Abstract
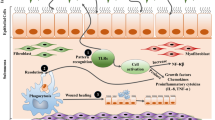
Interleukin (IL)-9 is a pleiotropic T helper 2-type cytokine that has been shown to be up-regulated in allergic airway disease, including asthma. IL-9 has been demonstrated to be a potent stimulus for the production and secretion of mucus from airway epithelial cells via induction of a calcium-activated chloride channel, hCLCA1. The objective of this study was to investigate the expression of IL-9 and hCLCA1 following allergen challenge in the nasal mucosa of allergic rhinitis patients. Nasal biopsies were obtained from allergic rhinitis patients out of allergen season both before (baseline) and after local antigen challenge with either ragweed or diluent (control). Immunohistochemistry and in situ hybridization were used to assess IL-9 protein and hCLCA1 messenger ribonucleic acid. Eosinophils and T cells were detected using immunohistochemistry. IL-9 and hCLCA1 were very low at baseline, and expression was significantly up-regulated following ragweed challenge. Whereas the number of eosinophils increased after allergen challenge, T-cell counts did not change significantly. The results of this study demonstrate the relationship between specific allergen challenge and expression of both IL-9 and hCLCA1, suggesting a possible mechanism for the increased production of mucus from airway epithelial cells in allergic rhinitis.
Similar content being viewed by others
Interleukin (IL)-9 is a pleiotropic T helper 2-type cytokine that has been shown to be associated with airway hyperresponsiveness and mucus hypersecretion in bronchial asthma [1, 2]. Animal studies using transgenic IL-9 overexpressing mice have demonstrated that elevated IL-9 levels lead to increased inflammatory cell infiltration (lymphocytes and eosinophils), goblet cell hyperplasia, and mucus over-production [3–5]. Instillation of exogenous IL-9 into the airway of B6 mice was associated with the specific up-regulation of MUC2 and MUC5AC mucin gene products [6]. In vitro, stimulation of airway epithelial cells with IL-9 led to up-regulation of chemokine expression and induction of several mucin genes, including MUC2 and MUC5AC [6–8].
It was shown in a previous study that the expression of the calcium-activated chloride channel hCLCA1 in human primary lung epithelial cells is up-regulated by IL-9[9]. Transfection of hCLCA1 into human mucoepidermoid cells resulted in up-regulation of the MUC5AC gene [10]. Intratracheal administration of adenovirus-expressing antisense ribonucleic acid (RNA) for gob-5 (mCLCA3, the murine counterpart of hCLCA1) into mice suppressed mucus overproduction following antigen challenge [10].
Toda and colleagues demonstrated increased protein levels of IL-9, IL-9 receptor, and messenger ribonucleic acid (mRNA) levels of hCLCA1 in the airways of asthmatic patients [11]. In that study, a strong correlation between IL-9, the IL-9 receptor, and hCLCA1 mRNA was observed [11]. These data strongly support the hypothesis that hCLCA1 is highly involved in mucus overproduction in airway inflammatory conditions.
Thus, given the important role that IL-9 plays in the maintenance of allergic responses and the association of the IL-9-induced chloride channel hCLCA1 with mucus overproduction, we sought to characterize the expression of IL-9 and hCLCA1 in the nasal mucosa of allergic patients following local specific allergen challenge. We propose that IL-9 and hCLCA1 expression is increased after allergen challenge.
Materials and methods
Allergen Challenge and Tissue Collection
Fourteen patients presenting with symptoms of allergic rhinitis with sensitization for seasonal allergens were recruited. Allergen sensitization was confirmed by skinprick test. Biopsies were obtained from the inferior nasal turbinate out of season (baseline). After 6 weeks, patients were challenged with either ragweed (n = 7) or diluent (saline, n = 7) by nasal spray. The second biopsies were taken 24 hours after challenge. Subjects showed typical clinical signs of late-phase response following specific allergen challenge, including sneezing, itchiness, and runny nose. Tissue was fixed in 4% paraformaldehyde, washed in a solution of 15% sucrose/phosphate-buffered saline, and blocked in optimal cutting temperature medium by snapfreezing in isopentane cooled in liquid nitrogen.
Probe Preparation
Sulphur 35 (S35)-labeled complementary RNA probe coding for the murine homologue of hCLCA1 mRNA was prepared from complementary deoxyribonucleic acid (cDNA) (Genaera Pharmaceuticals, Plymouth Meeting, PA), as described previously [12]. In brief, cDNA was inserted into expression vectors, linearized, and transcribed in vitro in the presence of S35-UTP, T7, and SP6 polymerase in either direction to produce antisense (complementary) and sense probes (identical to mRNA).
In Situ Hybridization
Sections of nasal mucosa were processed for in situ hybridization to identify hCLCA1 mRNA, according to the method of Hamid and colleagues [12, 13]. Briefly, after permeabilization with Triton X-100 and proteinase K solution (1 μg/mL), sections were prehybridized with 50% formamide in 2× standard sodium citrate for 15 minutes at 37°C. Hybridization was carried out overnight at 42°C with the hybridization mixture containing the appropriate S35-labeled sense or antisense probe (0.75 × 106 cpm/slide). Posthybridization involved high-stringency washings of the samples in decreasing concentrations of standard saline citrate at 42°C. To remove unbound RNA probes, the samples were washed with ribonuclease solution for 20 minutes at 42°C. The samples were then dehydrated with increasing concentrations of ethanol and air-dried. After this, the samples were dipped in Amersham LM-2 emulsion and then exposed for a period of 14 days. The autoradiographs were then developed in Kodak D-19 developer, fixed, and counterstained with periodic acid-Schiff (PAS). The samples were then mounted with a coverslip and examined under a graduated microscope for positive signals.
Immunohistochemistry
Immunohistochemistry was used to detect eosinophils, T cells, and IL-9 immunoreactivity within the sections. Immunostaining was performed with specific antibodies to eosinophils (anti-major basic protein [MBP], a gift from Dr. Moqbel), T cells (anti-CD3; Dako Diagnostics, Canada), and IL-9 (anti-IL-9, R&D Systems, Minneapolis, MN) and the alkaline phosphatase-antialkaline phosphatase technique, as previously described [14]. For negative control preparations, the primary antibody was replaced by either nonspecific immunoglobulin or TRIS-buffered saline solution.
Quantification
All slides were read blinded (coded), and positive cells were counted. PAS-positive epithelial cells with hCLCA1 mRNA signals were expressed in a quantification score from 0 (no positive cells) to 8 (all cells positive). IL-9-, CD3-, and MBP-immunoreactive cells were expressed as the number of positive cells per high-powered field (magnification × 200). The within-observer coefficient of variation for repeated measures was less than 5%.
Statistical Analysis
The difference in the number of cells expressing hCLCA1 mRNA, IL-9, CD3, or MBP before and after allergen or saline challenge was compared using Wilcoxon's signed rank test. Statistically significant differences between groups were subsequently analyzed with the Mann- Whitney U test (Systat, version 7.0, SPSS, Chicago, IL). The Results are expressed as means ± SD and were significant at a p value of less than .05.
Results
Baseline Characteristics
The numbers of T cells, eosinophils, IL-9-positive cells, and hCLCA1-positive epithelial cells were quantified before challenge. IL-9 immunoreactivity was localized to submucosal inflammatory cells. Expression of hCLCA1 was found in the epithelium in mucus-producing cells.
Most of the hCLCA1-positive epithelial cells stained positive for mucus (90%). Conversely, the majority of PAS-positive cells showed expression of hCLCA1 (95%). There was a strong significant correlation between hCLCA1 and mucus expression (r = .93; p < .05). There was no significant difference before challenge between the ragweed challenge group and the saline challenge group (p > .05) (Table 1).
Increase in Eosinophils, IL-9-Positive Cells, and hCLCA1 Expression Following Allergen Challenge
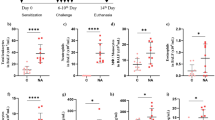
Following ragweed challenge, there was a sharp increase in eosinophils, IL-9-positive cells, and hCLCA1-positive mucous epithelial cells (p < .05) whereas no significant change occurred after saline challenge (p > .05) (Figures 1 to 3). The numbers of T cells did not change significantly after ragweed or saline challenge (p > .05) (Figure 4).
Interleukin (IL)-9-positive cells in the submucosa before and after challenge with either saline or ragweed. There is a significant increase in IL-9-positive cells after ragweed challenge but not after saline compared with baseline. Value pairs for each patient before and after challenge are shown by connected dots. Mean values are indicated by lines. *p < .01.
Expression of hCLCA1 in mucus-producing epithelial cells before and after challenge with either saline or ragweed. There is a significant increase in hCLCA1 expression after ragweed challenge but not after saline compared with baseline. Value pairs for each patient before and after challenge are shown by connected dots. Mean values are indicated by lines. *p < .01. Epithelial hCLCA1 expression is scored from 0 (no cells) to 8 (all cells).
Eosinophils in the submucosa before and after challenge with either saline or ragweed. There is a significant increase in eosinophils after ragweed challenge but not after saline compared with baseline. Value pairs for each patient before and after challenge are shown by connected dots. Mean values are indicated by lines. *p < .01.
CD3-positive T lymphocytes in the submucosa before and after challenge with either saline or ragweed. There is no significant change after either saline or ragweed challenge compared with baseline. Value pairs for each patient before and after challenge are shown by connected dots. Mean values are indicated by lines.
Discussion
In this study, we found that specific allergen challenge significantly increased eosinophils, IL-9-positive cells, and expression of hCLCA1 in allergic nasal mucosa whereas saline challenge did not have any significant effect.
We observed increased numbers of eosinophils after ragweed challenge but not after saline challenge. Eosinophilic influx after allergen challenge has been reported.15 No increase in CD3-positive T cells could be found in our study. However, activated T cells (CD25+) and T helper cells (CD4+) might be increased after allergen challenge, as previously shown [15].
The number of IL-9-positive cells was significantly increased following ragweed challenge whereas saline had no effect. IL-9 immunoreactivity was localized to submucosal inflammatory cells. Although we did not perform colocalization, most of the IL-9-positive cells might be either T cells (as demonstrated previously)[2] or eosinophils. No increase in T cells number was detected; thus, T cells needed to be activated to produce more IL-9 following allergen challenge. Eosinophils might also be partly responsible for this increased IL-9 expression. IL-9 was recently reported to be up-regulated following allergen challenge in a human skin model [16]. In that study, IL-9- positive mRNA cells were significantly elevated in the skin biopsies of atopic subjects at 48 hours after allergen challenge--a late-phase reaction [16]. However, IL-9 expression has not been studied in airway tissue following allergen challenge. Our study showed a marked increase in IL-9-positive cells at 24 hours, the late-phase reaction, specific to allergen in human airway mucosa.
IL-9 has been shown to induce mucus production [6, 7] by up-regulation of the expression of hCLCA1 [9]. However, IL-9-induced expression of hCLCA1 has been demonstrated only in mice and cell culture [9]. In our study, we found elevated expression of hCLCA1 associated with increased numbers of IL-9-positive cells following allergen challenge. These data support previous data showing IL-9-induced expression of hCLCA1 [9]. Moreover, our findings support the notion that hCLCA1 plays a role in mucus overproduction in allergic airway disease, especially after exposure to allergen.
Recently, mCLCA3 (the mouse homologue of hCLCA1) has been reported to be involved in JAK/STAT6 signalling following stimulation with IL-13[17]. In that study, niflumic acid, an inhibitor of hCLCA1, reduced airway inflammation and mucus expression. These data also show the potential role of hCLCA1 in allergic inflammation.
In conclusion, this report provides a link between allergen exposure and increased mucus production via IL-9-induced expression of hCLCA1. Further studies are warranted to investigate whether inhibition of either IL-9 or hCLCA1 offers a potential treatment option for mucus hypersecretion and tissue eosinophilia.
References
Nicolaides NC, Holroyd KJ, Ewart SL: Interleukin 9: a candidate gene for asthma. Proc Natl Acad Sci USA. 1997, 94: 13175-80. 10.1073/pnas.94.24.13175.
Shimbara A, Christodoulopoulos P, Soussi-Gounni A: IL-9 and its receptor in allergic and nonallergic lung disease: increased expression in asthma. J Allergy Clin Immunol. 1999, 104: 108-15.
McLane MP, Haczku A, van Rijn M: Interleukin-9 promotes allergen-induced eosinophilic inflammation and airway hyperresponsiveness in transgenic mice. Am J Respir Cell Mol Biol. 1998, 19: 713-20.
Dong Q, Louahed J, Vink A: IL-9 induces chemokine expression in lung epithelial cells and baseline airway eosinophilia in transgenic mice. Eur J Immunol. 1999, 29: 2130-9. 10.1002/(SICI)1521-4141(199907)29:07<2130::AID-IMMU2130>3.0.CO;2-S.
Temann UA, Ray P, Flavell RA: Pulmonary over-expression of IL-9 induces Th2 cytokine expression, leading to immune pathology. J Clin Invest. 2002, 109: 29-39.
Louahed J, Toda M, Jen J: Interleukin-9 upregulates mucus expression in the airways. Am J Respir Cell Mol Biol. 2000, 22: 649-56.
Longphre M, Li D, Gallup M: Allergeninduced IL-9 directly stimulates mucin transcription in respiratory epithelial cells. J Clin Invest. 1999, 104: 1375-82. 10.1172/JCI6097.
Little F, Cruikshank WW, Center DM: IL-9 stimulates release of chemotactic factors from human bronchial epithelial cells. Am J Respir Cell Mol Biol. 2001, 25: 347-52.
Zhou Y, Dong Q, Louahed J: Characterization of a calcium-activated chloride channel as a shared target of Th2 cytokine pathways and its potential involvement in asthma. Am J Respir Cell Mol Biol. 2001, 25: 486-91.
Nakanishi A, Morita S, Iwashita H: Role of gob-5 in mucus overproduction and airway hyperresponsiveness in asthma. Proc Natl Acad Sci USA. 2001, 98: 5175-80. 10.1073/pnas.081510898.
Toda M, Tulic MK, Levitt RC, Hamid Q: A calcium-activated chloride channel (hCLCA1) is strongly related to IL-9 expression and mucus production in bronchial epithelium of patients with asthma. J Allergy Clin Immunol. 2002, 109: 246-50. 10.1067/mai.2002.121555.
Hamid Q, Wharton J, Terenghi G: Localization of atrial natriuretic peptide mRNA and immunoreactivity in the rat heart and human atrial appendage. Proc Natl Acad Sci USA. 1987, 84: 6760-4. 10.1073/pnas.84.19.6760.
Hamid Q, Azzawi M, Ying S: Expression of mRNA for interleukin-5 in mucosal bronchial biopsies from asthma. J Clin Invest. 1991, 87: 1541-6. 10.1172/JCI115166.
Hamid Q, Springall DR, Riveros-Moreno V: Induction of nitric oxide synthase in asthma. Lancet. 1993, 342: 1510-3. 10.1016/S0140-6736(05)80083-2.
Varney VA, Jacobson MR, Sudderick RM: Immunohistology of the nasal mucosa following allergen-induced rhinitis. Identification of activated T lymphocytes, eosinophils, and neutrophils. Am Rev Respir Dis. 1992, 146: 170-6.
Ying S, Meng Q, Kay AB, Robinson DS: Elevated expression of interleukin-9 mRNA in bronchial mucosa of atopic asthmatics and allergen-induced cutaneous late-phase reaction: relationships to eosinophils, mast cells and T lymphocytes. Clin Exp Allergy. 2002, 32: 866-71. 10.1046/j.1365-2222.2002.01376.x.
Nakona T, Inoue H, Fukuyama S: Niflumic acid suppresses interleukin-13-induced asthma phenotypes. Am J Respir Crit Care Med. 2006,
Acknowledgements
This study was supported by Genaera Corporation. Hans-Peter Hauber is a recipient of a GSK/CLA/CIHR fellowship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Hauber, HP., Bergeron, C., Toda, M. et al. Up-Regulation of Interleukin-9 and the Interleukin-9-Associated Calcium-Activated Chloride Channel hCLCA1 in Nasal Mucosa Following In Vivo Allergen Challenge. All Asth Clin Immun 3, 19 (2007). https://doi.org/10.1186/1710-1492-3-1-19
Published:
DOI: https://doi.org/10.1186/1710-1492-3-1-19