics have been reported from several laboratories across India. This review aims to shed light on the increasing use of proteomics in India in a variety of biological conditions. It also highlights that India has the expertise and infrastructure needed for pursuing proteomics research in the country and to participate in global initiatives. Research in clinical proteomics is gradually picking up pace in India and its future seems very bright.
Similar content being viewed by others
Background
The beginning of the twentyfirst century, saw the birth of a new technology called ‘proteomics’ that emerged as a highly promising bioanalytical technique in various aspects of applied biological research. Though India could not play a role in human genome sequencing projects, the country has emerged as one of the frontrunners in global proteomics research and has come a long way from where it was, probably a decade ago [1–3]. The importance of proteomics research was realized when many Indian researchers initiated identifying proteins critical to the pathophysiology of various diseases. However, the actual momentum gained subsequently when several laboratories were successful in arranging sophisticated facilities for efficient proteome separation and detection using tissue as well as liquid samples. This was supported further with national efforts for network building and collaboration amongst various laboratories with a primary objective for early detection, diagnosis, and a way to therapeutics for common as well rare diseases.
Although the development of proteomics research in India was rather slow at the outset, the last decade or so has seen a dramatic expansion in the proteomics community [4]. Presently, there are over a hundred research laboratories in approximately 80 academic or research institutes across India involved in various proteome-level investigations. Thus it may safely be said that in the past few years, proteomics has emerged as a powerful tool for varied biological investigations in the country. Proteomics is also becoming an attractive method of choice for the identification and development of new biomarkers and potential therapeutic targets. Increasing financial and infrastructure support from the government is likely to take proteomics research in the country to new heights.
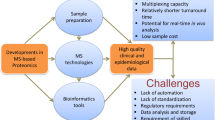
Proteomics is a scientific approach that attempts to completely characterize the proteome (or subproteome) of a cell or tissue. Various technologies can be employed during the three major steps involved in proteomic analysis: sample preparation, separation of proteins or peptides, identification and characterization of proteins, and the technologies can be mixed and matched to meet the needs to answer any particular biological or clinical question. The advancement of proteomic technologies now addresses the challenges associated with the pathophysiology of various diseases. Proteins are most widely affected in disease, response and recovery. The most important application of proteomics is believed to be discovery of disease biomarkers and drug targets which can lead to designing of products aimed at diagnosis and treatment of diseases like cancer, cardiovascular diseases, obesity and type 2 diabetes. This application of proteomic tools and knowhow to the field of medicine is called clinical proteomics (Fig. 1). It encompasses the translation of proteomic detection technologies and strategies towards the production of diagnostics and therapeutics for the direct improvement of human health [5]. Therefore, clinical proteomics methods hold special promise for the identification of new biomarkers that might improve disease staging, risk stratification, and the prognosis and treatment of diseases. The following review intends to look into the current developments and shortcomings in India’s clinical proteomics scene and the problems faced by the Indian scientific community in translational research. The review also deals with next generation proteomics methods and different proteomics databases developed in India.
Review
Proteomics in Indian academia and the emergence of Proteomics Society India
Proteomics is taking centre stage in biology and biomedical research in India. Several institutes are currently employing proteomics techniques in their research and at present, we have an enviable mass of proteomics scientists in the country. To foster entrepreneurship and skill development in proteomics research, many biotech parks have been established with public and private support. In recent years, proteomics and related disciplines have also been incorporated into academic curricula across India due to their increasing impact on clinical research. For instance, The Clinical Proteomics Remote Triggering Virtual Laboratory (http://iitb.vlab.co.in/?sub=41&brch=237) creates a virtual platform for beginners to acquire knowledge and a realistic experience of performing different techniques commonly applied in clinical proteomics research. These new e-learning resources in proteomics act as important global platforms for students, e-learners and researchers. With expansion of research efforts and introduction of modern technologies, it is important to facilitate interactions among the proteomics fraternity and help them disseminate knowledge for the holistic development of this discipline. “Proteomics Society India”, is thus a step in that direction that provides a platform to pursue an agenda that will meet this objective. The Society which is deeply involved towards imparting education in proteomics research through organizing periodic conferences, workshops and meetings, is still in its infancy. However, a lot is being done to make students and teachers familiar with proteomics. Consequently, the 3rd Annual Meeting of PSI—International Proteomics Conference on ‘OMICS MEETS DISEASE’, was held at Kolkata in December 2011. This was followed by a themed conference on ‘Medical Proteomics’ in Bangalore in November,2013. Both the events were well attended by clinicians, eminent scientists and students. This highlights India’s emergence as a global player in cutting-edge clinical proteomics research.
Methods adopted for clinical proteomics research in India
In India, clinical proteomics efforts initially relied upon the traditional 2DE-MS approach and its advanced variation, 2D-DIGE. However, these gel-based protein separation approaches offered limited proteome coverage. This resulted in a shift towards adopting alternative, high throughput shotgun approaches such as liquid chromatography–mass spectrometry (LC–MS or LC–MS/MS). This approach, having greater quantitative ability, enabled deeper proteome analysis generating huge protein datasets. This gave way to semi quantitative labeling technologies such as isobaric tags for relative and absolute quantitation (iTRAQ) and stable isotope labeling by amino acids in cell culture (SILAC). iTRAQ is a mass spectrometry based quantitative proteomics method that determines the amount of proteins from different sources in a single experiment using stable isotope labeled molecules that can be covalent bonded to the N-terminus and side chain amines of proteins [6–8]. Candidate tissue, serum and drug resistance biomarkers have been discovered using this technique. SILAC is another mass spectrometry based quantitative proteomic technique that detects differences in protein abundance among samples using non-radioactive isotopic labeling [9–12]. It has emerged as a very powerful method to study cell signaling, post-translational modifications such as phosphorylation, protein–protein interaction and regulation of gene expression [13, 14]. SILAC has become particularly important in the field of secretomics.
In general, mass spectrometry-a central tool used for monitoring, identifying and characterizing proteins has become an invaluable technique of choice for both protein profiling as well as for positive protein identification. With an increasing ability to correctly characterize reduced quantities of samples and more complex mixtures of proteins and peptides, mass spectrometry is quickly becoming a key tool in the discovery of alterations to the proteome thereby facilitating biomarker identification. Additionally, the possibility of using mass spectrometry directly in clinical assays is also becoming a reality because the technique has a low detection limit and can perform high throughput analyses. Though scientists prefer the antibody based targeted approaches (ELISAs, TMAs) for confirmation and validation of markers, targeted quantitative approaches like MRM are becoming quite popular in the proteomics community. MRM is highly reproducible and is able to replace expensive antibody-based quantification like Western blotting and ELISA. Many independent investigations have used this approach for targeted quantification. One such example is quantification of cancer related proteins in body fluids using targeted proteomics [15]. Multiple reaction monitoring (MRM) using mass spectrometry is a highly sensitive and selective method for the targeted quantitation of protein/peptide abundances in complex biological samples [16]. Parallel reaction monitoring (PRM) detects all product ions and identifies peptides with high confidence. Thus, interference is reduced and specificity is increased in PRM. Qquantification in PRM is done by extracting one or more fragment ions that are selected post-acquisition. However, data visualization and analysis in PRM is similar to MRM [17]. Compared to MRM, PRM provides data with better mass accuracy and removes noise of interfering signals. In addition, PRM based targeted proteomics reduces assay development time compared to MRM because in PRM, fragment ions can be selected post-acquisition. The high scan speed facilitates development of sequential window acquisition of all theoretical mass spectra (SWATH). Peptides are quantitated by targeted extraction of SWATH-MS data. However, direct proteome profiling has remained a major technical challenge for body fluid proteomics particularly serum and plasma. Hence, it may be concluded that conventional tools for doing proteomics research such as 2DE to separate a large number of proteins, spot picking, in-gel trypsin digestion of excised gel spots and MALDI–TOF–MS based identification are gradually giving way to more advanced proteomics tools.
Recent developments in next generation proteomics tools are yielding newer information in modern biology. MALDI imaging mass spectrometry (MALDI–IMS) is providing major contributions to the understanding of disease progression, improving diagnostics, and drug delivery. MALDI–IMS is a mass spectrometry based imaging technique which allows direct label free measurement of biomolecules, metabolites and drugs at one time in tissue sections without destroying the sample [18, 19]. Proteogenomics refers to use of proteomic information, generated from mass spectrometry, to improve gene annotations [20–22]. Proteogenomic studies can also provide information about the presence of programmed frameshifts, N-terminal methionine excision, signal peptides, proteolysis and other post-translational modifications [21]. Proteogenomics is becoming increasingly relevant in clinical research and biomarker discovery. Protein microarray is a sensitive and high throughput gel-free protein analysis method that is rapidly becoming powerful for protein detection, protein expression profiling, diagnostics, biomarker discovery, protein–protein interactions etc. These arrays are solid-phase ligand-binding assay systems using immobilized proteins of interest on surfaces which include membranes, glass, microtiter wells, mass spectrometer plates, beads etc. [23].
Studies in clinical proteomics carried out so far in India
As in other parts of the world, proteomics has been implemented by Indian researchers to elucidate biological mechanisms as well as to discover biomarkers for various diseases, enabled by advancements both in instrumentation and software. Considerable efforts have been made to explore the mechanism of disease pathogenesis using animal models as well as cell biology and molecular biology techniques. The publications from these studies have emerged from major research institutes and University laboratories from across the country. Several research groups from India are actively pursuing cutting-edge research on proteomics of different infectious and immune disorders, reproductive disorders, cardiovascular diseases, diabetes, eye disorders, human cancers and hematological disorders.
Infectious disease and immune disorders
Proteomics is employed as a study tool in tuberculosis which affects a large population in the country and is of major concern. Research institutes like National JALMA Institute for Leprosy & Other Mycobacterial Diseases, Agra and National Institute for Research in Tuberculosis, Chennai are the frontrunners in this area. The underlying objective is to characterize the various strains, decoding drug resistance and identifying biomarker candidates along with new therapeutic targets [24, 25]. Proteomics of clinical isolates of malarial parasites is undertaken by institutes such as IISc, Bangalore; ICGEB, New Delhi and IIT Powai, Mumbai. These efforts are providing insights into its physiology whereas proteomic analysis of serum from infected individuals gives preliminary information concerning potential biomarkers [26–29]. Several studies have also been conducted on clinical isolates of wild type and drug resistant varieties of Leishmania across different institutes of the country like NCCS, Pune; JNU, New Delhi; CDRI, Lucknow; IOB, Bangalore; AIIMS, New Delhi; NIPER, Mohali; BHU, Varanasi and IICB, Kolkata [30–44]. Interesting findings have also been reported on other infectious diseases like hepatitis, food poisoning, leprosy etc. [45–50]. Substantial research activities on some other immune disorders have also been carried out in centres like Bose Institute, Kolkata; PGIMER, Chandigarh; IOB, Bangalore; CDRI, Lucknow; IGIB, New Delhi and AIIMS, New Delhi using high throughput proteomic techniques [51–60].
Reproductive biology and respiratory disorder
Proteomic analysis of germ cells, seminal plasma and mammary epithelial cells is carried out at NDRI, Karnal to know about fertility and lactation in dairy animals [61–63]. Proteomics studies on reproductive disorders such as endometriosis and polycystic ovarian syndrome have also been reported from institutes like IIT Kharagpur and NIRRH, Mumbai [64–66]. The effect of hypobaric hypoxia occurring at high altitude is also being analyzed by a research group at DIPAS, DRDO, New Delhi through proteomics methods [67].
Metabolic diseases
Proteomic technologies have also been used to understand cardiovascular diseases like Cardiac Hypertrophy, CAD, MI, and RHD. Several renowned institutes like PGIMER, Chandigarh; IGIB, New Delhi; IICT, Hyderabad; Thrombosis Research Institute, Bangalore and IICB, Kolkata are involved in such initiatives [68–75]. The difference between stenotic and non stenotic human plasma proteins has been investigated in the context of rheumatic mitral valve disease which is prevalent in developing countries including India. Proteins, which manifest both inflammatory and thrombotic components have been identified in Rheumatic Mitral Stenosis at IICB, Kolkata using a label free proteomic approach, [75]. Very recently, iTRAQ based proteomic studies from IGIB, New Delhi have shown that downregulation of apolipoproteins and albumin might be responsible for impaired reverse cholesterol transport in stable CAD [76]. Interesting findings in Type 2 diabetes have emerged from proteomic studies carried out at NCL, Pune in human plasma and drug induced animal models [77–85]. One of the novel initiatives undertaken by this group is to study advanced glycation end products (AGE) in a diabetic animal model using MALDI–TOF–MS based proteomics [82].
Eye disease
Proteomics research on various eye disorders like diabetic retinopathy, mycotic keratitis, dry eye syndrome and retinoblastoma tumors have been undertaken across institutes like SRM University, Kattankulathur; Aravind Medical Research Foundation, Aravind Eye Care System, Madurai and Sankara Nethralaya, Chennai [86–92].
Cancers and hematological disorders
The above studies help us to understand that proteomics is now universally applied to study various types of circulating body fluids and other biological samples. For instance, proteome profiles of serum/plasma, other biological fluids and biopsy specimens from different types of cancers are currently being evaluated across centres like IISc, Bangalore; CCMB, Hyderabad; IOB, Bangalore; ACTREC, Mumbai and IIT Powai, Mumbai with necessary infrastructural support, scientific knowledge and technical expertise from the institutes as required [93–109]. Similar studies have been replicated for different hematological disorders at SINP, Kolkata [110–113]. These examples give an idea of some of the areas where proteomic tools are being currently used.
Newer separation methods for pre-fractionation and histochemical quantification of stained tissue sections are gaining popularity by the day [114–123]. Exosomes, are also becoming an attractive starting material for the analysis of circulatory biomarkers. The impact of these studies on basic as well as biomedical research will only be felt in the years to come.
Different proteomics databases developed in India
With the expansion of protein sequence databases and search engines designed for direct input of mass spectrometry data, the high-resolution mass spectrometers with greater sensitivities can provide rapid and accurate identification of proteins for any number of species for which complete or partial genomes have been sequenced. This has lured scientists and data analysts to use proteomic methods in biomedical investigations and generate vast amounts of biological and molecular data that are invaluable to the global proteomics community. Such diverse datasets help to identify enzyme-substrate, signaling and coordination networks involved in various biological processes, which in turn, can be helpful in molecular diagnostics and disease therapeutics. Progress in the field of proteomics will benefit substantially from the development of informatics programs which enable faster processing of complex datasets. Thus, bioinformatics is one of the pillars of proteomics. Researchers at IOB (http://ibioinformatics.org), Bangalore have developed sophisticated bioinformatics tools like HPRD (www.hprd.org), Human Proteinpedia (www.humanproteinpedia.org/) and Human Proteome Map, a database on pancreatic cancers and several human signaling pathways [124–126]. It is through creation of the ‘Human Proteome Map’ that India received global attention recently. Using high-resolution mass spectrometry, independent drafts of the ‘Human Proteome Maps’ have been published by two investigator led groups which only highlights the biological significance of the data [126]. NetPath (http://www.netpath.org)—the flagship database of IOB, contains manually curated data for 36 human signaling pathways [127–134]. Signaling pathways such as Delta-Notch, EGFR1, Hedgehog, TNF-alpha have been curated at IOB, Bangalore. Particularly, the Wnt signaling pathway has been selected and organized for Cancer Cell Map (http://cancer.cellmap.org/cellmap/), which is a database of pathways focusing on human cancer. NetSlim is a slimmer collective version of NetPath (http://www.netpath.org/netslim/). It contains a graphical network of major signaling reactions [135]. Research teams at Lab SurgPath, Mumbai have contributed significantly to the Human Protein Atlas (http://www.proteinatlas.org/).
At the IMTECH, Chandigarh a curated database of proteins associated with cervix cancer—CCDB, a database of anticancer peptides and proteins called CancerPPD, and a database of hemolytic and non-hemolytic peptides—Hemolytik have been developed [136, 137]. NCBS has also come up with the DOQCS, which represents the collection of basic models of different signaling pathways [138–141]. Researchers at IISc, Bangalore have developed quite a few protein databases related to structure and function of protein kinases like ‘KinG’, NrichD, PALI, PRODOC and MulPSSM [142–144]. A number of protein databases such as PepBind, Immune Epitope Prediction Database & Tools, SEDB, Clostridium-DT(DB), and VPDB have originated from the Center of Bioinformatics in Pondicherry University [145–147]. Databases of protein features, resources for signaling and metabolic pathways intrinsic to specific biological processes, and repositories of disease-specific molecular level changes will help researchers use systems biology approaches to understand disease mechanisms in greater detail.
Conclusion
Clinical proteomics is yielding some fruitful results for Indian proteomics researchers utilizing state-of-the-art experimental and analytical methodologies. All these will benefit the proteomics community in the long run. However, these efforts are not enough for the benefit of the huge Indian patient population. Despite some success stories, India is still some distance away from successful translation of laboratory findings into clinical practice. India needs focused, sustained policies to promote translational research through specialized mega projects. Establishment of tissue repositories or registries is the need of the hour. The country’s overall healthcare infrastructure and medical informatics system also needs both public and private sector support on a long term basis so as to facilitate more effective translation studies. Tie-ups with commercial companies to facilitate technological developments will be necessary. At present, most of the proteomic investigations are arising from individual labs in research institutes of national repute. Almost all the studies are in the discovery phase. Collaboration between scientists and clinicians will facilitate the execution of well-designed multi-institutional and multi-centric studies aimed at addressing the relevant health problems unique to India, evaluating disease outcomes and their validity.
Abbreviations
- 2DE:
-
two–dimensional electrophoresis
- 2D DIGE:
-
two-dimensional difference gel electrophoresis
- ACTREC:
-
Advanced Centre for Treatment, Research and Education in Cancer
- AIIMS:
-
All India Institute of Medical Sciences
- BHU:
-
Banaras Hindu University
- CAD:
-
coronary artery disease
- CCMB:
-
Centre for Cellular and Molecular Biology
- CDRI:
-
Central Drug Research Institute
- DIPAS:
-
Defence Institute of Physiology & Allied Sciences
- DOQCS:
-
Database of Quantitative Cellular Signaling
- DRDO:
-
Defence Research and Development Organization
- EGFR1:
-
epidermal growth factor receptor 1
- ELISAs:
-
enzyme linked immunosorbent assays
- HPRD:
-
Human Protein Reference Database
- ICGEB:
-
International Centre for Genetic Engineering and Biotechnology
- IGIB:
-
Institute of Genomics and Integrative Biology
- IICB:
-
Indian Institute of Chemical Biology
- IICT:
-
Indian Institute of Chemical Technology
- IISc:
-
Indian Institute of Science
- IIT:
-
Indian Institute of Technology
- IMTECH:
-
Institute of Microbial TECHnology
- IOB:
-
Institute of Bioinformatics
- LC–MS:
-
liquid chromatography–mass spectrometry
- LC–MS/MS:
-
liquid chromatography–tandem mass spectrometry
- MALDI-TOF MS:
-
matrix assisted laser desorption ionization-time of flight mass spectrometry
- MI:
-
myocardial infarction
- MRM:
-
multiple reaction monitoring
- NCBS:
-
National Centre for Biological Sciences
- NCCS:
-
National Centre for Cell Science
- NCL:
-
National Chemical Laboratory
- NIPER:
-
National Institute of Pharmaceutical Education and Research
- NIRRH:
-
National Institute for Research in Reproductive Health
- PepBind:
-
Peptide Binding Protein Database
- PGIMER:
-
Postgraduate Institute of Medical Education and Research
- R&D:
-
Research and Development
- SEDB:
-
Structural Epitope Database
- SINP:
-
Saha Institute of Nuclear Physics
- TMAs:
-
tissue microarrays
- VPDB:
-
Viral Protein Structural Database
References
Reddy PJ, Atak A, Ghantasala S, Kumar S, Gupta S, Prasad TS, et al. Proteomics research in India: an update. J Proteom. 2015;127:7–17.
Sirdeshmukh R. Indian proteomics efforts and human proteome project. J Proteom. 2015;127:147–51.
Ray S, Srivastava S. Trends and roadblocks in proteomics research in India. Nat India. 2015. doi:10.1038/nindia.2015.111.
Zingde SM. Has proteomics come of age in India? J Proteom. 2015;127:3–6.
Granger CB, Van Eyk JE, Mockrin SC, Anderson NL. NHLCI clinical proteomics working group report. Circulation. 2004;109:1697–703.
Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteom. 2004;3:1154–69.
Zieske LR. A perspective on the use of iTRAQ reagent technology for protein complex and profiling studies. J Exp Bot. 2006;57:1501–8.
Gafken PR, Lampe PD. Methodologies for characterizing phosphoproteins by mass spectrometry. Cell Commun Adhes. 2006;13:249–62.
Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate quantitation of protein expression and site-specific phosphorylation. Proc Natl Acad Sci USA. 1999;96:6591–6.
Jiang H, English AM. Quantitative analysis of the yeast proteome by incorporation of isotopically labeled leucine. J Proteome Res. 2002;1:345–50.
Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteom. 2002;1:376–86.
Zhu H, Pan S, Gu S, Bradbury EM, Chen X. Amino acid residue specific stable isotope labeling for quantitative proteomics. Rapid Commun Mass Spectrom. 2002;16:2115–23.
Ibarrola N, Molina H, Iwahori A, Pandey A. A novel proteomic approach for specific identification of tyrosine kinase substrates using 13C tyrosine. J Biol Chem. 2004;279:15805–13.
Ibarrola N, Kalume DE, Gronborg M, Iwahori A, Pandey A. A proteomic approach for quantitation of phosphorylation using stable isotope labeling in cell culture. Anal Chem. 2003;75:6043–9.
Huttenhain R, Soste M, Selevsek N, Röst H, Sethi A, Carapito C, et al. Reproducible quantification of cancer-associated proteins in body fluids using targeted proteomics. Sci Transl Med. 2014;4:142ra94.
Kondrat RW, McClusky GA, Cooks RG. Multiple reaction monitoring in mass spectrometry/mass spectrometry for direct analysis of complex mixtures. Anal Chem. 1978;1978(50):2017–21.
Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics. 2012;11:1475–88.
McDonnell LA, Heeren RM. Imaging mass spectrometry. Mass Spectrom Rev. 2007;26:606–43.
Walch A, Rauser S, Deininger SO, Hofler H. MALDI imaging mass spectrometry for direct tissue analysis: a new frontier for molecular histology. Histochem Cell Biol. 2008;130:421–34.
Faulkner S, Dun MD, Hondermarck H. Proteogenomics: emergence and promise. Cell Mol Life Sci. 2015;72:953–7.
Gupta N, Tanner S, Jaitly N, Adkins JN, Lipton M, Edwards R, et al. Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation. Genome Res. 2007;17:1362–77.
Ansong C, Purvine SO, Adkins JN, Lipton MS, Smith RD. Proteogenomics: needs and roles to be filled by proteomics in genome annotation. Brief Funct Genom Proteom. 2008;7:50–62.
Chandra H, Reddy PJ, Srivastava S. Protein microarrays and novel detection platforms. Expert Rev Proteom. 2011;8:61–79.
Kumar B, Sharma D, Sharma P, Katoch VM, Venkatesan K, Bisht D. Proteomic analysis of Mycobacterium tuberculosis isolates resistant to kanamycin and amikacin. J Proteome. 2013;94:68–77.
Anbarasu D, Raja CP, Raja A. Multiplex analysis of cytokines/chemokines as biomarkers that differentiate healthy contacts from tuberculosis patients in high endemic settings. Cytokine. 2013;61:747–54.
Acharya P, Pallavi R, Chandran S, Dandavate V, Sayeed SK, Rochani A, et al. Clinical proteomics of the neglected human malarial parasite Plasmodium vivax. PLoS ONE. 2011;6:e26623.
Ranjan R, Chugh M, Kumar S, Singh S, Kanodia S, Hossain MJ, et al. Proteome analysis reveals a large merozoite surface protein-1 associated complex on the Plasmodium falciparum merozoite surface. J Proteome Res. 2011;10:680–91.
Ray S, Renu D, Srivastava R, Gollapalli K, Taur S, Jhaveri T, et al. Proteomic investigation of falciparum and vivax malaria for identification of surrogate protein markers. PLoS ONE. 2012;7:e41751.
Singh M, Mukherjee P, Narayanasamy K, Arora R, Sen SD, Gupta S, et al. Proteome analysis of Plasmodium falciparum extracellular secretory antigens at asexual blood stages reveals a cohort of proteins with possible roles in immune modulation and signaling. Mol Cell Proteom. 2009;9:2102–18.
Jamdhade MD, Pawar H, Chavan S, Sathe G, Umasankar PK, Mahale KN, et al. Comprehensive proteomics analysis of glycosomes from Leishmania donovani. OMICS. 2015;19:157–70.
Singh AK, Pandey RK, Siqueira-Neto JL, Kwon YJ, Freitas-Junior LH, Shaha C, et al. Proteomic-based approach to gain insight into reprogramming of THP-1 cells exposed to Leishmania donovani over an early temporal window. Infect Immun. 2015;83:1853–68.
Singh AK, Roberts S, Ullman B, Madhubala R. A quantitative proteomic screen to identify potential drug resistance mechanism in α-difluoromethylornithine (DFMO) resistant Leishmania donovani. J Proteom. 2014;102:44–59.
Chawla B, Jhingran A, Panigrahi A, Stuart KD, Madhubala R. Paromomycin affects translation and vesicle-mediated trafficking as revealed by proteomics of paromomycin-susceptible-resistant Leishmania donovani. PLoS ONE. 2011;6:e26660.
Kumar A, Misra P, Sisodia B, Shasany AK, Sundar S, Dube A. Proteomic analyses of membrane enriched proteins of Leishmania donovani Indian clinical isolate by mass spectrometry. Parasitol Int. 2015;64:36–42.
Kumar A, Misra P, Sisodia B, Shasany AK, Sundar S, Dube A. Mass spectrometry-based proteomic analysis of Leishmania donovani soluble proteins in Indian clinical isolate. Pathog Dis. 2014;70:84–7.
Kumari S, Kumar A, Samant M, Sundar S, Singh N, Dube A. Proteomic approaches for discovery of new targets for vaccine and therapeutics against visceral leishmaniasis. Proteom Clin Appl. 2008;2:372–86.
Gupta SK, Sisodia BS, Sinha S, Hajela K, Naik S, Shasany AK, et al. Proteomic approach for identification and characterization of novel immunostimulatory proteins from soluble antigens of Leishmania donovani promastigotes. Proteomics. 2007;7:816–23.
Pawar H, Renuse S, Khobragade SN, Chavan S, Sathe G, Kumar P, et al. Neglected tropical diseases and omics science: proteogenomics analysis of the promastigote stage of Leishmania major parasite. OMICS. 2014;18:499–512.
Nirujogi RS, Pawar H, Renuse S, Kumar P, Chavan S, Sathe G, et al. Moving from unsequenced to sequenced genome: reanalysis of the proteome of Leishmania donovani. J Proteom. 2014;97:48–61.
Pawar H, Sahasrabuddhe NA, Renuse S, Keerthikumar S, Sharma J, Kumar GS, et al. A proteogenomic approach to map the proteome of an unsequenced pathogen—Leishmania donovani. Proteomics. 2012;12:832–44.
Rukmangadachar LA, Kataria J, Hariprasad G, Samantaray JC, Srinivasan A. Two-dimensional difference gel electrophoresis (DIGE) analysis of sera from visceral leishmaniasis patients. Clin Proteom. 2011;8:4.
Sharma S, Singh G, Chavan HD, Dey CS. Proteomic analysis of wild type and arsenite-resistant Leishmania donovani. Exp Parasitol. 2009;123:369–76.
Kumar S, Kumar D, Chakravarty J, Sundar S. Identification and characterization of a novel, 37-kilodalton Leishmania donovani antigen for diagnosis of Indian visceral leishmaniasis. Clin Vaccine Immunol. 2011;18:772–5.
Bag AK, Saha S, Sundar S, Saha B, Chakrabarti A, Mandal C. Comparative proteomics and glycoproteomics of plasma proteins in Indian visceral leishmaniasis. Proteome Sci. 2014;12:48.
Gouthamchandra K, Kumar A, Shwetha S, Mukherjee A, Chandra M, Ravishankar B, et al. Serum proteomics of hepatitis C virus infection reveals retinol-binding protein 4 as a novel regulator. J Gen Virol. 2014;95:1654–67.
Sengupta N, Alam SI. In vivo studies of Clostridium perfringens in mouse gas gangrene model. Curr Microbiol. 2011;62:999–1008.
Alam SI, Bansod S, Kumar RB, Sengupta N, Singh L. Differential proteomic analysis of Clostridium perfringens ATCC13124; identification of dominant, surface and structure associated proteins. BMC Microbiol. 2009;9:162.
Gupta N, Shankernarayan NP, Dharmalingam K. Serum proteome of leprosy patients undergoing erythema nodosum leprosum reaction: regulation of expression of the isoforms of haptoglobin. J Proteome Res. 2007;6:3669–79.
Selvan LD, Sreenivasamurthy SK, Kumar S, Yelamanchi SD, Madugundu AK, Anil AK, et al. Characterization of host response to Cryptococcus neoformans through quantitative proteomic analysis of cryptococcal meningitis co-infected with HIV. Mol BioSyst. 2015;11:2529–40.
Kumar A, Singh S, Ahirwar SK, Nath G. Proteomics-based identification of plasma proteins and their association with the host–pathogen interaction in chronic typhoid carriers. Int J Infect Dis. 2014;19:59–66.
Ghosh N, Sircar G, Saha B, Pandey N, Gupta-Bhattacharya S. Search for allergens from the pollen proteome of sunflower (Helianthus annuus L.): a major sensitizer for respiratory allergy patients. PLoS ONE. 2015;10:e0138992.
Sircar G, Chakrabarti HS, Saha B, Gupta-Bhattacharya S. Identification of aero-allergens from Rhizopus oryzae: an immunoproteomic approach. J Proteom. 2012;77:455–68.
Rani L, Minz RW, Arora A, Kannan M, Sharma A, Anand S, et al. Serum proteomic profiling in granumomatosis with polyangiitis using two-dimensional gel electrophoresis along with matrix assisted laser desorption ionization time of flight mass spectrometry. Int J Rheum Dis. 2014;17:910–9.
Pinto SM, Nirujogi RS, Rojas PL, Patil AH, Manda SS, Subbannayya Y, et al. Quantitative phosphoproteomic analysis of IL-33-mediated signaling. Proteomics. 2015;15:532–44.
Saxena R, Gupta S, Singh K, Mitra K, Tripathi AK, Tripathi RK. Proteomic profiling of SupT1 cells reveal modulation of host proteins by HIV-1 Nef variants. PLoS ONE. 2015;10:e0122994.
Mahajan L, Gautam P, Dodagatta-Marri E, Madan T, Kishore U. Surfactant protein SP-D modulates activity of immune cells: proteomic profiling of its interaction with eosinophilic cells. Expert Rev Proteom. 2014;11:355–69.
Kumar S, Verma AK, Sharma A, Kumar D, Tripathi A, Chaudhari BP, et al. Phytohemagglutinins augment red kidney bean (Phaseolus vulgaris L.) induced allergic manifestations. J Proteom. 2013;93:50–64.
Biswas S, Sharma S, Saroha A, Bhakuni DS, Malhotra R, Zahur M, et al. Identification of novel autoantigen in the synovial fluid of rheumatoid arthritis patients using an immunoproteomics approach. PLoS ONE. 2013;8:e56246.
Gautam P, Sundaram CS, Madan T, Gade WN, Shah A, Sirdeshmukh R, et al. Identification of novel allergens of Aspergillus fumigatus using immunoproteomics approach. Clin Exp Allergy. 2007;37:1239–49.
Tiwari V, Vashistt J, Kapil A, Moganty RR. Comparative proteomics of inner membrane fraction from carbapenem-resistant Acinetobacter baumannii with a reference strain. PLoS ONE. 2012;7:e39451.
Tripathi UK, Aslam MKM, Pandey S, Nayak S, Chhillar S, Srinivasan A, et al. Differential proteomic profile of spermatogenic and sertoli cells from peri-pubertal testes of three different bovine breeds. Front Cell Dev Biol. 2014;2:24.
Aslam MKM, Kumaresan A, Sharma VK, Tajmul M, Chhillar S, Chakravarty AK, et al. Identification of putative fertility markers in seminal plasma of crossbred bulls through differential proteomics. Theriogenology. 2014;82:1254–62.
Janjanam J, Jamwal M, Singh S, Kumar S, Panigrahi AK, Hariprasad G, et al. Proteome analysis of functionally differentiated bovine (Bos indicus) mammary epithelial cells isolated from milk. Proteomics. 2013;13:3189–204.
Dutta M, Subramani E, Taunk K, Gajbhiye A, Seal S, Pendharkar N, et al. Investigation of serum proteome alterations in human endometriosis. J Proteom. 2015;114:182–96.
Bhagwat SR, Redij T, Phalnikar K, Nayak S, Iyer S, Gadkar S, et al. Cell surfactomes of two endometrial epithelial cell lines that differ in their adhesiveness to embryonic cells. Mol Reprod Dev. 2014;81:326–40.
Ambekar AS, Kelkar DS, Pinto SM, Sharma R, Hinduja I, Zaveri K, et al. Proteomics of follicular fluid from women with polycystic ovary syndrome suggests molecular defects in follicular development. J Clin Endocrinol Metab. 2015;100:744–53.
Ahmad Y, Sharma NK, Ahmad MF, Sharma M, Garg I, Bhargava K. Proteomic identification of novel differentiation plasma protein markers in hypobaric hypoxia-induced rat model. PLoS ONE. 2014;9:e98027.
Kaul D, Kaur R, Baba I, Singh D. Functional proteomics of receptor-Ck in the developmental stages of human atherosclerotic arterial wall. Indian Heart J. 2003;55:252–5.
Mitra A, Basak T, Ahmad S, Datta K, Datta R, Sengupta S, et al. Comparative proteome profiling during cardiac hypertrophy and myocardial infarction reveals altered glucose oxidation by differential activation of pyruvate dehydrogenase E1 component subunit β. J Mol Biol. 2015;427:2104–20.
Basak T, Varshney S, Hamid Z, Ghosh S, Seth S, Sengupta S. Identification of metabolic markers in coronary artery disease using an untargeted LC–MS based metabolomic approach. J Proteomics. 2015;127:169–77.
Chowdhury D, Tangutur AD, Khatua TN, Saxena P, Banerjee SK, Bhadra MP. A proteomic view of isoproterenol induced cardiac hypertrophy: prohibitin identified as a potential biomarker in rats. J Transl Med. 2013;11:130.
Nair J, Ghatge M, Kakkar VV, Shanker J. Network analysis of inflammatory genes and their transcriptional regulators in coronary artery disease. PLoS ONE. 2014;9:e94328.
Vangala RK, Ravindran V, Ghatge M, Shanker J, Arvind P, Bindu H, et al. Integrative bioinformatics analysis of genomic and proteomic approaches to understand the transcriptional regulatory program in coronary artery disease pathways. PLoS ONE. 2013;8:e57193.
Poduri A, Bahl A, Talwar KK, Khullar M. Proteomic analysis of circulating human monocytes in coronary artery disease. Mol Cell Biochem. 2012;360:181–8.
Mukherjee S, Jagadeeshaprasad MG, Banerjee T, Ghosh SK, Biswas M, Dutta S, et al. Proteomic analysis of human plasma in chronic rheumatic mitral stenosis reveals proteins involved in the complement and coagulation cascade. Clin Proteom. 2014;11:35.
Basak T, Tanwar VS, Bhardwaj G, Bhardwaj N, Ahmad S, Garg G, et al. Plasma proteomic analysis of stable coronary artery disease indicates impairment of reverse cholesterol pathway. Sci Rep. 2016;6:28042.
Chougale AD, Bhat SP, Bhujbal SV, Zambare MR, Puntambekar S, Somani RS, et al. Proteomic analysis of glycated proteins from streptozotocin-induced diabetic rat kidney. Mol Biotechnol. 2012;50:28–38.
Bhonsle HS, Korwar AM, Kote SS, Golegaonkar SB, Chougale AD, Shaik ML, et al. Low plasma albumin levels are associated with increased plasma protein glycation and HbA1c in diabetes. J Proteome Res. 2012;11:1391–6.
Bhonsle HS, Korwar AM, Kesavan SK, Bhosale SD, Bansode SB, Kulkarni MJ. “Zoom-ln”—a targeted database search for identification of glycation modifications analyzed by untargeted tandem mass spectrometry. Eur J Mass Spectrom. 2012;18:475–81.
Bansode SB, Chougale AD, Joshi RS, Giri AP, Bodhankar SL, Harsulkar AM, et al. Proteomic analysis of protease resistant proteins in the diabetic rat kidney. Mol Cell Proteom. 2013;12:228–36.
Bhonsle HS, Korwar AM, Chougale AD, Kote SS, Dhande NL, Shelgikar KM, et al. Proteomic study reveals downregulation of apolipoprotein A1 in plasma of poorly controlled diabetes: a pilot study. Mol Med Rep. 2013;7:495–8.
Kesavan SK, Bhat S, Golegaonkar SB, Jagadeeshaprasad MG, Deshmukh AB, Patil HS, et al. Proteome wide reduction in AGE modification in streptozotocin induced diabetic mice by hydralazine mediated transglycation. Sci Rep. 2013;3:2941.
Singh P, Jayaramaiah RH, Agawane SB, Vannuruswamy G, Korwar AM, Anand A, et al. Potential dual role of eugenol in inhibiting advanced glycation end products in diabetes: proteomic and mechanistic insights. Sci Rep. 2016;6:18798.
Jagadeeshaprasad MG, Batkulwar KB, Meshram NN, Tiwari S, Korwar AM, Unnikrishnan AG, et al. Targeted quantification of N-1-(carboxymethyl) valine and N-1-(carboxyethyl) valine peptides of β-hemoglobin for better diagnostics in diabetes. Clin Proteom. 2016;13:7.
Bhat S, Jagadeeshaprasad MG, Patil YR, Shaikh ML, Regin BS, Mohan V, et al. Proteomic insight reveals elevated levels of albumin in circulating immune complexes in diabetic plasma. Mol Cell Proteom. 2016;15:2011–20.
Gopalakrishnan V, Purushothaman P, Bhaskar A. Proteomic analysis of plasma proteins in diabetic retinopathy patients by two dimensional electrophoresis and MALDI–TOF–MS. J Diabetes Complic. 2015;29:928–36.
Ananthi S, VenkateshPrajna N, Lalitha P, Valarnila M, Dharmalingam K. Pathogen induced changes in the protein profile of human tears from Fusarium keratitis patients. PLoS One. 2013;8:e53018.
Aluru SV, Agarwal S, Srinivasan B, Iyer GK, Rajappa SM, Tatu U, et al. Lacrimal proline rich 4 (LPRR4) protein in the tear fluid is a potential biomarker of dry eye syndrome. PLoS ONE. 2012;7:e51979.
Mallikarjuna K, Sundaram CS, Sharma Y, Deepa PR, Khetan V, Gopal L, et al. Comparative proteomic analysis of differentially expressed proteins in primary retinoblastoma tumors. Proteom Clin Appl. 2010;4:449–63.
Saijyothi AV, Angayarkanni N, Syama C, Utpal T, Shweta A, Bhaskar S, et al. Two dimensional electrophoretic analysis of human tears: collection method in dry eye syndrome. Electrophoresis. 2010;31:3420–7.
Ananthi S, Chitra T, Bini R, Prajna NV, Lalitha P, Dharmalingam K. Comparative analysis of the tear protein profile in mycotic keratitis patients. Mol Vis. 2008;14:500–7.
Selvam RM, Nithya R, Devi PN, Shree RSB, Nila MV, Demonte NL, et al. Exoproteome of Aspergillus flavus corneal isolates and saprophytes: identification of proteoforms of an oversecreted alkaline protease. J Proteomics. 2014;115C:23–35.
Kumar DM, Patil V, Ramachandran B, Nila MV, Dharmalingam K, Somasundaram K. Temozolomide modulated glioma proteome: role of interleukin-1receptor-associated kinase-4 (IRAK4) in chemosensitivity. Proteomics. 2013;13:2113–24.
Chumbalkar VC, Subhashini C, Dhople VM, Sundaram CS, Jagannadham MV, Kumar KN, et al. Differential protein expression in human gliomas and molecular insights. Proteomics. 2005;5:1167–77.
Polisetty RV, Gupta MK, Nair SC, Ramamoorthy K, Tiwary S, Shiras A, et al. Glioblastoma cell secretome: analysis of three glioblastoma cell lines reveal 148 non-redundant proteins. J Proteom. 2011;74:1918–25.
Polisetty RV, Gautam P, Sharma R, Harsha HC, Nair SC, Gupta MK, et al. LC-MS/MS analysis of differentially expressed glioblastoma membrane proteome reveals altered calcium signaling and other protein groups of regulatory functions. Mol Cell Proteom. 2012;11(M111):013565.
Gautam P, Nair SC, Gupta MK, Sharma R, Polisetty RV, Uppin MS, et al. Proteins with altered levels in plasma from glioblastoma patients as revealed by iTRAQ-based quantitative proteomic analysis. PLoS ONE. 2012;7:e46153.
Gupta MK, Polisetty RV, Ramamoorthy K, Tiwary S, Kaur N, Uppin MS, et al. Secretome analysis of glioblastoma cell line–HNGC-2. Mol BioSyst. 2013;9:1390–400.
Fulzele A, Malgundkar SA, Govekar RB, Patil A, Kane SV, Chaturvedi P, et al. Proteomic profile of keratins in cancer of the gingivo buccal complex: consolidating insights for clinical applications. J Proteomics. 2013;91:242–58.
Govekar RB, D’Cruz AK, Alok Pathak K, Agarwal J, Dinshaw KA, Chinoy RF, et al. Proteomic profiling of cancer of the gingivo-buccal complex: identification of new differentially expressed markers. Proteom Clin Appl. 2009;3:1451–62.
Shukla S, Govekar RB, Sirdeshmukh R, Sundaram CS, D’Cruz AK, Pathak KA, et al. Tumor antigens eliciting autoantibody response in cancer of gingivo-buccal complex. Proteom Clin Appl. 2007;1:1592–604.
Kashyap MK, Harsha HC, Renuse S, Pawar H, Sahasrabuddhe NA, Kim MS, et al. SILAC-based quantitative proteomic approach to identify potential biomarkers from the esophageal squamous cell carcinoma secretome. Cancer Biol Ther. 2010;10:796–810.
Pawar H, Kashyap MK, Sahasrabuddhe NA, Renuse S, Harsha HC, Kumar P, et al. Quantitative tissue proteomics of esophageal squamous cell carcinoma for novel biomarker discovery. J Proteom. 2011;74:1918–25.
Marimuthu A, Chavan S, Sathe G, Sahasrabuddhe NA, Srikanth SM, Renuse S, et al. Identification of head and neck squamous cell carcinoma biomarker candidates through proteomic analysis of cancer cell secretome. Biochim Biophys Acta. 2013;1834:2308–16.
Sahasrabuddhe NA, Barbhuiya MA, Bhunia S, Subbannayya T, Gowda H, Advani J, et al. Identification of prosaposin and transgelin as potential biomarkers for gallbladder cancer using quantitative proteomics. Biochem Biophys Res Commun. 2014;446:863–9.
Syed P, Gupta S, Choudhary S, Pandala NG, Atak A, Richharia A, et al. Autoantibody profiling of glioma serum samples to identify biomarkers using human proteome arrays. Sci Rep. 2015;5:13895.
Sharma S, Ray S, Moiyadi A, Sridhar E, Srivastava S. Quantitative proteomic analysis of meningiomas for the identification of surrogate protein markers. Sci Rep. 2014;4:7140.
Bansal N, Gupta A, Sankhwar SN, Mahdi AA. Low- and high-grade bladder cancer appraisal via serum-based proteomics approach. Clin Chim Acta. 2014;436:97–103.
Hariprasad G, Hariprasad R, Kumar L, Srinivasan A, Kola S, Kaushik A. Apolipoprotein A1 as a potential biomarker in the ascitic fluid for the differentiation of advanced ovarian cancers. Biomarkers. 2013;18:532–41.
Chakrabarti A, Halder S, Karmakar S. Erythrocyte & platelet proteomics in haematological disorders. Proteom Clin Appl. 2016;10:403–14.
Karmakar S, Banerjee D, Chakrabarti A. Platelet proteomics in thalassemia: factors responsible for hypercoagulation. Proteom Clin Appl. 2016;10:239–47.
Karmakar S, Saha S, Banerjee D, Chakrabarti A. Differential proteomics study of platelets in asymptomatic constitutional macrothrombocytopenia: altered levels of cytoskeletal proteins. Eur J Haematol. 2015;94:43–50.
Basu A, Saha S, Karmakar S, Chakravarty S, Banerjee D, Dash BP. Chakrabarti A.2D DIGE based proteomics study of erythrocyte cytosol in sickle cell disease: altered proteostasis and oxidative stress. Proteomics. 2013;13:3233–42.
Gadewal NS, Zingde SM. Database and interaction network of genes involved in oral cancer: version II. Bioinformation. 2011;6:169–70.
Akhoon BA, Singh KP, Varshney M, Gupta SK, Shukla Y, Gupta SK. Understanding the mechanism of atovaquone drug resistance in Plasmodium falciparum cytochrome bmutationY268S using computational methods. PLoS ONE. 2014;9:e110041.
Kumar D, Mondal AK, Yadav AK, Dash D. Discovery of rare protein-coding genes in model methylotroph Methylobacterium extorquens AM1. Proteomics. 2014;14:2790–4.
Sinha A, Nagarajaram HA. Nodes occupying central positions in human tissue specific PPI networks are enriched with many splice variants. Proteomics. 2014;14:2242–8.
Prasanna RR, Sidhik S, Kamalanathan AS, Bhagavatula K, Vijayalakshmi MA. Affinity selection of histidine-containing peptides using metal chelate methacrylate monolithic disk for targeted LC–MS/MS approach in high-throughput proteomics. J Chromatogr B Anal Technol Biomed Life Sci. 2014;955–956:42–9.
Alam M, Ghosh W. Optimization of a phenol extraction-based protein preparation method amenable to downstream 2DE and MALDI–MS based analysis of bacterial proteomes. Proteomics. 2014;14:216–21.
Yadav AK, Bhardwaj G, Basak T, Kumar D, Ahmad S, Priyadarshini R, et al. A systematic analysis of eluted fraction of plasma post immunoaffinity depletion: implications in biomarker discovery. PLoS ONE. 2011;6:e24442.
Mandlik V, Shinde S, Singh S. Molecular evolution of the enzymes involved in the sphingolipid metabolism of Leishmania: selection pressure in relation to functional divergence and conservation. BMC Evol Biol. 2014;14:142.
Varghese F, Bukhari AB, Malhotra R, De A. IHC profiler: an open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS ONE. 2014;9:e96801.
Swamidoss IN, Kårsnäs A, Uhlmann V, Ponnusamy P, Kampf C, Simonsson M, et al. Automated classification of immunostaining patterns in breast tissue from the human protein atlas. J Pathol Inform. 2013;4:S14.
Mathivanan S, Ahmed M, Ahn NG, Alexandre H, Amanchy R, Andrews PC, et al. Human proteinpedia enables sharing of human protein data. Nat Biotechnol. 2008;26:164–7.
Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, et al. A draft map of the human proteome. Nature. 2014;509:575–81.
Wilhelm M, Schlegl J, Hahne H, MoghaddasGholami A, Lieberenz M, Savitski MM, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–7.
Radhakrishnan A, Raju R, Tuladhar N, Subbannayya T, Thomas JK, et al. A pathway map of prolactin signaling. J Cell Commun Signal. 2012;6:169–73.
Subbannayya Y, Anuja K, Advani J, Ojha UK, Nanjappa V, George B, et al. A network map of the gastrin signaling pathway. J Cell Commun Signal. 2014;8:165–70.
Subbannayya T, Balakrishnan L, Sudarshan G, Advani J, Kumar S, Mahmood R, et al. An integrated map of corticotropin-releasing hormone signaling pathway. J Cell Commun Signal. 2013;7:295–300.
Raju R, Palapetta SM, Sandhya VK, Sahu A, Alipoor A, Balakrishnan L, et al. A network map of FGF-1/FGFR signaling system. J Signal Transduct. 2014;2014:962962.
Sandhya VK, Raju R, Verma R, Advani J, Sharma R, Radhakrishnan A, et al. A network map of BDNF/TRKB and BDNF/p75NTR signaling system. J Cell Commun Signal. 2013;7:301–7.
Nanjappa V, Raju R, Muthusamy B, Sharma J, Thomas JK, Nidhina PAH, et al. A comprehensive curated reaction map of leptin signaling pathway. J Proteom Bioinform. 2011;4:184–9.
Dey G, Radhakrishnan A, Syed N, Thomas JK, Nadig A, Srikumar K, et al. Signaling network of oncostatin M pathway. J Cell Commun Signal. 2013;7:103–8.
Raju R, Balakrishnan L, Nanjappa V, Bhattacharjee M, Getnet D, Muthusamy B, et al. A comprehensive manually curated reaction map of RANKL/RANK signaling pathway. Database (Oxford). 2011. doi:10.1093/database/bar021.
Raju R, Nanjappa V, Balakrishnan L, Radhakrishnan A, Thomas JK, Sharma J, et al. NetSlim: high-confidence curated signaling maps. Database (Oxford). 2011. doi:10.1093/database/bar032.
Agarwal SM, Raghav D, Singh H, Raghava GP. CCDB: a curated database of genes involved in cervix cancer. Nucleic Acids Res. 2011;39:D975–9.
Tyagi A, Tuknait A, Anand P, Gupta S, Sharma M, Mathur D, et al. CancerPPD: a database of anticancer peptides and proteins. Nucleic Acids Res. 2015;43:D837–43.
Bhalla US, Iyengar R. Robustness of the bistable behavior of a biological signaling feedback loop. Chaos. 2001;11:221–6.
Bhalla US, Iyengar R. Functional modules in biological signalling networks. Novartis Found Symp. 2001;239:4–13.
Bhalla US, Iyengar R. Emergent properties of networks of biological signaling pathways. Science. 1999;283:381–7.
Sivakumaran S, Hariharaputran S, Mishra J, Bhalla US. The database of quantitative cellular signaling: management and analysis of chemical kinetic models of signaling networks. Bioinformatics. 2003;19:408–15.
Krupa A, Abhinandan KR, Srinivasan N. KinG: a database of protein kinases in genomes. Nucleic Acids Res. 2004;32:D153–5.
Mudgal R, Sandhya S, Kumar G, Sowdhamini R, Chandra NR, Srinivasan N. NrichD database: sequence databases enriched with computationally designed protein-like sequences aid in remote homology detection. Nucleic Acids Res. 2015;43:D300–5.
Gowri VS, Krishnadev O, Swamy CS, Srinivasan N. MulPSSM: a database of multiple position-specific scoring matrices of protein domain families. Nucleic Acids Res. 2006;34:D243–6.
Sharma OP, Das AA, Krishna R, Suresh Kumar M, Mathur PP. Structural Epitope Database (SEDB): a web-based database for the epitope, and its intermolecular interaction along with the tertiary structure information. J Proteom Bioinform. 2012;5:084–9.
Jadhav A, Ezhilarasan V, Prakash Sharma O, Pan A. Clostridium-DT(DB): a comprehensive database for potential drug targets of Clostridium difficile. Comput Biol Med. 2013;43:362–7.
Sharma OP, Jadhav A, Hussain A, Kumar MSVPDB. Viral Protein Structural Database. Bioinformation. 2011;6:324–6.
Authors’ contributions
SM conducted experiments, analysed data and drafted the manuscript. AB conceived the study, participated in the design and coordination and helped drafting the manuscript. Both authors read and approved the final manuscript.
Authors’ information
Arun Bandyopadhyay is working as a Senior Principal Scientist in CSIR-Indian Institute of Chemical Biology, Kolkata. His team is involved in clinical proteomics research for understanding pathophysiology of coronary artery disease and rheumatic heart disease. His group has made valuable contribution on rheumatic heart disease which is a major health concern in India and is prevalent among people of poor socioeconomic strata. Using human sample this group has been successful in finding a circulatory marker which might be useful to predict the severity of rheumatic valve disorders.
Acknowledgements
None.
Competing interests
The authors declare that they have no competing interests.
Funding
This work is supported by the grants from CSIR new Delhi (MLP113 and BSC 0118).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Mukherjee, S., Bandyopadhyay, A. Proteomics in India: the clinical aspect. Clin Proteom 13, 21 (2016). https://doi.org/10.1186/s12014-016-9122-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12014-016-9122-0