Abstract
Background
Porcine circovirus type 2 (PCV2) and porcine reproductive and respiratory syndrome virus (PRRSV) have been present for the last 2 decades in Croatia, causing large economical losses in the pig production. The clinical features of the infections are mostly manifested by the development of respiratory problems, weight loss and poor growth performance, as well as reproductive failure in pregnant sows. Even though the infections are continuously recognized in some regions in Croatia, the heterogeneity of the detected viral strains from 2012 has not yet been investigated. The objective of this study was to compare virus strains of PCV2 and PRRSV detected until 2008 in Croatia with strains isolated in 2012 to gain a better epidemiological understanding of these two infections.
Results
PCV2 and PRRSV strains detected in 2012 in fattening pigs from regions where these two diseases have been previously described were compared to strains that have been detected in the same regions within the past two decades. The phylogenetic analysis revealed that the circulating PCV2 and PRRSV strains are distantly related to the previously described Croatian viral strains. However, when compared to known isolates from the GenBank a high genetic identity of PRRSV isolates with isolates from Hungary, Denmark and the Netherlands was found.
Conclusion
The results of this study reveal that even though PCV2 and PRRSV are constantly present in the investigated regions in Croatia, the viral strains found in 2012 genetically differ from those detected in earlier years. This indicates that new entries into the pig population appeared with regard to both infections, probably as a result of pig trade.
Similar content being viewed by others
Background
Porcine circovirus (PCV) is a small, non-enveloped virus with a single-stranded circular DNA of 1.76 kb [1, 2] belonging to the genus Circovirus within the family Circoviridae, along with other animal viruses such as chicken anaemia virus and beak and feather disease virus [3]. PCV genome contains three open reading frames (ORF), ORF1 encoding a replicase, ORF2 encoding the capsid protein and ORF3 encoding a protein related to cell apoptosis [4]. Two types of PCV have been described, namely PCV1 and PCV2. Their DNA sequence homology is 68 to 76% [5]. While PCV1 is not pathogenic, PCV2 has been recognised as the causative agent of several pathological syndromes in swine causing great economical losses in the pig industry worldwide. PCV2 isolates are genetically divided into three groups, PCV2a, PCV2b and PCV2c, isolates of most of which are distributed among groups PCV2b and PCV2a, respectively [6, 7]. Boisseson et al. [8] found that the variations among the PCV2 genomic sequences are mainly due to the variability within ORF2 (91.2-100% nucleotide identity), while the ORF1 is highly conserved (97.8-100% nucleotide identity).
Porcine reproductive and respiratory syndrome virus (PRRSV) is a small enveloped positive-stranded RNA virus that belongs to the Arteriviridae family, genus Arterivirus[9]. Its genome is composed of approximately 15 kb [9] containing 9 ORF’s [10, 11]. According to the genome characteristics, PRRSV is divided into two genotypes, the European, or type 1 virus, also reffered to as the Lelystad virus (LV), and the American, or type 2 virus. They share a sequence identity of 50-70% [12, 13]. Since PRRSV has the highest calculated rate of nucleotide substitution reported so far for a RNA virus, a significant genetic heterogeneity exists even within the two main groupings and the sequence analysis allows sub-typing of PRRSV to geographical levels [12, 14]. According to a new proposal, based on recent sequence information, the European PRRSV-1 genotype should consist of 4 subtypes [14].
The wide variability of PRRSV strains causes several potential problems in the diagnosis of PRRS [15]. Therefore, molecular methods accompanied by sequencing represent a more sensitive and specific tool in PRRS diagnosis. Genetic analysis of ORF7 is mostly used for revealing the genetic relationships among PRRSV strains [16].
Both, PRRS [17] and PCV2 [18] infections have been described in Croatia in 1997 and 2004, respectively, on large, small and medium size pig farms in several Croatian counties. Since then, limited data is available on the predominant genotypes and eventual genetic changes of both viruses, especially after introducing vaccination.
Therefore, the objective of this study was to compare isolates of PCV2 and PRRSV collected until 2008 with the newly detected strains from 2012 that have been the cause of severe losses in the pig industry in Croatia. Direct sequencing of the amplified part of PCV2 ORF1 and PRRSV ORF7 was performed to determine genetic heterogeneity of the virus isolates and to carry out the comparison with previously described isolates.
Materials and methods
Sample collection
Serum samples from domestic pigs 3 to 6 months of age with severe signs of PCV2 clinical disease from two large pig farms located in Osijek-Baranja County with a history of PCV2 infection were collected during 2012. Prior to the extraction of total DNA, samples were pooled (5–10 samples per pool) and kept frozen at −70°C. A total of 10 pools, 5 cointaining samples collected from one PCV2 affected farm (CRO-PCV2-1 to CRO-PCV2-5) and 5 pools from the second PCV2 affected farm (CRO-PCV2-6 to CRO-PCV2-10) were examined for the presence of PCV2 DNA.
In addition, serum samples collected from domestic pigs with clinical signs of PRRS from large farms located in Brod-Posavina as well as Osijek-Baranja County were also included in this study.
Prior to the extraction of total RNA, samples that originated from 3–6 month old pigs from the same farm were pooled and kept frozen at −70°C. A total of 11 pools containing 5–10 samples each from Brod-Posavina (samples CRO-PRRSV-1 to CRO-PRRSV-3) and Osijek-Baranja County (samples CRO-PRRSV-4 to CRO-PRRSV-11) were examined for the presence of both viruses, PCV2 and PRRSV.
Sample preparation and viral DNA and RNA extraction
Sera were separated from cellular elements by centrifuging coagulated blood (the blood clots were rimmed with a sterile glass stick to facilitate separation) for 15 min at 1000 × g. An amount of 140 μl of each serum sample was used for viral RNA purification by using the QIAamp® Viral RNA Mini Kit (Qiagen, USA) according to the manufacturer’s instructions. An amount of 200 μl of each serum sample was used for viral DNA purification by using the QIAamp® DNA Mini kit (Qiagen, USA) according to the manufacturer’s instructions.
Detection of PCV2 by polymerase chain reaction
For detection of PCV2 DNA a specific primer pair for the amplification of a 360 bp long sequence of ORF1 region [19] was used. The reaction mixtures (50 μl) consisted of 6 μl of viral template DNA, 5 μl of 10x PCR buffer, 1.5 mM MgCl2, 200 μM of each deoxinucleotide triphosphates (dNTP), 400 nM of each primer, 5U of Platinum Taq DNA polymerase (Invitrogen, USA) and nuclease free water. Reaction mixtures without template DNA were used as negative controls. Croatian PCV2-positive samples were used as positive controls. The thermal profile of the amplifications contained an initial denaturation step at 94°C for 2 min, which was followed by 30 cycles of denaturation at 94°C for 30 sec, annealing at 56°C for 30 sec, and primer extension at 72°C for 30 sec. The amplifications were finished by a final extension at 72°C for 3 min. Eventually, the reaction mixtures were maintained at 4°C until they were removed from the device.
Detection of PRRSV by RT-PCR
For detection of PRRSV RNA a specific primer pair positioned in the ORF7 region (660 bp) [20] was used. RNA samples were reverse transcribed by SuperScript III reverse transcriptase and random hexamers (Invitrogen Life Technologies, USA), following the manufacturer’s instructions. The generated cDNA was used immediately for PCR amplification or stored at −20°C. The reaction mixtures were prepared as described for PCV2 mixtures. Reaction mixtures without template RNA were used as negative controls. Croatian PRRSV-positive samples were used as positive controls.
The thermal profile of the amplifications contained an initial denaturation step at 94°C for 3 min, which was followed by 30 cycles of denaturation at 94°C for 45 sec, annealing at 55°C for 45 sec, and primer extension at 72°C for 45 sec. The amplifications were finished by a final extension at 72°C for 7 min. Eventually, the reaction mixtures were maintained at 4°C until they were removed from the device.
Gel electrophoresis
Amplification products were separated by agarose gel electrophoresis in 1.5% agarose gel stained with ethidium bromide and visualized by UV transillumination.
Nucleotide sequencing and phylogenetic analysis
Prior to sequencing PCR products were excised from 2% agarose gels and then purified using Wizard SV Gel and PCR Clean-Up System (Promega, USA). Purified samples were sent for direct sequencing in both directions to Macrogen Inc; Amsterdam, the Netherlands.
The first comparisons of sequence data with the reference strains from the GenBank were performed by algorithm BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). For nucleotide sequence determination chromatograms were analysed by the Sequencer 4.6. programme (http://www.genecodes.com, Genes Codes Corporation). Sequences characterized in this study were aligned with published reference strains using the ClustalX programme. For the reconstruction of phylogenetic trees the Neighbor-Joining (NJ) method with Kimura-2 Parameter Model followed by MEGA 5 were used. The clustering stability of the NJ tree was evaluated by bootstrap analysis with 1000 replicates. The GenBank accession numbers of all reference sequences used for phylogenetic comparisons are indicated in Table 1. All PCV2 and PRRSV sequences displaying nucleotide differences obtained in the present study were deposited in the GenBank under accession numbers KF498717-KF498720 (CRO_PCV2_1, 3, 4, 6) and KF498721-KF498727 (CRO_PRRSV_1, 2, 3, 4, 7, 8, 9).
Results
The amplification of PCV2 genome regions from samples originating from the two farms with clinical signs of PCV2 (CRO-PCV2-1 to CRO-PCV2-10) resulted in clear PCR products with the expected molecular size. RT-PCR assay, using a primer pair specific for a sequence within the PRSSV ORF7, resulted in strong amplification signals with the expected molecular size in all pooled samples collected from swine with clinical signs of PRRS (CRO-PRRSV-1 to CRO-PRRSV-11).
No co-infections with these two viruses were detected in the tested samples. Amplifications were never detected in negative controls.
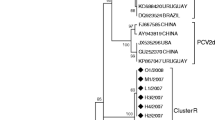
The PCR products of 360 bp were sequenced and confirmed to be PCV2 specific. The phylogenetic relationship analysis of the obtained sequences, including referent ones, showed that all analysed PCV2 sequences clustered into phylogenetic group 1 or PCV2b group (Figure 1). The seven PCV2 sequences (CRO-PCV2-1, CRO-PCV-2, CRO-PCV2-5, CRO-PCV2-7 to CRO-PCV2-10) were found to be 100% identical among themselves in the 297 nt ORF1 region. Sequences CRO-PCV2-3 and CRO-PCV2-4 differed in one (0.33%), whereas sequence CRO-PCV2-6 differed in two (0.66%) nucleotides. The robustness of the tree topology was supported by high bootstrap values. The obtained phylogenetic clustering shows that the PCV2 isolates differ from the published Croatian isolates from the year 2008 (HQ591366, HQ591367) in 2.3% and 3.0% of nucleotides, respectively. They differ in 4.4% nucleotides from the published Croatian isolates HQ591365, HQ591370 (from the year 2008), HQ591368 and HQ591369 (from the year 2007).
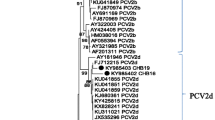
The PCR products of 660 bp were sequenced and confirmed to be PRRSV specific. The phylogenetic relationship analysis of the obtained sequences, including referent ones, showed that all analysed PRRSV sequences clustered into European type (type 1), subtype 1 (Figure 2). Sequences CRO-PRRSV-4, CRO-PRRSV-5 and CRO-PRRSV-6 (group 1) were found to be 100% identical among themselves in the 375 nt ORF7 region, while sequences CRO-PRRSV-9, CRO-PRRSV-10 and CRO-PRRSV-11 (group 2) were 100% identical. Sequences CRO-PRRSV-1, CRO-PRRSV-2, CRO-PRRSV-3, CRO-PRRSV-7 and CRO-PRRSV-8 (group 3) differed in one (0.26%) to six (1.60%) nucleotides. Sequences belonging to group 1 differed from the sequences belonging to group 3 in 31 (8.26%) to 35 (9.33%). The lowest sequence identity was found in sequences from group 2 when compared to the ones from group 3 where 39 (10.40%) to 43 (11.47%) nucleotides differed. Sequences from group 1 differed from the sequences from group 2 in 37 (9.87%) nucleotides. The obtained phylogenetic clustering shows that the PRRSV isolates differ from the published Croatian isolates from the year 2007, JQ043211 in the range of 3.7-9.2%, JQ043210 in the range of 4.7-10.0%, JQ043212 in the range 8.0-11.7%. They differ from the Croatian isolate from the year 2008 (JQ043213) in the range of 4.7-9.0%. The Croatian PRRSV isolates from 2012 showed a high genetic identity with Danish (92.78%), Hungarian (96.51%) and Dutch (99.00%) isolates.
Discussion and conclusions
Due to their potentially high economic impact, PRRSV [21] and PMWS [22] are two of the most important diseases affecting pig production in swine producing countries. It is thought that the PRRSV and PCV2 (the causative agent of PMWS) have been present in pig herds for decades if not centuries before the diseases emerged, and the viruses slowly mutated into disease-causing forms. Evidence from field surveys suggests that co-infections of pigs with these two viruses is common [23]. Preliminary studies have shown that both PCV2 and PRRSV were present in the livers of sows with PRRS-associated hepatitis and both PRRSV and PCV2 antigen or genome have been detected in tissues from the majority of PMWS cases [24]. However, the detection of PCV2 and PRRSV in the same animals may only be co-incidental since both are present in a high prevalence in commercial pig populations. In our study, no co-infection has been found.
Porcine circovirus type 2 (PCV2) has been detected in pigs with various clinical conditions on pig farms in different regions in Croatia [18]. The signs that predominated in the affected pigs were skin lesions, loss of weight, respiratory and digestive disorders followed by an increase in the mortality rate. Phylogenetic analyses from the year 1997 and 2002 showed clustering of PCV2 isolates into PCV2a and PCV2b groups, respectively [18]. Isolates from the year 2012 clustered into the PCV2b group and were related to those from year 2002, even though nucleotide differences occur suggesting the appearance of novel strains. No isolates from group PCV2a were found in 2012 indicating that this viral group has spontaneously decreased in its spread during the last decade or has been eliminated from the tested farms as a result of vaccination. Similar findings were reported and recognised in countries neighbouring Croatia, such as Slovenia [25], Hungary [26], Austria [27], Serbia [28, 29], and Italy [30]. Vaccination against PCV2 infection in Croatia was introduced rather late, in 2010. Only commercially available inactivated and subunit vaccines are registered, therefore no chimeric PCV2 were expected to be found during this study, as it was recorded in Canada [31].
First cases of reproductive failures of higher intensity caused by PRRSV on several pig farms in Croatia date from the year 1995 [17]. The same clinical signs of acute respiratory disease, mortality among growing pigs, reproductive and/or respiratory disorders in sows were reported at the same time in countries neighbouring Croatia, such as Slovenia [15], Serbia [32], Hungary [26, 33], and Austria [34]. Phylogenetic analysis of strains detected in the year 2007 showed clustering of PRRSV isolates into the European type [35]. The same results have been gained when isolates from Croatia collected in 2012 were genetically analysed. However, new subgroups of PRRSV strains were identified in 2012 when compared to the ones previously described in Croatia. Therefore, a possibility of new PRRSV entries within the last decades is possible. The high genetic identity shown among the 2012 PRRSV isolates from Croatia and isolates from Hungary, Denmark and the Netherlands could indicate that the novel strains have entered through trade. Vaccination against PRRS started in Croatia in 2004. It was massively applied until 2008. Two commercially available vaccines are registered, of which only one (based on an attenuated PRRSV strain) was actually in use. However, vaccination has drastically decreased after 2008 and only 750 doses were sold in 2012 and 1500 in 2013. All isolates in this study were sequenced and vaccinal strains were not found.
The detection of new PCV2 and PRRSV isolates in this study expanded the known diversity of detected strains in Croatia. This study also confirmed that the molecular-based reverse transcription polymerase chain reaction method and the routine direct sequencing of PCR products are important tools for rapid recognition of new PRRSV field strains.
References
Tischer L, Gelderblom H, Vettermann W, Koch MA: A very small porcine virus with circular single stranded DNA. Nature. 1982, 295: 64-66. 10.1038/295064a0.
Crowther RA, Berriman JA, Curran WL, Allan GM, Todd D: Comparison of the structures of three circoviruses: chicken anemia virus, porcine circovirus type 2, and beak and feather virus. J Virol. 2003, 77: 13036-13041. 10.1128/JVI.77.24.13036-13041.2003.
Todd D, Bendinelli M, Biagini P, Hino S, Mankertz A, Mishiro S, Niel C, Okamoto H, Raidal S, Ritchie BW, Teo GC: Circoviridae. Ball virus taxonomy, Eighth report of the international committee on taxonomy of viruses. Edited by: Fauguet MA, Mayo J, Maniloff U, Desselberger LA. 2005, San Diego, CA: Elsevier Academic Press, 327-334.
Mankertz A, Caliskan R, Hatterman K, Hillenbrand B, Kurzendoerfer P, Mueller B, Schmitt C, Steinfeldt T, Finsterbusch T: Molecular biology of porcine circovirus: analyses of gene expression and viral replication. Vet Microbiol. 2004, 98: 81-88. 10.1016/j.vetmic.2003.10.014.
Meehan BM, McNeilly F, Todd D, Kennedy S, Jewhurst VA, Ellis JA, Hassard LE, Clark EG, Haines DM, Allan GM: Characterisation of novel circovirus DNAs associated with wasting syndromes in pigs. J Gen Virol. 1998, 79: 2171-2179.
Opriessnig T, Meng XJ, Halbur PG: Porcine circovirus type 2-associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J Vet Diagn Invest. 2007, 19: 591-615. 10.1177/104063870701900601.
Olvera A, Cortey M, Segales J: Molecular evolution of porcine circovirus type 2 genomes: phylogeny and clonality. Virology. 2007, 357: 175-185. 10.1016/j.virol.2006.07.047.
Boisseson C, Beven V, Bigarre L, Thiery R, Rose N, Eveno E, Madec F, Jestin A: Molecular characterisation of porcine cyrcovirus type 2 isolates from post-weaning multysistemic wasting syndrome-affected and non-affected pigs. J Gen Virol. 2004, 85: 293-304. 10.1099/vir.0.19536-0.
Meulenberg JJ, Hulst MM, de Meijer EJ, Moonen PL, den Besten A, de Kluyver EP, Wensvoort G, Moormann RJ: Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS) is related to LDV and EAV. Virology. 1993, 192: 62-74. 10.1006/viro.1993.1008.
Meng XJ, Paul PS, Halbur PG: Molecular cloning and nucleotide sequencing of the 3'-terminal genomic RNA of the porcine reproductive and respiratory syndrome virus. J Gen Virol. 1994, 75: 1795-1801. 10.1099/0022-1317-75-7-1795.
Suarez P, Diaz-Guerra M, Prieto C, Esteban M, Castro JM, Nieto A, Ortin J: Open reading frame 5 of porcine reproductive and respiratory syndrome virus as a cause of virus-induced apoptosis. J Virol. 1996, 70: 2876-2882.
Meng XJ, Paul PS, Halbur PG, Lum MA: Phylogenetic analyses of the putative M (ORF6) and N (ORF7) genes of porcine reproductive and respiratory syndrome virus (PRRSV): Imolication for the existence of two genotypes of PRRSV in the USA and Europe. Arch Virol. 1995, 140: 745-755. 10.1007/BF01309962.
Murtaugh MP, Elam MR, Kakach LT: Comparison of the structural protein coding sequences of the VR-2332 and Lelystad virus. Arch Virol. 1995, 140: 1451-1460. 10.1007/BF01322671.
Stadejek T, Oleksiewicz MB, Potapchuk D, Podgorska K: Porcine reproductive and respiratory syndrome virus strains of exceptional diversity in eastern Europe support the definition of new genetic subtypes. J Gen Virol. 2006, 87: 1835-1841. 10.1099/vir.0.81782-0.
Toplak I, Rihtarič D, Hostnik P, Grom J, Štukelj M, Valenčak Z: Identification of genetically diverse sequence of porcine reproductive and respiratory syndrome virus in Slovenia and the importance on the sensitivity of four molecular tests. J Virol Methods. 2012, 179: 51-56. 10.1016/j.jviromet.2011.09.019.
Kang SY, Yun SI, Park HS, Park CK, Choi HS, Lee YM: Molecular characterisation of p L97–1, the first Korean isolate of porcine reproductive and respiratory syndrome virus. Virus Res. 2004, 104: 165-179. 10.1016/j.virusres.2004.04.003.
Lipej Z, Šoštarić B, Roić B, Lojkić M, Karačić V: Porcine reproductive and respiratory syndrome in Croatia. Vet Arch. 1997, 67: 113-130.
Jemeršić L, Cvetnić Ž, Toplak I, Špičić S, Grom J, Barlič-Maganja D, Terzić S, Hostnik P, Lojkić M, Humski A, Habrun B, Krt B: Detection and genetic characterisation of porcine circovirus type 2 (PCV2) in pigs from Croatia. Res Vet Sci. 2004, 77: 171-175. 10.1016/j.rvsc.2004.04.003.
Yang JS, Song DS, Kim SY, Lyoo S, Park BK: Detection of porcine circovirus type 2 in feces of pigs with or without enteric disese by polymerase chain recation. J Vet Diagn Invest. 2003, 15: 369-373. 10.1177/104063870301500412.
Oleksiewicz MB, Bøtner A, Madsen KG, Storgaard T: Sensitive detection and typing of porcine reproductive and respiratory syndrome virus by RT-PCR amplification of whole viral genes. Vet Microbiol. 1998, 64: 7-22. 10.1016/S0378-1135(98)00254-5.
Lunney JK, Benfield DA, Rowland RR: Porcine reproductive and respiratory syndrome virus: An update on an emerging and re-emerging viral disease of swine. Virus Research. 2010, 154: 1-6. 10.1016/j.virusres.2010.10.009.
Baekbo P, Kistensen CS, Larsen LE: A review of PMWS. Transbound Emerg Dis. 2012, 59: 60-67.
Allan GM, Ellis J, Krakowa S, Meehan B, McNair I, Kennedy S: Experimental infection of colostrum deprived piglets with porcine circovirus 2 (PCV2) aand porcine reproductive and respiratory syndrome virus (PRRSV) potentiates PCV2 replication. Arch Virol. 2000, 145: 2421-2429. 10.1007/s007050070031.
Harms PA, Sorden SD, Halbur PG, Bolin SR, Lager KM, Morozov I, Paul PS: Experimental reproduction of severe disease in CD/CD pigs currently infected with type 2 porcine circovirus and porcine reproductive and respiratory syndrome virus. Vet Pathol. 2001, 38: 528-539. 10.1354/vp.38-5-528.
Toplak I, Grom J, Hostnik P, Barlič-Maganja D: Detection and differentiation of porcine circovirus types 1 and 2 in Slovenia using ‘multiplex PCR’. 2002, Portorož: 3rd Slovenian Veterinary Congress, 43-
Kiss I, Sámi L, Kecskeméti S, Hanada K: Genetic variation of the prevailing porcine respiratory and reproductive syndrome viruses occurring on a pig farm upon vaccination. Arch Virol. 2006, 151: 2269-2276. 10.1007/s00705-006-0805-0.
Schmoll F, Opriessnig T, Schilcher F, Leeb B, Bauder B, Schuh M: First description of postweaning multisystemic wasting syndrome (PMWS) in Austria. 2002, Wiener Tierärztliche Monatsschrift 89, 50-56. in German, with English abstract
Becskei Z, Aleksić-Kovačević S, Rusvai M, Balka G, Jakob C, Petrović T, Knežević M: Distribution of porcine circovirus 2 cap antigen in the lymphoid tissue of pigs affected by postweaning multysistemic wasting syndrome. Acta Vet Hung. 2010, 58: 483-498. 10.1556/AVet.58.2010.4.9.
Savić B, Miličević V, Jakić-Dimić J, Bojkovski J, Prodanović R, Kureljušić B, Potkonjak A, Savić B: Genetic characterization and phylogenetic analysis of porcine circovirus type 2 (PCV2) in Serbia. Arch Virol. 2012, 157: 21-28. 10.1007/s00705-011-1130-9.
Martelli P, Terreni M, Amenna N, Morvan H, Cavirani S: Involvement of PCV2 in an outbreak of postweaning multisystemic wasting syndrome (PMWS) in Italy. 2000, Melbourne, Australia: Proceedings of the 16th International Pig Veterinary Society Congress, 534-
Gagnon CA, Music N, Fontaine G, Tremblay D, Harel J: Emergence of a new type of porcine circovirus in swine (PCV): a type 1 and type 2 PCV recombinant. Vet Microbiol. 2010, 144: 18-23. 10.1016/j.vetmic.2009.09.072.
Lazić S, Petrović T: Exploring possibilities of diagnosing, control and eradication of porcine respiratory and reproductive syndrome (PRRS). Biotechnol Anim Husbandry. 2007, 23: 391-401. 10.2298/BAH0701391L.
Medveczky I, Bálint Á, Makranszky L, Steverink P, Jacobs L: Sequence analysis of the membrane protein gene and nucleocapsid gene of porcine reproductive and respiratory syndrome virus isolated from a swine herd in Hungary. Acta Vet Hung. 2001, 49: 237-244. 10.1556/AVet.49.2001.2.14.
Indik S, Schmoll F, Sipos W, Klein D: Genetic variability of PRRS virus in Austria: consequences for molecular diagnosis and viral quantification. Vet Microbiol. 2005, 107: 171-178. 10.1016/j.vetmic.2005.01.024.
Stadejek T, Balka G, Chabros K, Lipej Z, Miličević V, Novosel D, Stankevicius A: Genetic diversity of European genotype PRRSV in Central and Eastern Europe- An update. Edited by: Proceedings of the 21st International Pig Veterinary Society Congress/ Sylvie D'Allaire and Robert friendship. 2010
Acknowledgements
We thank Ms. Ivana Prajdić for her technical assistance. This study was supported by the Croatian Ministry of Science, Education, and Sports (project no. 048-0481186-1183).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JP: Designer and performer of the study and author of the manuscript; TK, TB, DB, ŽC, BR, LJ: Contributed to the design, performing and revising of the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Prpić, J., Keros, T., Bedeković, T. et al. Phylogenetic comparison of porcine circovirus type 2 (PCV2) and porcine reproductive respiratory syndrome virus (PRRSV) strains detected in domestic pigs until 2008 and in 2012 in Croatia. Ir Vet J 67, 9 (2014). https://doi.org/10.1186/2046-0481-67-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/2046-0481-67-9