Abstract
Halomonas zincidurans strain B6T was isolated from a deep-sea heavy metal rich sediment from the South Atlantic Mid-Ocean Ridge. The strain showed significant resistance to heavy metals, especially to zinc. Here we describe the genome sequence and annotation, as well as the features, of the organism. The genome contains 3,325 protein-coding genes (2,848 with predicted functions), 61 tRNA genes and 6 rRNA genes. H. zincidurans strain B6T encodes 31 genes related to heavy metal resistance. And HGT may play an important role in its adaption to the heavy metal rich environment. H. zincidurans strain B6T may have potential applications in the bioremediation of heavy metal-contaminated environments.
Similar content being viewed by others
Introduction
Heavy metals, either essential (e.g. Mn, Zn, Cu, Co, Ni and Mo) or toxic (e.g. Hg, Ag and Cd), are generally harmful to microbial cells even at low concentrations, as to other living organisms [1, 2]. However, some microorganisms are able to resist to certain kinds and concentrations of heavy metals through several mechanisms, such as incorporating or precipitating heavy metals into complexes, oxidizing or reducing metals to less toxic valence states, and direct transporting metals out of the cell [3, 4]. These heavy metal resistant microorganisms have been attracting great interests because of their potential biotechnological applications in bio-mining of expensive heavy metals and bioremediation of heavy metal-contaminated environment [2].
Halomonas, the largest genus of the family Halomonadaceae, can be found in most saline environments, including marine environments, salterns, saline lakes and soils, as well as salty foods, etc. [5, 6]. Halomonas zincidurans strain B6T, a moderately halophilic bacterium, was isolated from a deep-sea sediment from the South Atlantic Mid-Ocean Ridge [5]. The strain was able to grow in medium containing high concentrations of heavy metals, especially Zn2+ ion, which is not detected in the reference strains and other moderately halophiles [5, 7]. Therefore, the novel isolate was named as H. zincidurans due to its particular resistance to zinc ion [5]. Here, we present a summary classification and a set of features of H. zincidurans strain B6T, together with the description of the genomic sequencing and annotation.
Organism information
A deep-sea sediment sample, TVG10, was collected from the South Atlantic Mid-Ocean Ridge (Table 1). There were many small hard orange red-colored lumps mixed in the sediment sample, which might be the particles containing ferric oxide and diffusing with hydrothermal plumes [8]. Not surprisingly, the concentrations of heavy metals in sample TVG10 were much higher than those in the samples collected from deep-sea seamount sediment [9], offshore sediment [10] and continental crust [11] (Additional file 1: Table S1), including Fe (98.99 mg/g), Mn (42.48 mg/g), Cu (0.839 mg/g), Ni (0.338 mg/g), Zn (0.285 mg/g), Cr (0.195 mg/g) and Co (0.064 mg/g). With consideration of the heavy metal rich environment, marine broth 2216 medium (MB, BD) containing 20 mM Mn2+ was used to isolate heavy metal resistant strains. Subsequently a strain named B6T was obtained [5].
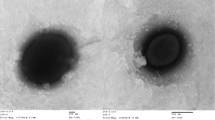
H. zincidurans strain B6T is a Gram-stained negative, rod-shaped (Figure 1), moderately halophilic bacterium growing at 0.5-15% (w/v) NaCl (Table 1). Strain B6T exhibited the highest 16S rRNA gene sequence similarity with H. xinjiangensis (96.1%). Phylogenetic analysis based on 16S rRNA gene sequences showed that strain B6T and H. xinjiangensis clustered together in a distinct branch within the genus Halomonas with a high bootstrap value (Figure 2). Strain B6T was able to resist high concentrations of heavy metals in liquid HM medium, including Mn2+ (200 mM), Co2+ (1.0 mM), Cu2+ (2.5 mM) and Zn2+ (14 mM). Its resistance to Zn2+ could be much higher (30 mM) when incubated on marine agar 2216 medium (MA, BD) [5], comparing to only 1 mM Zn2+ resisted by H. xinjiangensis TRM0175T. And the maximum zinc resistance concentration for 250 moderately halophilic bacteria, reported by Nieto et al., was only 2.5 mM [7]. Therefore, H. zincidurans strain B6T is of significant interest due to its prominent resistance to zinc.
Phylogenetic tree highlighting the position of H. zincidurans strain B6T relative to phylogenetically closely related type strains within the family Halomonadaceae. The sequences were aligned using Clustal W [26], and the neighbor-joining tree [27] was constructed based on kimura 2-parameter distance model [28] by using MEGA5 [29]. Bootstrap values above 60% are shown obtained from 1,000 bootstrap replications. Bar, 0.05 substitutions per nucleotide position. The corresponding GenBank accession numbers are displayed in parentheses.
Genome sequencing information
Genome project history
The next-generation shotgun-sequencing and quality assurance was performed at the Beijing Genome Institute (BGI, Shenzhen). The gap closure and annotation processes were performed by the authors. The Whole Genome Shotgun project of H. zincidurans strain B6T has been deposited at DDBJ/EMBL/GenBank under the accession JNCK00000000. The version described in this paper is version JNCK01000000. Table 2 presents the project information and its association with MIGS version 2.0 compliance [12].
Growth conditions and DNA isolation
H. zincidurans strain B6T was aerobically cultivated in MB medium at 30°C. Total genomic DNA was extracted using the method described by Marmur [32]. The quality and quantity of the genomic DNA was determined by 0.6% agarose gel electrophoresis with λ-Hind III digest DNA marker (TaKaRa, Dalian, China) and by a Qubit® fluorometer (Invitrogen, CA, USA) with Qubit dsDNA BR Assay kit (Invitrogen, CA, USA). About 350 μg DNA with a concentration of 450 ng/μl was obtained.
Genome sequencing and assembly
Whole-genome shotgun DNA sequencing of H. zincidurans strain B6T was performed using Solexa paired-end sequencing technology (HiSeq2000 system, Illumina, USA) [33]. Two libraries with insert size 494 bp and 2,586 bp were constructed and a total of 519 Mb and 416 Mb raw data were produced before filtering. After removing the adapter, duplicated reads and short inserts from the data of large library, there remained 433 Mb (~120-folds genome coverage) and 328 Mb (~90-folds genome coverage) clean data from the small and large libraries for assembling, respectively. Then these sequences were assembled into 15 contigs using the SOAPdenovo v.1.05 [30], the contig N50 length of which was 1,864,365 bp. PCR primers for gap closure were designed by Primer Premier v.5. PCR reactions were performed with PrimeSTAR HS Polymerase (TaKaRa, Dalian, China) and the amplicons were sequenced using Sanger and primer walking technologies. The sequenced fragments were subsequently assembled with the contigs using SeqMan of the Lasergene package (DNAstar, Madison, WI) into 2 contigs.
Genome annotation
The whole genomic tRNAs were identified using tRNAscan-SE v.1.21 [34] with bacterial model, and rRNAs were found by RNAmmer v.1.2 Server [35]. ORFs were predicted using Glimmer v.3.0 [31]. The predicted ORFs were translated and analyzed using the NCBI nonredundant, Swiss-Prot [36] and COG [37] databases, as well as RAST server online [38] for genome annotation. KAAS [39] was used to assign the predict proteins into KEGG pathway [40] with BBH method. Genes with signal peptides and transmembrane helices were predicted using TMHMM server v.2.0 [41] and SignalP server v.4.1 [42], respectively. The G+C content, G+C content at the third-codon position and RSCU were calculated by CodonW v.1.4.4.
Genome properties
The genome was assembled into 2 contigs, one with a size of 3,546,937 bp and the other with 7,823 bp (Table 3). The G+C content determined based on the total 3,554,760 bp sequences was 66.41%. A total of 3,392 genes were predicted, including 3,325 protein-coding genes, 61 tRNA genes and two copies of 16S-23S-5S rRNA gene operons (Table 4 and Figure 2). Among the protein coding genes, 2,848 were assigned to putative functions, and the remaining was annotated as hypothetical proteins. In total, 1,938 and 442 protein coding genes were assigned to KEGG and subsystems, respectively. The detailed properties and the statistics of the genome as well as the distribution of genes into COG functional categories are summarized in Tables 3, 4 and 5, Figure 3 and Additional file 2: Table S2.
Circular map of the chromosome of H. zincidurans strain B6T . Labeling from the outside to the inside circle: ORFs on the forward strand (colored by COG categories), ORFs on the reverse strand (colored by COG categories), RNA genes (tRNAs red, rRNAs blue), G+C content (peaks out/inside the circle indicate values higher or lower than the average G+C content, respectively), GC skew (calculated as (G-C)/(G+C), green/purple peaks out/inside the circle indicates values higher or lower than 1, respectively).
Insights into the genome
The genome of H. zincidurans strain B6T contains 31 genes related to heavy metal resistance, especially to zinc resistance (Table 6). Zinc is an essential but also toxic metal for living being [2, 43]. The concentration of zinc inside bacterial cells is maintained by importing limitation, efflux, accumulation and sequestration [44, 45]. H. zincidurans strain B6T possesses four heavy metal translocating P-type ATPases (HALZIN_733, HALZIN_1240, HALZIN_2196 and HALZIN_2262), which may participate in the transport of Zn2+, Mn2+, Cu2+, Cd2+, Pb2+, Ag + and Hg2+ against the concentration gradient to the periplasm [2, 44]. Especially the two ZntA P-type ATPases (HALZIN_733 and HALZIN_2196) may mediate resistance to Zn2+, Cd2+ and Pb2+ [46, 47]. Zn2+, Co2+, Cu2+, Cd2+ and Ni2+ are able to be transported by RND family efflux transporter protein (HALZIN_54, HALZIN_1411, HALZIN_2047, HALZIN_2208 and HALZIN_2209) from both the cytoplasm and the periplasm to outside [2, 44]. Usually the P-type ATPases are regulated by MerR family regulators responding to the intracellular heavy metal concentration [44, 48, 49]. Six analogues of MerR family regulators (HALZIN_399, HALZIN_922, HALZIN_2261, HALZIN_2264, HALZIN_2469 and HALZIN_2675) were found in the genome of H. zincidurans strain B6T. Additionally, a zinc uptake regulation protein ZUR (HALZIN_1413), which is a repressor regulator during zinc uptake, is also detected [44, 50]. The presence of these genes is accordance with zinc resistance phenotype of H. zincidurans strain B6T.
Among the 31 ORFs related to heavy metal resistance, it is noteworthy of two mer-operons. One mer-operon encodes a mercuric transport protein (MerE, HALZIN_916) for organic mercury uptake [51], a transcriptional regulator (MerD, HALZIN_917), three alkylmercury lyases (MerB, HALZIN_918-920) catalyzing organomercurials yielding Hg2+ [52] and a transcriptional regulator (MerR, HALZIN_922). The other one encodes a transcriptional regulator (MerR, HALZIN_2469), two mercuric transport proteins (MerT and MerP, HALZIN_2470-2471) for inorganic mercury uptake [51] and a mercuric reductase (MerA, HALZIN_2472) catalyzing Hg2+ to Hg0 [53]. According to the genomic data, H. zincidurans strain B6T is able to survive in both inorganic and organic mercury environments. Interestingly, the four ORFs of the inorganic mer-operon showed the highest sequence identities to those of Halomonas lutea. Nevertheless, all the six ORFs of the organic mer-operon did not show the highest sequence identities to those of the genus Halomonas, but to the genera Burkholderia, Pseudomonas, Gladiecola and Stenotrophomonas, which indicates that the organic mer-operon might be acquired by HGT. Of special interest are the three alkylmercury lyases (MerB, HALZIN_918-920), which had obvious differences between the G+C content (56.6%; 57.1, 56.6 and 56.0% for these three gene sequences, respectively) as well as the G+C content at the third-codon positions (60.3%; 60.4, 61.0 and 59.4% for these three gene sequences, respectively) and those of the total protein-coding genes (65.4 and 82.8%, respectively). Besides, the RSCUs of nearly half of the 59 codons used by the three genes (23, 27 and 26 codons for HALZIN_918-920, respectively) change more than 2 folds, compared with those used by total protein-coding genes. 13 of the 31 ORFs (41.9%) were not related to Halomonadaceae genes according to the gene sequence similarity analysis, 9 of the 13 ORFs had RSCU change larger than 2 folds in more than 25% codons. These results indicated the existence of HGT events among the heavy metal resistance-related genes. Thus, HGT events might be an important way for H. zincidurans strain B6T to acquire heavy metal resistant ability and to adapt to the heavy metal rich environment.
Conclusion
The draft genome sequence of the heavy metal resistant bacteria H. zincidurans strain B6T isolated from the South Atlantic Mid-Ocean Ridge provide an insight into the genomic basis of its heavy metal resistance ability. And HGT may play an important role in its adaption to the heavy metal rich environment. On the basis of analysis and characterization of genome, H. zincidurans strain B6T might be resistant more kinds of heavy metal than we tested, such as Hg2+, Cd2+, Pb2+, Ni2+ and Ag+, etc. And it may have the potential for the bioremediation of multi-metal-contaminated environments. In addition, further analysis will be performed to confirm its resistant ability to other heavy metals and determine the mechanism of heavy metal resistance that we don’t know yet.
Abbreviations
- HGT:
-
Horizontal gene transfer
- RSCU:
-
Relative synonymous codon usage.
References
Valls M, de Lorenzo V: Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution. FEMS Microbiol Rev 2002,26(4):327–338. 10.1111/j.1574-6976.2002.tb00618.x
Nies DH: Microbial heavy-metal resistance. Appl Microbiol Biotechnol 1999,51(6):730–50. 10.1007/s002530051457
Özdemir S, Kilinc E, Poli A, Nicolaus B, Güven K: Cd, Cu, Ni, Mn and Zn resistance and bioaccumulation by thermophilic bacteria. Geobacillus toebii subsp. decanicus and Geobacillus thermoleovorans subsp. stromboliensis. World J Microbiol Biotechnol 2012,28(1):155–163.
Teitzel GM, Parsek MR: Heavy metal resistance of biofilm and planktonic Pseudomonas aeruginosa . Appl Environ Microbiol 2003,69(4):2313–2320. 10.1128/AEM.69.4.2313-2320.2003
Xu L, Xu X-W, Meng F-X, Huo Y-Y, Oren A, Yang J-Y, Wang C-S: Halomonas zincidurans sp. nov., a heavy-metal-tolerant bacterium isolated from the deep-sea environment. Int J Syst Evol Microbiol 2013,63(Pt 11):4230–4236. 10.1099/ijs.0.051656-0
Arahal DR, Ventosa A: The family Halomonadaceae . In The Prokaryotes: a Handbook on the Biology of Bacteria, Volume 6. 3rd edition. Edited by: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. New York: Springer; 2006:811.
Nieto JJ, Fernandez-Castillo R, Marquez MC, Ventosa A, Quesada E, Ruiz-Berraquero F: Survey of metal tolerance in moderately halophilic eubacteria. Appl Environ Microbiol 1989,55(9):2385–90.
Feely RA, Geiselman TL, Baker ET, Massoth GJ, Hammond SR: Distribution and composition of hydrothermal plume particles from the ASHES Vent Field at Axial Volcano, Juan de Fuca Ridge. J Geophys Res: Solid Earth 1990,95(B8):12855–12873. 10.1029/JB095iB08p12855
Huo Y, Cheng H, Anton FP, Wang C, Jiang X, Pan J, Wu M, Xu X: Ecological functions of uncultured microorganisms in the cobalt-rich ferromanganese crust of a seamount in the central Pacific are elucidated by fosmid sequencing. Acta Oceanologica Sinica 2014. in press
Zhao Q: Ocean Geochemistry. Beijing: The Geological Publishing House; 1988.
Hans WK: The composition of the continental crust. Geochim Cosmochim Acta 1995,59(7):1217–1232. 10.1016/0016-7037(95)00038-2
Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, Ashburner M, Axelrod N, Baldauf S, Ballard S, Boore J, Cochrane G, Cole J, Dawyndt P, De Vos P, dePamphilis C, Edwards R, Faruque N, Feldman R, Gilbert J, Gilna P, Glöckner FO, Goldstein P, Guralnick R, Haft D, Hancock D, et al.: The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008,26(5):541–7. 10.1038/nbt1360
Woese CR, Kandler O, Wheelis ML: Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A 1990,87(12):4576–9. 10.1073/pnas.87.12.4576
Garrity GM, Bell JA, Lilburn T: Phylum XIV. Proteobacteria phyl. nov. In Bergey's Manual of Systematic Bacteriology, Volume 2, Part B. 2nd edition. Edited by: Garrity GM, Brenner DJ, Krieg NR, Staley JT. New York: Springer; 2005:1.
Validation of publication of new names and new combinations previously effectively published outside the IJSEM Int J Syst Evol Microbiol 2005,55(6):2235–2238.
Garrity GM, Bell JA, Lilburn T: Class III. Gammaproteobacteria class. nov. In Bergey's Manual of Systematic Bacteriology, Volume 2, Part B. 2nd edition. Edited by: Garrity GM, Brenner DJ, Krieg NR, Staley JT. New York: Springer; 2005:1.
Garrity GM, Bell JA, Lilburn T: Order VIII. Oceanospirillales ord. nov. In Bergey's Manual of Systematic Bacteriology, Volume 2. 2nd edition. Edited by: Brenner DJ, Krieg NR, Staley JT, Garrity GM. Springer, New York: Part B; 2005:270.
Franzmann PD, Wehmeyer U, Stackebrandt E: Halomonadaceae fam. nov., a new family of the class Proteobacteria to accommodate the genera Halomonas and Deleya . Syst Appl Microbiol 1988,11(1):16–19. 10.1016/S0723-2020(88)80043-2
Ntougias S, Zervakis GI, Fasseas C: Halotalea alkalilenta gen. nov., sp. nov., a novel osmotolerant and alkalitolerant bacterium from alkaline olive mill wastes, and emended description of the family Halomonadaceae Franzmann et al . 1989, emend. Dobson and Franzmann 1996. Int J Syst Evol Microbiol 2007,57(9):1975–1983. 10.1099/ijs.0.65078-0
Ben Ali Gam Z, Abdelkafi S, Casalot L, Tholozan JL, Oueslati R, Labat M: Modicisalibacter tunisiensis gen. nov., sp. nov., an aerobic, moderately halophilic bacterium isolated from an oilfield-water injection sample, and emended description of the family Halomonadaceae Franzmann et al . 1989 emend Dobson and Franzmann 1996 emend. Ntougias et al . 2007. Int J Syst Evol Microbiol 2007,57(10):2307–2313. 10.1099/ijs.0.65088-0
NOTES: Validation of the publication of new names and new combinations previously effectively published outside the IJSB: List No. 29†. Int J Syst Bacteriol 1989,39(2):205–206.
Dobson SJ, Franzmann PD: Unification of the genera Deleya (Baumann et al . 1983), Halomonas (Vreeland et al . 1980), and Halovibrio (Fendrich 1988) and the species Paracoccus halodenitrificans (Robinson and Gibbons 1952) into a single genus, Halomonas , and placement of the genus Zymobacter in the Family Halomonadaceae . Int J Syst Bacteriol 1996,46(2):550–558. 10.1099/00207713-46-2-550
Vreeland RH, Litchfield CD, Martin EL, Elliot E: Halomonas elongata , a new genus and species of extremely salt-tolerant bacteria. Int J Syst Bacteriol 1980,30(2):485–495. 10.1099/00207713-30-2-485
Mellado E, Moore ERB, Nieto JJ, Ventosa A: Phylogenetic inferences and taxonomic consequences of 16S ribosomal DNA sequence comparison of Chromohalobacter marismortui , Volcaniella eurihalina , and Deleya salina and reclassification of V. eurihalina as Halomonas eurihalina comb. nov. Int J Syst Bacteriol 1995,45(4):712–716. 10.1099/00207713-45-4-712
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G: Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000,25(1):25–9. 10.1038/75556
Thompson JD, Higgins DG, Gibson TJ: CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994,22(22):4673–4680. 10.1093/nar/22.22.4673
Saitou N, Nei M: The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987,4(4):406–425.
Kimura M: A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980,16(2):111–120. 10.1007/BF01731581
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S: MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011,28(10):2731–2739. 10.1093/molbev/msr121
Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K: De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 2010,20(2):265–272. 10.1101/gr.097261.109
Delcher AL, Bratke KA, Powers EC, Salzberg SL: Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007,23(6):673–679. 10.1093/bioinformatics/btm009
Marmur J: A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 1961,3(2):208–218. 10.1016/S0022-2836(61)80047-8
Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Cheetham RK, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, et al.: Accurate whole human genome sequencing using reversible terminator chemistry. Nature 2008,456(7218):53–9. 10.1038/nature07517
Lowe TM, Eddy SR: tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997,25(5):955–64. 10.1093/nar/25.5.0955
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW: RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007,35(9):3100–3108. 10.1093/nar/gkm160
Bairoch A, Boeckmann B: The SWISS-PROT protein sequence data bank. Nucleic Acids Res 1992,20(Suppl):2019–22. 10.1093/nar/20.suppl.2019
Tatusov RL, Galperin MY, Natale DA, Koonin EV: The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 2000,28(1):33–6. 10.1093/nar/28.1.33
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O: The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008, 9: 75. 10.1186/1471-2164-9-75
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M: KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic acids res 2007,35(suppl 2):W182–5.
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M: The KEGG resource for deciphering the genome. Nucleic Acids Res 2004,32(Database issue):D277–80.
Krogh A, Larsson B, von Heijne G, Sonnhammer ELL: Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol 2001,305(3):567–580. 10.1006/jmbi.2000.4315
Dyrløv Bendtsen J, Nielsen H, von Heijne G, Brunak S: Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 2004,340(4):783–795. 10.1016/j.jmb.2004.05.028
McCall KA, C-c H, Fierke CA: Function and mechanism of zinc metalloenzymes. J Nutr 2000,130(5):1437S-1446S.
Choudhury R, Srivastava S: Zinc resistance mechanisms in bacteria. Cur Sci 2001,81(7):768–775.
Blencowe DK, Morby AP: Zn (II) metabolism in prokaryotes. FEMS Microbiol Rev 2003,27(2‒3):291–311.
Rensing C, Sun Y, Mitra B, Rosen BP: Pb(II)-translocating P-type ATPases. J Biol Chem 1998,273(49):32614–32617. 10.1074/jbc.273.49.32614
Rensing C, Mitra B, Rosen BP: The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc Natl Acad Sci U S A 1997,94(26):14326–14331. 10.1073/pnas.94.26.14326
Brown NL, Stoyanov JV, Kidd SP, Hobman JL: The MerR family of transcriptional regulators. FEMS Microbiol Rev 2003,27(2–3):145–63.
Brocklehurst KR, Hobman JL, Lawley B, Blank L, Marshall SJ, Brown NL, Morby AP: ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli . Mol Microbiol 1999,31(3):893–902. 10.1046/j.1365-2958.1999.01229.x
Hantke K: Bacterial zinc uptake and regulators. Curr Opin Microbiol 2005,8(2):196–202. 10.1016/j.mib.2005.02.001
Sone Y, Pan-Hou H, Nakamura R, Sakabe K, Kiyono M: Roles played by MerE and MerT in the transport of inorganic and organic mercury compounds in Gram-negative bacteria. J Health Sci 2010,56(1):123–127. 10.1248/jhs.56.123
Pitts KE, Summers AO: The roles of thiols in the bacterial organomercurial lyase (MerB). Biochemistry 2002,41(32):10287–96. 10.1021/bi0259148
Felske A, Fehr W, Pauling B, von Canstein H, Wagner-Dobler I: Functional profiling of mercuric reductase ( mer A) genes in biofilm communities of a technical scale biocatalyzer. BMC Microbiol 2003,3(1):22. 10.1186/1471-2180-3-22
Acknowledgements
This work was supported by the China Ocean Mineral Resources R & D Association (COMRA) Special Foundation (No. DY125-15-R-03); the National Natural Science Foundation of China (No. 41276173); the Zhejiang Provincial Natural Science Foundation of China (No. LQ13D060002) and the Scientific Research Fund of the Second Institute of Oceanography, SOA (No. JT1305).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YH designed and performed experiments, analyzed data and wrote the paper; ZL performed experiments; HC analyzed genome data; CW analyzed data; XX designed the experiments and wrote the paper. All authors read and approved the final manuscript.
Electronic supplementary material
40793_2014_23_MOESM1_ESM.doc
Additional file 1: Table S1: Concentrations of heavy metals in deep-sea sediment collected from the South Atlantic Mid-Ocean Ridge (1) and the sediments from the Central Pacific seamount (2), offshore sediment (3) and continental crust (4). (DOC )
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Huo, YY., Li, ZY., Cheng, H. et al. High quality draft genome sequence of the heavy metal resistant bacterium Halomonas zincidurans type strain B6T . Stand in Genomic Sci 9, 30 (2014). https://doi.org/10.1186/1944-3277-9-30
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1944-3277-9-30