Abstract
Background
The association between MHTFR Ala222Val polymorphism and breast cancer (BC) risk are inconclusive. To derive a more precise estimation of the relationship, a systematic review and meta-analysis was performed.
Methods
A comprehensive search was conducted through researching MEDLINE, EMBASE, PubMed, Web of Science, Chinese Biomedical Literature database (CBM) and China National Knowledge Infrastructure (CNKI) databases before August 2012. Crude odds ratios (ORs) with 95% confidence intervals (CIs) were calculated to estimate the strength of the association.
Results
A total of 51 studies including 20,907 cases and 23,905 controls were involved in this meta-analysis. Overall, significant associations were found between MTHFR Ala222Val polymorphism and BC risk when all studies pooled into the meta-analysis (Ala/Ala vs Val/Val: OR=0.870, 95%CI=0.789–0.958,P=0.005; Ala/Val vs Val/Val: OR=0.895, 95%CI=0.821–0.976, P=0.012; dominant model: OR=0.882, 95%CI=0.808–0.963, P=0.005; and recessive model: OR = 0.944, 95%CI=0.898–0.993, P=0.026; Ala allele vs Val allele: OR = 0.935, 95%CI=0.887–0.986, P=0.013). In the subgroup analysis by ethnicity, the same results were found in Asian populations, while no significant associations were found for all comparison models in other Ethnicity populations.
Conclusion
In conclusion, our meta-analysis provides the evidence that MTHFR Ala222Val gene polymorphisms contributed to the breast cancer development.
Virtual slides
The virtual slide(s) for this article can be found here: http://www.diagnosticpathology.diagnomx.eu/vs/1966146911851976
Similar content being viewed by others
Introduction
Breast cancer is the most common cancer and the main cause of cancer mortality in women. The etiology towards to this disease is poorly understood, some risk factors including familial history of the disease, age of menarche and of menopause, diet, reproductive history, high estrogen exposure as well as genetic factors may contribute to its development[1, 2]. Studies suggest that the effect determined by low-penetrance genes, may provide a plausible explanation for BC susceptibility. Polymorphisms in genes are associated with a risk or protection against the disease. 5,10-methylenetetrahydrofolate reductase (MTHFR) is one important genes located at 1p36.3[3]. MTHFR Ala222Val polymorphism has become the most commonly studied one, which has been considered to influence the enzyme activity of MTHFR[4]. The MTHFR 222Val/Val (homozygote) genotype results in 30% enzyme activity in vitro compared with the Ala/Ala wild-type[5]. Numerous epidemiological studies have evaluated the association between the MTHFR Ala222Val polymorphisms and BC risk. However, these studies have yielded conflicting results, partially because of the possible small effect of the polymorphism on BC risk and the relatively small sample size in each of published studies. The aim of this study is to derive a more precise estimation of these associations by performing this meta-analysis.
Materials and methods
Literature search
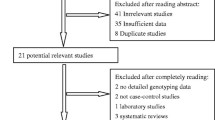
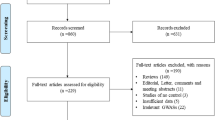
All studies that examined the association between the MFTHR Ala222Val polymorphism and BC were identified. A comprehensive search was conducted through researching MEDLINE, EMBASE, PubMed, Web of Science, China Biomedical Literature database (CBM) and China National Knowledge Infrastructure (CNKI) databases before August 2012. The search strategy included the combination of “breast cancer,” “breast neoplasm,” “methylene-tetrahydrofolate reductase,” “MTHFR,” “Ala222Val”, “rs1801133”, “variant,” and “polymorphism.” References of the retrieved articles were also screened. Non-familial case–control studies were eligible if they determined the distribution for this polymorphism in unrelated patients with breast cancer and in a concurrent control group of healthy subjects using molecular methods for genotyping. Of the studies with the same or overlapping data by the same investigators, we selected the most recent ones with the most subjects. We evaluated all associated publications to retrieve the most eligible literatures. The reference lists of reviews and retrieved articles were hand searched at the same time. We did not include abstracts or unpublished reports. When overlapping data of the same patient population were included in more than one publication, only the most recent or complete study was used in this meta-analysis.
Inclusion and exclusion criteria
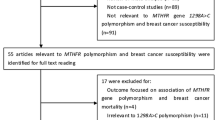
The following inclusion criteria were used to select literatures for the meta-analysis: (1) information on the evaluation of MFTHR Ala222Val polymorphism and BC susceptibility; (2)Only the cohort and case-control studies were considered;(3) sufficient genotype data were presented to calculate the OR with 95% CI. Major reasons for exclusion of studies were: (1) none-case–control studies; (2) reviews and duplication of the previous publication; (3) control population including malignant tumor patients; (4) no usable data reported.
Data extraction
Two investigators reviewed and extracted information from all eligible publications independently, according to the inclusion and exclusion criteria listed above. An agreement was reached by discussion between the two reviewers whenever there was a conflict. The following items were collected from each study: first author’s surname, year of publication, ethnicity, total number of cases and controls with Ala/Ala, Ala/Val, and Val/Val genotypes, respectively. Different descents were categorized as Caucasians, Asians, and Mixed populations which included more than one ethnic descent. For case–control studies, data were extracted separately for each group whenever possible.
Statistical analysis
The strength of the association between MFHTR Ala222Val polymorphism and BC risk was measured by ORs, whereas a sense of the precision of the estimate was given by 95% Cls. The significance of the summary OR was determined with a Z-test. We first examined MFHTR Ala222Val genotypes using co-dominant model (homogeneous co-dominant model: Ala/Ala vs Val/Val, heterogeneous co-dominant model: Ala/Val vs Val/Val), recessive (Ala/Ala vs Ala/Val + Val/Val), and dominant (Ala/Ala + Ala/Val vs Val/Val) genetic models. Then, the relationship between the allele and susceptibility to BC was examined (addictive model: Ala allele vs Val allele). Stratified analyses were also performed by ethnicities. A chi-square-based Q-statistic test and an I2-test test were both performed to evaluate the between-study heterogeneity of the studies.
Two models including the fixed-effects model and the random-effects model of meta-analysis were applied for dichotomous outcomes. The fixed-effects model assumes that studies are sampled from populations with the same effect size, making an adjustment to the study weights according to the in-study variance. The random-effects model assumes that studies are taken from populations with varying effect sizes, calculating the study weights both from in-study and between-study variances, considering the extent of variation, or heterogeneity. A P-value ≥0.10 for the Q-test indicated lack of heterogeneity among the studies, and so the summary OR estimate of each study was calculated by the fixed-effects modelm[6]. Otherwise, the random-effects model (DerSimonian and Laird method) was used[7]. I2 statistic can be used to quantify heterogeneity irrespective of the number of studies. The significance of the pooled OR was determined by the Z-test and P<0.05 was considered as statistically significant. Subgroup analyses were performed by ethnicity to explore the reasons of heterogeneity. Sensitivity analyses were performed to assess the stability of the results. To investigate whether publication bias might affect the validity of the estimates, funnel plot were constructed. An asymmetric plot suggests a possible publication bias. Funnel plot asymmetry was assessed by the method of Egger’s linear regression test, a linear regression approach to measure funnel plot asymmetry on the natural logarithm scale of OR. The significance of the intercept was determined by the t-test suggested by Egger (P<0.05 was considered representative of statistically significant publication bias). All statistical tests were performed with Stata (Version 12.0, Stata Corporation, College Station, TX), using two-sided P-values.
Results
Eligible studies
51 eligible studies on MTHFR Ala222Val genotypes and colorectal cancer were identified through literature search and selection based on the inclusion and exclusion criteria [8–58]. The publishing year of the studies was from 2002 to 2012. There were 25 studies of Caucasian, 19 studies of Asians and 7 studies of Mixed populations. In total, 20,907 BC cases and 23,905 controls were included in the meta-analysis. The selected study characteristics were summarized in Table 1.
Meta-analysis results
Overall, there was statistically significant difference in BC risk between the patients with Ala/Ala genotype and those with Val/Val genotype (OR=0.870, 95%CI=0.789-0.958, P=0.005; Figure1). Similarly, significant associations were also found in the recessive model comparison (OR=0.944, 95%CI=0.898-0.993, P=0.026; Table2) and dominant model comparison (OR=0.882, 95%CI=0.808-0.963, P=0.005; Table2). Moreover, we found significant association between Ala222Val polymorphism and BC when examining the contrast of Ala versus Val (OR=0.935, 95%CI=0.887-0.986, P=0.013; Figure2). In the stratified analysis by ethnicity, there was significant association between Ala222Val polymorphism and BC risk for Ala/Ala vs Val/Val comparison (OR=0.787, 95%CI=0.645-0.961, P=0.019; Figure3), recessive model comparison (OR=0.890, 95%CI=0.799-0.991, P=0.034; Table2), dominant model comparison (OR=0.826, 95%CI=0.703-0.972, P=0.021; Table2) and Ala allele versus Val allele comparison (OR=0.877, 95%CI=0.801-0.960, P=0.008; Figure4) among Asian populations. For Caucasian and Mixed populations, there was no significant association between Ala222Val polymorphism and breast cancer risk (Table2).
Sensitivity analysis
In order to compare the difference and evaluate the sensitivity of the meta-analyses, we conducted one-way sensitivity analysis to evaluate the stability of the meta-analysis. The statistical significance of the results was not altered when any single study was omitted, confirming the stability of the results. Hence, results of the sensitivity analysis suggest that the data in this meta-analysis are relatively stable and credible.
Publication bias
Begg’s funnel plot and Egger’s test were performed to assess the publication bias. The shape of funnel plots did not reveal any evidence of obvious asymmetry in all comparison models, and the Egger’s test was used to provide statistical evidence of funnel plot symmetry. The results of Begg’s test did not show any evidence of publication bias.
Discussion
Breast cancer is currently the most frequently occurring cancer and the leading causes of cancer-related death among women in the world. Single nucleotide polymorphism (SNP) is the most common form of human genetic variation, and may contribute to individual’s susceptibility to cancer, however, the underlying molecular mechanism is unknown. Previous study suggested that some variants, especially those in the promoter regions of genes, may affect either the expression or activity levels of enzymes[59–61] and therefore may be mechanistically associated with cancer risk. Previous studies on the relationship between MTHFR Ala222Val polymorphisms and BC risk were contradictory. These inconsistent results are possibly because of a small effect of the polymorphism on BC risk or the relatively low statistical power of the published studies. Hence, the meta-analysis was needed to provide a quantitative approach for combining the results of various studies with the same topic, and for estimating and explaining their diversity.
Meta analysis has great power for elucidating genetic factors in cancer. On the bases of the character of cancer, the effect of one genetic component on the development of the disease can be easily masked by other genetic and environmental factors. A meta-analysis potentially investigates a large number of individuals and can estimate the effect of a genetic factor on the risk of the disease[62, 63]. The present study included data from 51 association studies that had investigated the relationship between the MTHFR Ala222Val polymorphism and BC.
This present meta-analysis, including 20,907 cases and 23,905 controls, concerned the Ala222Val polymorphism of MTHFR gene and BC risk. In the meta-analysis, we found that the variant genotypes of the MTHFR Ala222Val polymorphisms were significantly associated with BC risk. Simultaneously, the same results presented in stratified analysis by ethnicity. We found that the variant genotype of the MTHFR Ala222Val polymorphism, in Asian populations, was associated with significant increase in BC risk. Although the MTHFR Ala222Val polymorphism may be associated with DNA repair activity, no significant association of the variant genotype with BC risk was found in Caucasian and Mixed populations, suggesting the influence of the genetic variant may be masked by the presence of other as-yet unidentified causal genes involved in colorectal cancer.
Some limitations of this meta-analysis should be acknowledged. First, our result was based on unadjusted estimates, while a more precise analysis should be conducted adjusted by other factors like diet habit, smoking, drinking status, environmental factors and so on. Second, in the subgroup analyses by ethnicity, relatively limited study numbers to perform ethnic subgroup analysis of mixed populations. Moreover, there are no American and African-American descent populations. Thus, additional studies are warranted to evaluate the effect of this functional polymorphism on BC risk in different ethnicities, especially in American, African-American and Mixed populations. In addition, our analysis did not consider the possibility of gene-gene or SNP-SNP interactions or the possibility of linkage disequilibrium between polymorphisms.
Despite of some limitations, this meta-analysis provided evidence of the association between the MTHFR Ala222Val polymorphisms and BC risk, supporting the hypothesis that MTHFR Ala222Val polymorphism contributes to overall BC risk. In subgroup analysis, the same results were found in Asian populations. In order to verify our findings, well-designed studies including different ethnic groups with a careful matching between cases and controls should be considered in future association studies to confirm the results from our meta-analysis. Moreover, further evaluating the effect of gene-gene and gene-environment interactions on the Ala222Val polymorphism and BC risk are necessary.
Abbreviations
- BC:
-
Breast cancer
- HWE:
-
Hardy–Weinberg equilibrium
- OR:
-
Odds ratio
- CI:
-
Confidence interval
- MTHFR:
-
Methylenetetrahydrofolate reductase.
References
Hankinson SE, Colditz GA, Willett WC: Towards an integrated mod breast cancer etiology. The lifelong interplay of genes, lifestyle, and hormones. Breast Cancer Res. 2004, 6: 213-218. 10.1186/bcr921.
Umitrescu RG, Cotarla I: Understanding breast cancer risk—where do we stand in 2005. J Cell Mol Med. 2005, 9: 208-221. 10.1111/j.1582-4934.2005.tb00350.x.
Rosenberg N, Murata M, Ikeda Y, Opare-Sem O, Zivelin A, Geffen E, Seligsohn U: The frequent 5,10-methylenetetrahydrofolate reductase C677T polymorphism is associated with a common haplotype in Whites, Japanese, and Africans. Am J Hum Genet. 2002, 70: 758-762. 10.1086/338932.
Vander Put NM: A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects. Am J Hum Genet. 1998, 62: 1044-1051. 10.1086/301825.
Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, Boers GJ, Den Heijer M, Kluijtmans LA, Van den Heuvel LP: A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995, 10: 111-113. 10.1038/ng0595-111.
Mantel N, Haenszel W: Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959, 22: 719-748.
DerSimonian R, Laird N: Meta-analysis in clinical trials. Control Clin Trials. 1986, 7: 177-188. 10.1016/0197-2456(86)90046-2.
Sharp L, Little J, Schofield AC, Pavlidou E, Cotton SC, Miedzybrodzka Z, Baird JO, Haites NE, Heys SD, Grubb DA: Folate and breast cancer: the role of polymorphisms in methylenetetrahydrofolate reductase (MTHFR). Cancer Lett. 2002, 181: 65-71. 10.1016/S0304-3835(02)00030-7.
Campbell IG, Baxter SW, Eccles DM, Choong DY: Methylenetetrahydrofolate reductase polymorphism and susceptibility to breast cancer. Breast Cancer Res. 2002, 4 (6): R14-10.1186/bcr457.
Semenza JC, Delfino RJ, Ziogas A, Anton-Culver H: Breast cancer risk and methylenetetrahydrofolate reductase polymorphism. Breast Cancer Res Treat. 2003, 77: 217-223. 10.1023/A:1021843019755.
Langsenlehner U, Krippl P, Renner W, Yazdani-Biuki B, Wolf G, Wascher TC, Paulweber B, Weitzer W, Samonigg H: The common 677C>T gene polymorphism of methylenetetrahy- drofolate reductase gene is not associated with breast cancer risk. Breast Cancer Res Treat. 2003, 81: 169-172. 10.1023/A:1025752420309.
Chen J, Gammon MD, Chan W, Palomeque C, Wetmur JG, Kabat GC, Teitelbaum SL, Britton JA, Terry MB, Neugut AI, Santella RM: One-carbon metabolism, MTHFR polymorphisms, and risk of breast cancer. Cancer Res. 2005, 65: 1606-1614. 10.1158/0008-5472.CAN-04-2630.
Shrubsole MJ, Gao YT, Cai Q, Shu XO, Dai Q, Hebert JR, Jin F, Zheng W: MTHFR polymorphisms, dietary folate intake, and breast cancer risk: results from the Shanghai Breast Cancer Study. Cancer Epidemiol Biomarkers Prev. 2004, 13: 190-196. 10.1158/1055-9965.EPI-03-0273.
Forsti A, Angelini S, Festa F, Sanyal S, Zhang Z, Grzybowska E, Pamula J, Pekala W, Zientek H, Hemminki K, Kumar R: Single nucleotide polymorphisms in breast cancer. Oncol Rep. 2004, 11: 917-922.
Lee SA, Kang D, Nishio H, Lee MJ, Kim DH, Han W, Yoo KY, Ahn SH, Choe KJ, Hirvonen A, Noh DY: Methylenete-trahydrofolate reductase polymorphism, diet, and breast cancer in Korean women. Exp Mol Med. 2004, 36: 116-121.
Grieu F, Powell B, Beilby J, Iacopetta B: Methylenete-Trahydrofolate reductase and thymidylate synthase polymorphisms are not associated with breast cancer risk or phenotype. Anticancer Res. 2004, 24: 3215-3219.
Lin WY, Chou YC, Wu MH, Huang HB, Jeng YL, Wu CC, Yu CP, Yu JC, You SL, Chu TY, Chen CJ, Sun CA: The MTHFR C677T polymorphism, estrogen exposure and breast cancer risk: a nested case–control study in Taiwan. Anticancer Res. 2004, 24: 3863-3868.
Qi J, Miao XP, Tan W, Yu CY, Liang G, Lu WF, Lin DX: Association between genetic polymorphisms in methylenetetrahydrofolate reductase and risk of breast cancer. Chin J Oncol. 2004, 26: 287-289.
Justenhoven C, Hamann U, Pierl CB, Rabstein S, Pesch B, Harth V, Baisch C, Vollmert C, Illig T, Bruning T, Ko Y, Brauch H: One-carbon metabolism and breast cancer risk: no association of MTHFR, MTR, and TYMS polymorphisms in the GENICA study from Germany. Cancer Epidemiol Biomark Prev. 2005, 14: 3015-3018. 10.1158/1055-9965.EPI-05-0592.
Kalemi TG, Lambropoulos AF, Gueorguiev M, Chrisafi S, Papazisis KT, Kotsis A: The association of p53 mutations and p53 codon 72, Her 2 codon 655 and MTHFR C677T polymorphisms with breast cancer in Northern Greece. Cancer Lett. 2005, 222: 57-65. 10.1016/j.canlet.2004.11.025.
Deligezer U, Akisik EE, Dalay N: Homozygosity at the C677T of the MTHFR gene is associated with increased breast cancer risk in the Turkish population. In Vivo. 2005, 19: 889-893.
Ergul E, Sazci A, Utkan Z, Canturk NZ: Polymorphisms in the MTHFR gene are associated with breast cancer. Tumour Biol. 2003, 24: 286-290. 10.1159/000076460.
Chou YC, Wu MH, Yu JC, Lee MS, Yang T, Shih HL, Wu TY, Sun CA: Genetic polymorphisms of the methylenete-trahydrofolate reductase gene, plasma folate levels and breast cancer susceptibility: a case–control study in Taiwan. Carcinogenesis. 2006, 27: 2295-2300. 10.1093/carcin/bgl108.
Kalyankumar C, Jamil K: Methylene tetrahydofolate reductase (MTHFR) C677T and A1298C polymorphisms and breast cancer in South Indian population. Int J Cancer Res. 2006, 2: 143-151.
Xu X, Gammon MD, Zhang H, Wetmur JG, Rao M, Teitelbaum SL, Britton JA, Neugut AI, Santella RM, Chen J: Polymorphisms of one-carbon-metabolizing genes and risk of breast cancer in a population-based study. Carcinogenesis. 2007, 28: 1504-1509. 10.1093/carcin/bgm061.
Hekim N, Ergen A, Yaylim I, Yilmaz H, Zeybek U, Ozturk O, Isbir T: No association between methylenetetrahydrofolate reductase C677T polymorphism and breast cancer. Cell Biochem Funct. 2007, 25: 115-117. 10.1002/cbf.1274.
Macis D, Maisonneuve P, Johansson H, Bonanni B, Botteri E, Iodice S, Santillo B, Penco S, Gucciardo G, D’Aiuto G, Rosselli Del Turco M, Amadori M, Costa A, Decensi A: Methylenetetrahydrofolate reductase (MTHFR) and breast cancer risk: a nested-case–control study and a pooled meta-analysis. Breast Cancer Res Treat. 2007, 106: 263-271. 10.1007/s10549-006-9491-6.
Yu CP, Wu MH, Chou YC, Yang T, You SL, Chen CJ, Sun CA: Breast cancer risk associated with multigenotypic poly-Morphisms in folate-metabolizing genes: a nested case–control study in Taiwan. Anticancer Res. 2007, 27: 1727-1732.
Reljic A, Simundic AM, Topic E, Nikolac N, Justinic D, Stefanovic M: The methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and cancer risk: the Croatian case–control study. Clin Biochem. 2007, 40: 981-985. 10.1016/j.clinbiochem.2007.05.005.
Inoue M, Robien K, Wang R, Van Den Berg DJ, Koh WP, Yu MC: Green tea intake, MTHFR/TYMS genotype and breast cancer risk: the Singapore Chinese Health Study. Carcinogenesis. 2008, 29: 1967-1972. 10.1093/carcin/bgn177.
Kotsopoulos J, Zhang WW, Zhang S, McCready D, Trudeau M, Zhang P, Sun P, Narod SA: Polymorphisms in folate metabolizing enzymes and transport proteins and the risk of breast cancer. Breast Cancer Res Treat. 2008, 112: 585-593. 10.1007/s10549-008-9895-6.
Suzuki T, Matsuo K, Hirose K, Hiraki A, Kawase T, Watanabe M, Yamashita T, Iwata H, Tajima K: One-carbon metab-olism-related gene polymorphisms and risk of breast cancer. Carcinogenesis. 2008, 2: 356-362. 10.1093/carcin/bgm295.
Cheng CW, Yu JC, Huang CS, Shieh JC, Fu YP, Wang HW, Wu PE, Shen CY: Polymorphism of cytosolic serine hydroxymethyltransferase, estrogen and breast cancer risk among Chinese women in Taiwan. Breast Cancer Res Treat. 2008, 111: 145-155. 10.1007/s10549-007-9754-x.
Langsenlehner T, Renner W, Yazdani-Biuki B, Langsenlehner U: Methylenetetrahydrofolatereductase (MTHFR) and breast cancer risk: a nested-case–control study and a pooled meta-analysis. Breast Cancer Res Treat. 2008, 107: 459-460. 10.1007/s10549-007-9564-1.
Ericson U, Sonestedt E, Ivarsson MI, Gullberg B, Carlson J, Olsson H, Wirfalt E: Folate intake, methylenetetrahydrofolate reductase polymorphisms, and breast cancer risk in women from the Malmo¨ Diet and Cancer cohort. Cancer Epidemiol Biomarkers Prev. 2009, 18: 1101-1110. 10.1158/1055-9965.EPI-08-0401.
Gao CM, Tang JH, Cao HX, Ding JH, Wu JZ, Wang J, Liu YT, Li SP, Su P, Matsuo K, Takezaki T, Tajima K: MTHFR polymorphisms, dietary folate intake and breast cancer risk in Chinese women. J Hum Genet. 2009, 54: 414-418. 10.1038/jhg.2009.57.
Ma E, Iwasaki M, Kobayashi M, Kasuga Y, Yokoyama S, Onuma H, Nishimura H, Kusama R, Tsugane S: Dietary intake of folate, vitamin B2, vitamin B6, vitamin B12, genetic polymorphism of related enzymes, and risk of breast cancer: a case–control study in Japan. Nutr Cancer. 2009, 61: 447-456. 10.1080/01635580802610123.
Platek ME, Shields PG, Marian C, McCann SE, Bonner MR, Nie J, Ambrosone CB, Millen AE, Ochs-Balcom HM, Quick SK, et al.: Alcohol consumption and genetic variation in Methylenetetrahydrofolate reductase and 5-methyltetrahydrofolate homocysteine methyltransferase in relation to breast cancer risk. Cancer Epidemiol Biomark Prev. 2009, 18: 2453-2459. 10.1158/1055-9965.EPI-09-0159.
Henrıquez-Hernandez LA, Murias-Rosales A, Hernandez Gon-zalez A, Cabrera De Leon A, Dıaz-Chico BN, Mori De Santiago M, Fernandez Perez L: Gene polymorphisms in TYMS, MTHFR, p53 and MDR1 as risk factors for breast cancer: a case–control study. Oncol Rep. 2009, 22: 1425-1433.
Cam R, Eroglu A, Egin Y, Akar N: Dihydrofolate reduc-Tase (DHRF) 19-bp intron-1 deletion and methylenetetrahydrofolate reductase (MTHFR) C677T polymorphisms in breast cancer. Breast Cancer Res Treat. 2009, 115: 431-432. 10.1007/s10549-008-0054-x.
Maruti SS, Ulrich CM, Jupe ER, White E: MTHFR C677T and postmenopausal breast cancer risk by intakes of one-carbon metabolism nutrients: a nested case–control study. Breast Cancer Res. 2009, 11: R91-10.1186/bcr2462.
Ma E, Iwasaki M, Junko I, Hamada GS, Nishimoto IN, Carvalho SM, Motola J, Laginha FM, Tsugane S: Dietary intake of folate, vitamin B6, and vitamin B12, genetic polymorphism of related enzymes, and risk of breast cancer: a case–control study in Brazilian women. BMC Cancer. 2009, 9: 122-10.1186/1471-2407-9-122.
Li WD, Chen SQ: Association of methylenetetrahydrofolate reductase C677T polymorphism and breast cancer risk. J Prac Med. 2009, 25: 2031-2033.
Yuan H, Xu XY, Wang ZL: The relation between polymorphisms of methylenetetrahydrofolate reductase C677T and the risk of breast cancer. J MuDanJiang Med Univ. 2009, 30: 2-4.
Jin ZZ, Lu Q, Ge DH, Zong M, Zhu QH: Effect of the methylenetetrahydrofolate reductase gene C677T polymorphism on C-erbB-2 methylation status and its association with cancer. Mol Med Rep. 2009, 2: 283-289.
Bentley AR, Raiszadeh F, Stover PJ, Hunter DJ, Hankinson SE, Cassano PA: No association between cSHMT genotypes and the risk of breast cancer in the Nurses’ Health Study. Eur J Clin Nutr. 2010, 64: 108-110. 10.1038/ejcn.2009.104.
Alshatwi AA: Breast cancer risk, dietary intake, and methylenetetrahydrofolate reductase (MTHFR) single nucleotide polymorphisms. Food Chem Toxicol. 2010, 48: 881-1885.
Sangrajrang S, Sato Y, Sakamoto H, Ohnami S, Khuhaprema T, Yoshida T: Genetic polymorphisms in folate and alcohol metabolism and breast cancer risk: a case–control study in Thai women. Breast Cancer Res Treat. 2010, 123: 885-893. 10.1007/s10549-010-0804-4.
Weiner AS, Boyarskih UA, Voronina EN, Selezneva IA, Sinkina TV, Lazarev AF, Petrova VD, Filipenko ML: Polymorphic Variants of Folate Metabolizing Genes (C677T and A1298C MTHFR and C1420T SHMT1 and G1958A MTHFD) are Not Associated with the Risk of Breast Cancer in the West Siberian Region of Russia. Mol Biol. 2010, 44 (5): 720-727. 10.1134/S0026893310050067.
Vidudala V.T.S. Prasad Harpreet Wilkhoo: Association of the Functional Polymorphism C677T in the Methylenetetrahydrofolate Reductase Gene with Colorectal, Thyroid, Breast, Ovarian, and Cervical Cancers. Onkologie. 2011, 34: 422-426. 10.1159/000331131.
Batschauer AP, Cruz NG, Oliveira VC, Coelho FF, Santos IR, Alves MT, Fernandes AP, Carvalho MG, Gomes KB: HFE, MTHFR, and FGFR4 genes polymorphisms and breast cancer in Brazilian women. Mol Cell Biochem. 2011, 357: 247-253. 10.1007/s11010-011-0895-1.
Mohammad NS, Rupasree Y, Addepalli P, Gottumukkala SR, Digumarti RR, Kutala VK: Aberrations in one-carbon metabolism induce oxidative DNA damage in sporadic breast cancer. Mol Cell Biochem. 2011, 349: 159-167. 10.1007/s11010-010-0670-8.
Mohammad NS, Addepalli P, Digumarti RR, Gottumukkala SR, Kutala VK: Epistatic interactions between loci of one-carbon metabolism modulate susceptibility to breast cancer. Mol Biol Rep. 2011, 38: 4893-4901. 10.1007/s11033-010-0631-z.
Ja SMina Ziva C, Vida STG, KSeniJa GSK, Kovic SJn: Lack of association between methylenetetrahydrofolate reductase genetic polymorphisms and postmenopausal breast cancer risk. Molecular Medicine Reports. 2011, 4: 175-179.
Muhammad A, Malik FA, Mahmood Akhtar K: Mutational Analysis of the MTHFR Gene in Breast Cancer Patients of Pakistani Population. Asian Pacific J Cancer Pre. 2012, 13: 1599-1603. 10.7314/APJCP.2012.13.4.1599.
de Cassia Carvalho Barbosa R, Debora Costa M, Thiago Fernando Vasconcelos F, Diogo Campos S, Victor Hugo Medeiros A, Silvia Helena Barem R: Associations of polymorphisms of folate cycle enzymes and risk of breast cancer in a Brazilian population are age dependent. Mol Biol Rep. 2012, 39: 4899-4907. 10.1007/s11033-011-1285-1.
Lajin B, Sakur AA, Ghabreau L, Alachkar A: Association of polymorphisms in one-carbon metabolizing genes with breast cancer risk in Syrian women. Tumor Biol. 2012, 33: 1133-1139. 10.1007/s13277-012-0354-y.
Jakubowska A, Rozkrut D, Antoniou A, et al.: Association of PHB 1630 C4T and MTHFR 677 C>T polymorphisms with breast and ovarian cancer risk in BRCA1/2 mutation carriers: results from a multicenter study. British Journal of Cancer. 2012, 106: 2016-2024. 10.1038/bjc.2012.160.
Skoog T, Van't Hooft FM, Kallin B, Jovinge S, Boquist S, Nilsson J, Eriksson P, Hamsten A: A common functional polymorphism (C-->A substitution at position −863) in the promoter region of the tumor necrosis factor-alpha (TNF-alpha) gene associated with reduced circulating levels of TNF-alpha. Hum Mol Genet. 1999, 8: 1443-1449. 10.1093/hmg/8.8.1443.
Momparler RL, Bovenzi V: DNA methylation and cancer. J Cell Physiol. 2000, 183: 145-154. 10.1002/(SICI)1097-4652(200005)183:2<145::AID-JCP1>3.0.CO;2-V.
Shen H, Wang L, Spitz MR, Hong WK, Mao L, Wei Q: A novel polymorphism in human cytosine DNA-methyltransferase-3B promoter is associated with an increased risk of lung cancer. Cancer Res. 2002, 62: 4992-4995.
Fujisawa T, Ikegami H, Kawaguchi Y: Meta-analysis of association of insertion/deletion polymorphism of angiotensin I-converting enzyme gene with diabetic nephropathy and retinopathy. Diabetologia. 1998, 41 (1): 47-53. 10.1007/s001250050865.
Liwei L, Chunyu L, Ruifa H: Association between manganese superoxide dismutase gene polymorphism and risk of prostate cancer: a meta-analysis. Urology. 2009, 74 (4): 884-888. 10.1016/j.urology.2008.11.016.
Acknowledgements
This work was not supported by any kind of fund.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interest
Both authors declared that they have no conflict interest in relation to this study.
Authors’ contributions
LY drafted the manuscript, and carried out the molecular genetic studies, participated in the sequence alignment and JC drafted the manuscript, carried out the molecular genetic studies, participated in the sequence alignment and reviewed the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Yu, L., Chen, J. Association of MHTFR Ala222Val (rs1801133) polymorphism and breast cancer susceptibility: An update meta-analysis based on 51 research studies. Diagn Pathol 7, 171 (2012). https://doi.org/10.1186/1746-1596-7-171
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1746-1596-7-171