Abstract
Background
Pseudoalteromonas is an important genus widespread in marine environment, and a lot of psychrophilic Pseudoalteromonas strains thrive in deep sea and polar sea. By now, there are only a few genetic systems for Pseudoalteromonas reported and no commercial Pseudoalteromonas genetic system is available, which impedes the study of Pseudoalteromonas, especially for psychrophilic strains. The aim of this study is to develop a heterologous expression system for psychrophilic Pseudoalteromonas.
Results
A cryptic plasmid pSM429 isolated from psychrophilic Pseudoalteromonas sp. BSi20429 from the Arctic sea ice, was sequenced and characterized. The plasmid pSM429 is 3874 bp in length, with a G+C content of 28%. Four putative open reading frames (ORFs) were identified on pSM429. Based on homology, the ORF4 was predicted to encode a replication initiation (Rep) protein. A shuttle vector (Escherichia coli, Pseudoalteromonas), pWD, was constructed by ligating pSM429 and pUC19 and inserting a chloramphenicol acetyl transferase (CAT) cassette conferring chloramphenicol resistance. To determine the minimal replicon of pSM429 and to check the functionality of identified ORFs, various pWD derivatives were constructed. All derivatives except the two smallest ones were shown to allow replication in Pseudoalteromonas sp. SM20429, a plasmid-cured strain of Pseudoalteromonas sp. BSi20429, suggesting that the orf4 and its flanking intergenic regions are essential for plasmid replication. Although not essential, the sequence including some repeats between orf1 and orf2 plays important roles in segregational stability of the plasmid. With the aid of pWD-derived plasmid pWD2, the erythromycin resistance gene and the cd gene encoding the catalytic domain of a cold-adapted cellulase were successfully expressed in Pseudoalteromonas sp. SM20429.
Conclusions
Plasmid pSM429 was isolated and characterized, and the regions essential for plasmid replication and stability were determined, helping the development of pSM429-based shuttle vectors. The shuttle vectors pWD and its derivatives could be used as cloning vectors for Pseudoalteromonas, offering new perspectives in the genetic manipulation of Pseudoalteromonas strains. With the aid of pWD-derived vector and its host, the erythromycin resistance gene and the cd gene of a cold-adapted protein were successfully expressed, indicating that the potential use of this system for recombinant protein production, especially for cold-adapted proteins.
Similar content being viewed by others
Introduction
Cold-adapted bacteria are excellent candidates for investigations of protein evolution and molecular adaptations to extreme conditions. Moreover, cold-adapted enzymes from psychrophiles offer novel opportunities for biotechnological applications [1, 2]. Native plasmids from cold-adapted bacteria are of particular interest, because plasmids contribute directly in the adaptation of bacteria to their natural environments and provide easy model systems for investigation of basic molecular processes [3]. Furthermore, native plasmids can be used in developing genetic systems.
Pseudoalteromonas is a genus of gamma-proteobacteria that is widespread in the world's oceans, and a lot of psychrophilic Pseudoalteromonas strains thrive in deep sea and polar sea. The genus Pseudoalteromonas contains over 30 marine species [4], which play an important role in marine ecosystem. Pseudoalteromonas has attracted significant interest for two reasons. First, Pseudoalteromonas species are important for investigation of microbe-host interactions in the sea. Second, many species synthesize biologically active molecules, such as extracellular antibiotics, toxins, extracellular enzymes and polysaccarides [4–8]. However, by now there are only a few genetic systems for Pseudoalteromonas reported [9–12] and no commercial Pseudoalteromonas genetic system is available, which impedes the study of Pseudoalteromonas, especially for psychrophilic strains.
In our laboratory, psychrophilic Pseudoalteromonas strains have been studied for over ten years on their cold-adaptation mechanism and the proteases they secreted [5, 13–18]. To further study the cold-adaptation mechanism of psychrophilic Pseudoalteromonas and protease expression, development of genetic systems for gene knockout and cold-adapted protein expression is essential. In this article, a small cryptic plasmid from psychrophilic Pseudoalteromonas sp. BSi20429 (BSi20429 thereafter), designated pSM429, was isolated and characterized. A shuttle vector pWD was constructed based on pSM429. The regions responsible for replication and stability on pWD were determined by studying the replication of pWD-derived plasmids in Pseudoalteromonas sp. SM20429 (SM20429 thereafter), a plasmid-cured strain of BSi20429. A stable pWD-derived plasmid, pWD2, was successfully used as an express vector to express the erythromycin resistance gene and the cd gene of a cold-adapted cellulase in SM20429.
Materials and methods
Bacterial strains and plasmid isolation
Pseudoalteromonas strains were previously isolated from the sea-ice samples collected using a MARKII ice auger during the Second Chinese National Arctic Research Expedition cruise of the USCGC icebreaker Xue Long into the Canada Basin in August 2003. 102 Pseudoalteromonas strains were screened for the presence of plasmids. Alkaline SDS lysis and DNA preparation method was used to isolate plasmids as described by Sambrook et al. [19]. Pseudoalteromonas sp. BSi20429 grown at 20°C in 2216E broth was found to carry a plasmid. Strain BSi20429, a gram negative bacterium, grows well in aerobic conditions at temperatures from 4°C to 34°C in 2216E broth (Difco) or marine LB broth (10 g peptone, 5 g yeast extract, 1 l artificial seawater, pH 7.5). The optimal temperature for BSi20429 growing is 20-25°C. Escherichia coli DH5α [(supE 44, △lac U169 (θ80lacZ△M15) endA1, recA1, hsdR17, thi-1 l- gyrA96, relA1] was grown at 37°C in LB medium [19] and used as a host for gene cloning and plasmid construction. Pseudoalteromonas transformants were selected in the presence of 8 μg/ml chloramphenicol and 25 μg/ml ampicillin, and further propagated in broth medium containing 20 μg/ml choramphenicol. Antibiotics were used in E. coli at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml. Plasmids used in this study were listed in Table 1.
Plasmid sequencing and analysis
The plasmid isolated from BSi20429 was termed as pSM429. Analysis of restriction digests on agarose gel showed that pSM429 contains singular restriction site for Hin dIII and Nde I, respectively. The plasmid was linearised by these two restriction enzymes, and restriction fragments of different sizes were cloned into pGEM-T Easy and sequenced by dideoxy chain termination method using an Applied Biosystems DNA sequencer. The nucleotide sequence of pSM429 was determined by using a combination of subcloning and primer walking. The whole plasmid sequence was covered at least three times by single read. The nucleotide sequence, amino acid sequence and the distribution of direct and inverted repeated sequences were analyzed using DNASTAR. The predicted protein-coding regions were initially defined by searching for ORFs longer than 80 codons. The potential coding regions were then confirmed with prokaryotic gene finder GeneMark.hmm 2.4 for Prokaryotes [20] at http://opal.biology.gatech.edu/GeneMark/gmhmm2_prok.cgi using Pseudoalteromonas haloplanktis TAC125 chromosome I genome as a model. The functional protein domain analysis of predicted ORFs was performed by the InterProScan program at the European Bioinformatics Institute http://www.ebi.ac.uk/InterProScan/. Similarity searches were carried out using Blast http://www.ncbi.nlm.nih.gov/blast/[21].
The nucleotide sequence of pSM429 was deposited in the GenBank database under the accession number EU627679.
Determination of plasmid copy number
The copy number of plasmid pSM429 in strain BSi20429 was determined using the method described by Qin et al. [22]. Strain BSi20429 was grown in marine LB broth at 20°C and total genome DNA was isolated by using PowerMax™Soil DNA Isolation Kit (MO BIO). The genome DNA was diluted by 10 and 100 times in TE Buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.5), and then was separated in equal volumes on a 0.7% agarose gel by electrophoresis at 50 V for 2.5 h. By comparing the intensity of the diluted chromosomal DNA band with the non-diluted plasmid DNA band using the image analysis software Gene Tools (Syngene), the copy number of pSM429 was calculated as follows:

(N c , copy number of the plasmid; S c , chromosomal DNA size (4.5 M); S p , plasmid DNA size (3.78 kb); I p , intensity of plasmid DNA band; I c , intensity of chromosomal DNA band).
Curing of pSM429 from BSi20429
BSi20429 was grown for at least 8 subcultures (approximately 160 generations) in 2216E broth containing 0.07% SDS. An aliquot of culture was serially diluted and spread onto 2216E agar. Colonies were screened for the presence of pSM429 as described above. Putative cured isolates were checked for plasmid integration into the genome by PCR with two pairs of primers covering different regions of pSM429 (RepF & RepR, ORF3F & ORF3R). Primer sequences used in this study were shown in Table 2.
Construction of the shuttle vector pWD and its derivatives
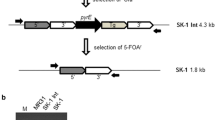
Plasmids pSM429 and pUC19 were isolated as described above. The complete DNA sequence of pSM429 was amplified by PCR with primers AF (containing a Sal I site) and AR (containing a Bam HI site). The 3.8-kb linearised pSM429 was digested with Sal I and Bam HI and cloned in pUC19. The chloramphenicol acetyl transferase (CAT) cassette was amplified by PCR from pBT (Stratagene), using primers CB (containing a Bam HI site) and CE (containing an Eco RI site). The 1.3-kb product was double-digested using Bam HI and Eco RI and ligated to the constructed pUC19+pSM429 treated with the same restriction enzymes, and then transformed into E. coli DH5α. The constructed shuttle vector was designated as pWD.
Derivatives of pWD (pWD1, pWD2, pWD3, pWD4, pWD5, pWD6) were constructed as outlined in Figure 1. Purified DNA of pSM429 was used as a template in PCR. All primers were designed with sites for Sal I or Bam HI at their 5' ends to allow direct cloning of the amplicons digested with these two enzymes into the Sal I and Bam HI sites of pWD. In new constructs, the complete fragment of pSM429 was replaced by the amplicons with different length. New constructed derivatives were transformed into E. coli DH5α. Plasmids isolated from E. coli DH5α and verified by sequencing were electroporated into SM20429.
Map of shuttle vector pWD (A) and schematic diagram of the pWD derivatives (B). Sal I and Bam HI sites were introduced with the oligonucleotide primers used for amplification. Primers BF/AR, BF/BR, BF/CR, BF/DR, BF/ER and CF/DR were used to amplify pSM429 derivatives pWD 1-6, respectively. Arrows denote direction and approximate length of the different ORFs.
Electroporation
The protocol to electroporate plasmids into SM20429 was based on the method described by Kurusu et al. [23] with some modification. A 1% inoculum from over-night culture of SM20429 in 2216E medium was inoculated into 20 ml of the same culture and grown at 20°C. After incubation for 4 h, the logarithmic-phase cells were harvested and washed twice with chilled ddH2O. The cells were suspended in a final volume of 100 μl ddH2O containing 20 mM glucose, and mixed with 1 μg plasmid DNA. Electroporation was performed with a Gene Pulser (Bio-Rad) at a capacity of 50 μF and a voltage of 2.0 kV. After one pulse, the cells were grown in 2216E medium supplemented with glucose (20 mM) for 3 h, and then were spread on selective plates with chloramphenicol.
Analysis of plasmid stability
SM20429 strains with plasmid pWD or other derivative plasmids were grown in 2216E broth for about 24 generations by being serially transferred to fresh broth medium. Samples were removed from each transfer and serial dilutions of the sample were plated on 2216E agar medium with or without antibiotics. The fraction of chloramphenicol-resistant (CmR) cells in the total population was determined to calculate the segregational stability of the constructed plasmid vectors. Finally, presence of plasmid DNA in CmR cells was confirmed by plasmid isolation and transformation to E. coli DH5α.
Construction of expression system in SM20429
The erythromycin resistance gene (ermC) from plasmid pMG36e [24] was amplified by PCR using the primers EMF (containing Pst I restriction site) and EMR (containing Sal I restriction site) to construct an ermC-expression vector. The 735-bp PCR product was cloned into vector pMD19T simple (Takara). The fragment of the promoter-less ermC gene in the pMD19T was separated with Pst I and Sal I and was used in the construction of the ermC-expression vector with the smallest stable plasmid pWD2.
The promoter-less erythromycin resistance gene was cloned into pWD2 downstream of the Plac of pUC19. The constructed ermC-expression vector was designated as pWD-ermC. This expression vector was electroporated into SM20429. The transformed SM20429 cells were cultured at 25°C in 2216E broth for 24 h. To test the gene expression in SM20429, the total cell lysates of recombinant strain SM20429 expressing ermC under the control of Plac were analyzed by 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). To test whether the recombinant ErmC was active, the transformed SM20429 cells harboring pWD2 and the transformed SM20429 cells harboring pWD-ermC were inoculated in 2216E medium supplemented with 30 μg/ml or 60 μg/ml erythromycin (EM), respectively, and then cultured at 25°C.
Gene cloning and expression of the catalytic domain of the cold-adapted cellulase from P. sp. BSw20308 and cellulase assay
A gene encoding a cold-adapted cellulase Cel308 (GenBank Accession No. HQ997897) has been cloned from the psychrophilic strain Pseudoalteromonas sp. BSw20308 isolated from the Arctic sea ice in our lab. The DNA fragment encoding the catalytic domain (CD) of the cellulase was amplified by PCR from BSw20308, and then ligated into pWD2 to construct the expression vector pWD-cd. The vector pWD-cd was introduced into SM20429 by electroporation for the production of the CD. The transformed SM20429 cells were cultured at 25-30°C in 2216E broth. To analyze the expression of the CD of the cellulase, the cellulase activity in the transformed SM20429 cells was assayed. After grown at 25-30°C for 48 h, the SM20429 cells (3 ml) were harvested and resuspended in 5 ml PBS buffer (0.02 M, pH 7.5), and then sonicated in an ice-water bath. After centrifugation, the cellulase activity in the supernatant was measured as described by Bhat et al. [25]. A mixture containing 100 μl of the supernatant and 900 μl of 1% sodium carboxyl methyl cellulose was incubated at 35°C for 30 min. After incubation, the released amount of reducing sugar was measured using the dinitrosalicylic acid (DNS) method with glucose as standard [26]. One unit of the enzyme activity was defined as the amount of enzyme to release 1 μg of glucose per min.
Results
Characterization of the cryptic plasmid from BSi20429
A cryptic plasmid, designed as pSM429, was isolated from Pseudoalteromonas sp. BSi20429. The copy number of pSM429 in BSi20429 was about 30 per cell (Additional file 1, Figure S1). The complete sequence of pSM429 consists of 3874 bp with a G+C content of 28%, which is lower than that of the chromosomal DNA (39-44%) of Pseudoalteromonas, as well as that of other published Pseudoalteromonas plasmids, such as pPS1M3 (37%) [23], pMtBL (39%) [27] and pKW1 (43%) [28]. Sequence analysis revealed that the plasmid pSM429 contains four open reading frames (ORFs) with more than 80 amino acid residues (Table 3).
Searches in the databases revealed that ORF4 showed moderate similarity (25-26% identity, 50% similarity) with the Rep proteins of pFKN [29] and the other plasmids belonging to pPT23A family from Pseudomonas syringae[30, 31]. It also showed some similarity (with identity lower than 26%) with the Rep proteins of the ColE2-related plasmids, as well as the Rep proteins of plasmids isolated from the bacteria of different genus. As plasmids are generally classified based on the amino acid sequence similarity between their replication initiator proteins [32], a phylogenetic tree was constructed using Rep amino acid sequences of some related plasmids as shown in Figure 2. pFKN and pAV505 isolated from Pseudomonas syringae were selected from pPT23A group and the other plasmids were found by homology search using putative Rep amino acid sequence of pSM429. All of the 12 Rep proteins in the phylogenetic tree contain two conserved domains [33], a replicase domain (Pfam Accession No. PF03090) believed to be involved in DNA binding [34], and an alpha helical domain (Pfam Accession No. PF08708) that was found at the C terminus of primases [35]. This suggests that these Rep proteins might be involved in the replication with the similar mechanism. However, pSM429 did not cluster with other plasmids in the phylogenetic tree, implying that pSM429 is a novel plasmid.
Phylogenetic analysis of the Rep proteins from pSM429 and related plasmids. The unrooted phylogenetic tree was constructed by the sequence distance method using the neighbor-joining algorithm. The GenBank accession numbers of the Rep proteins are as follows: ColE2-CA42 from Escherichia coli, BAA0694; pEI2 from Edwardsiella ictaluri, NP_061811; pUb6060 from Plesiomonas shigelloides, CAB56518; pEMCJH03 from Moraxella catarrhalis, NP_957539; pAsal1 from Aeromonas salmonicida, NP_710167; pAV512 from Pseudomonas syringae, AAZ29772; pBC1 from Bifidobacterium catenulatum, YP_241107; pDOJH10s from Bifidobacterium longum, NP_694604; pRBL1 from Brevibacterium linens, AAB03568; pJD1 from Neisseria gonorrhoeae, NC_040411.
Construction of the shuttle vector pWD and transformation system
The shuttle vector pWD, based on plasmid pSM429, was constructed for replication in E. coli and Pseudoalteromonas strains (Figure 1). The 3.8-kb plasmid pSM429 was linearised and cloned in pUC19. In addition, the 1.3-kb chloramphenicol resistance gene was inserted into the resulted plasmid as selection marker to construct the shuttle vector pWD. The presence of the pUC19 ori and pSM429 ori in pWD enabled pWD to replicate in both E. coli and BSi20429. The constructed plasmid pWD could be purified from E. coli in a large amount.
To construct the host strain for the plasmid transformation, the plasmid pSM429 was cured from BSi20429 using the SDS-treated method. Screening of 40 colonies yielded 4 plasmid-cured colonies, showing an efficiency of 10%, which is much more effective than the method used in Pseudoalteromonas strains previously reported [23]. One plasmid-cured strain showing a similar phenotype to the parental strain was designated as SM20429, which was used for further study. The total DNA of SM20429 was purified and used as template for PCR checking. The results showed that the plasmid did not integrate into the genome of SM20429. As SM20429 was sensitive to erythromycin and chloramphenicol (data not shown), chloramphenicol was used for the direct selection of Pseudoaltermonas transformants. The constructed shuttle vector pWD was introduced into SM20429 by electroporation. Chloramphenicol-resistant transformants of SM20429 were obtained at the frequency of 5.0x 102/μg DNA. To confirm that the Pseudoalteromonas transformants harbored plasmids, several transformants were subjected to plasmid analysis. Electrophoresis showed that plasmids were prepared from all of the selected transformants, and the prepared plasmids showed the same electroporetic DNA pattern as that of pWD (Additional file 2, Figure S2). Moreover, pWD DNA prepared from the Pseudoalteromonas transformants could be transformed into SM20429 at a higher frequency (104/μg DNA) than that prepared from E. coli transformants, suggesting the presence of DNA restriction and modification system in BSi20429.
Properties of the shuttle vector pWD and its derivatives
The shuttle vector pWD constructed above harbors the complete sequence of plasmid pSM429. To check the functionality of the identified ORFs on pSM429 and delete the unnecessary regions on pWD for an expression vector, six plasmids derived from pWD with different deletions were constructed (Figure 1). Each of the six derivatives was checked in E. coli, and then was introduced into SM20429 by electroporation. Except for the two smallest derivatives, pWD5 and pWD6, transformants of the other four derived plasmids were all obtained, indicating that pWD5 and pWD6 were not capable of replication in SM20429. Since pWD5 and pWD6 lack the upstream or the downstream sequences of the rep gene (orf4) compared to pWD4, the results indicated that the fragment containing rep gene and its flanking sequences are the minimal replication region of pSM429 and all of orf1, orf2 and orf3 are not essential for plasmid replication.
The segregational stability of pWD and its four derivatives were tested twice and counts were done in duplicate. Average results were presented in Figure 3. In the absence of selective pressure, nearly 100% of the cells retain plasmids in the first 40 generations of SM20429. However, after 40 generations, the smaller derivatives, pWD3 and pWD4, exhibited less segregational stability than pWD and the other bigger derivatives, pWD1 and pWD2, in SM20429. Compared to pWD2, pWD3 lacks some repeat sequences located between orf1 and orf2, as shown in Figure 4. The above result suggests that these repeat sequences are essential for the segregational stability of the plasmid and possibly related to plasmid partition.
Segregational stability of pWD and its derivatives in SM20429. Average results of two independent experiments for each construct were presented. Strains harboring the constructs were cultured in the absence of selective pressure, plated under the same conditions, and assayed for plasmid maintenance by replica-plating onto antibiotic-containing media at 40-, 80-, 120-, and 160-generation intervals. The presence of plasmids was finally checked by plasmid preparation and gel electrophoresis.
Nucleotide sequence of the 200-bp region between orf1 and orf2 in pSM429 related to plasmid segregational stability. The region lacking in pWD3 compared to pWD2 is shown between two vertical lines. The four imperfect direct repeats are marked with single arrows, and the inverted repeat is marked with double arrows. The deduced amino acid sequence of ORF2 is shown and the initiation coden is shown in bold.
Construction of an ermC-expression vector and expression of ErmC in SM20429
The erythromycin resistance gene ermC was cloned into pWD2 downstream of the Plac promoter to construct an ermC-expression vector, pWD-ermC. This vector was introduced into SM20429 by electroporation and the ermC gene was expressed under the control of Plac promoter. SDS-PAGE analysis showed that the ErmC protein was expressed in SM20429 cells, and the expression level of ErmC in SM20429 was lower than that in E. coli DH5α (Figure 5A). To test whether the expressed ErmC protein is active, the SM20429 strain harboring pWD2 and the SM20429 strain harboring pWD-ermC were inoculated in 2216E medium supplemented with erythromycin (EM), respectively. The pWD2-harboring strain that is sensitive to EM as the wild strain BSi20429 could not grow in the medium supplemented with 30 μg/ml EM, while the pWD-ermC-harboring strain grew well in the medium supplemented with 60 μg/ml EM (Figure 5B). Therefore, the pWD-ermC-harboring strain obtained the ability resistant to EM, indicating that the ErmC protein expressed in SM20429 was active. These results suggest that Plac promoter is efficient for heterologous gene expression in SM20429.
Expression of the recombinant ErmC protein in SM20429 and E. coli with the constructed vector pWD-ermC. (A) SDS-PAGE analysis of the expressed ErmC protein in the total-cell lysates of recombinant SM20429 and E. coli. Lane 1, SM20429 harboring pWD2, used as a negative control; Lane2, SM20429 harboring pWD-ermC; Lane3, E. coli DH5α harboring pWD-ermC, used as a positive control. The SDS-PAGE gel was stained with Coomassie blue. Arrow denotes the band of the expressed ErmC. (B) Analysis of the ErmC-resistance of the transformed SM20429. 1, pWD-ermC-harboring SM20429 inoculated in 2216E medium supplemented with 30 μg/ml EM; 2, pWD2-harboring SM20429 inoculated in 2216E medium supplemented with 30 μg/ml EM; 3, pWD-ermC-harboring SM20429 inoculated in 2216E medium supplemented with 60 μg/ml EM; 4, pWD2-harboring SM20429 inoculated in 2216E medium supplemented with 60 μg/ml.
Expression of cold-adapted protein with the constructed expression system
In our previous work, a cellulase named Cel308 was purified from the Arctic sea ice bacterium Pseudoalteromonas sp. BSw 20308 (Additional file 3, Figure S3), and the gene encoding Cel308 was cloned and sequenced. Character analysis showed that Cel308 is a cold-adapted enzyme (Additional file 3, Figure S4). The cd gene encoding the catalytic domain of Cel308 was cloned into pWD2 to construct a cd-expression vector pWD-cd. The plasmid pWD-cd was transformed into SM20429 by electroporation. After cultivation for 48 h, the intracellular cellulase activity of the transformed strain was detected using a quantitative assay. The results showed that 60 units of cellulase enzyme were expressed in 1 ml culture of SM20429. In comparison, no cellulase activity was detected in the cells of SM20429 harboring pWD2 (Figure 6). The results indicated that the constructed expression system consisting of strain SM20429 and vector pWD2 could be used to express cold-adapted protein.
Discussion
As the first step to develop an effective genetic system for Pseudoalteromonas strains, especially the psychrophilic ones, the Pseudoalteromonas strains isolated from the Arctic sea ice were screened for plasmids. Only one small plasmid, pSM429, was obtained out of over one hundred strains, suggesting that small plasmids from the Arctic sea ice isolates of Pseudoalteromonas are less common. This finding is consistent with the fact that only a few reports have been published about the isolation and characterization of plasmids from cold-adapted Pseudoalteromonas strains, including Pseudoalteromonas sp. PS1M3 [23], Pseudoalteromonas haloplanktis TAC125 [27] and Pseudoalteromonas sp. 643A [36]. pSM429 has much lower G+C content than other Pseudoalteromonas plasmids, implying its cold-adaptation. The small plasmid pSM429 is valuable for expanding the knowledge about the genetics of extrachromosomal elements in cold-adapted Pseudoalteromonas strains and opens the possibility to construct a genetic system for Pseudoalteromonas strains.
Genetic systems require stable cloning and expression vectors that allow efficient cloning and expression of homologous and heterologous genes. To develop a genetic system of psychrophlic Pseudoalteromonas, a plasmid-cured strain of BSi20429 was obtained to be used as an expression host, and a shuttle vector for Pseudoalteromonas-E. coli was constructed based on pSM429. Sequential deletions were used to study the functionality of ORFs in plasmid pSM429 and to analyze their effect on plasmid stability. The results showed that the rep gene is essential for the autonomous replication of pSM429 in SM20429. This is consistent with the finding that plasmids need specific replication protein to recognize and cut the double-stranded origin of replication for the process to begin [37]. Furthermore, deletion of surrounding sequences of rep gene caused the loss of replication ability of the plasmid in SM20429, probably because these regions contain the promoter sequence of rep gene and the replication origin where the Rep protein binds. Although not essential, an intergenic fragment including some repeat sequences on pSM429 markedly affects the segregational stability. Accordingly, the stable construction pWD2 containing the replication region and the intergenic region related to segregational stability was used as an E. coli- Pseudoalteromonas shuttle vector and for construction of expression vectors.
An expression system in cold-adapted bacteria may circumvent the problem of limited stability of the product that is always encountered during heterogonous expression of psychrophilic proteins in mesophilic hosts [15, 38]. A few low-temperature expression systems have been reported [39, 40]. However, there are still many cold-adapted proteins could not be successfully expressed by a recombinant protein expression system. The alternative systems would be important for the study of cold-adapted proteins. In order to check the usefulness of plasmid pWD2 and its host SM20429 as expression system, the promoter-less ermC gene was cloned into the vector pWD2 and introduced into SM20429. The results showed that the ermC gene was expressed in SM20429 and the recombinant strain displayed resistance to erythromycin, suggesting that the Plac promoter was efficient in SM20429. Then, with this system, the cd gene encoding the catalytic domain of a cold-adapted cellulase from a psychrophilic strain was successfully expressed, suggesting the potential use of this system for overexpression of genes from psychrophiles. Indeed, this low-temperature expression system is still in an early stage of development. Further improvement, such as increasing the yield of proteins and controlling the expression of proteins, is being underway.
Conclusion
In this paper, we report the characterization and use of a plasmid pSM429 from the Arctic Pseudoalteromonas strain BSi20429. The plasmid pSM429 is 3874 bp in length, with a very low G+C content (28%). The ORF4 on pSM429 was predicted to encode a Rep protein. Phylogenetic analysis implied that pSM429 is a novel plasmid. The regions essential for plasmid replication and stability were determined. Accordingly, the stable shuttle vectors based on pSM429 for E. coli and Pseudoalteromonas strains, pWD, pWD1 and pWD2, were developed, which can be used as cloning vectors for the Pseudoalteromonas strain we studied. With the aid of pWD2 and its Pseudoalteromonas host, the heterologous expressions of the erythromycin resistance gene and the cold-adapted cellulase gene were successful, indicating the potential use of this system for recombinant protein production, especially for cold-adapted proteins. These results are helpful for Pseudoalteromonas study.
References
Cavicchioli R, Siddiqui KS, Andrews D, Sowers KR: Low-temperature extremophiles and their applications. Curr Opin Biotechnol. 2002, 13: 253-261. 10.1016/S0958-1669(02)00317-8.
Feller G, Gerday C: Psychrophilic enzymes: hot topics in cold adaptation. Nat Rev. 2003, 1: 200-208. 10.1038/nrmicro773. 10.1038/nrmicro773.
Ochman H, Lawrence JG, Groisman EA: Lateral gene transfer and the nature of bacterial innovation. Nature. 2000, 405: 299-304. 10.1038/35012500.
Holmstrom C, Kjelleberg S: Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol Ecol. 1999, 30: 285-293.
Chen XL, Zhang YZ, Gao PJ, Luan XW: Two different proteases produced by a deep-sea psychrotrophilc bacterial strain, Pseudoaltermonas sp. SM9913. Mar Biol. 2003, 143: 989-993. 10.1007/s00227-003-1128-2. 10.1007/s00227-003-1128-2.
Cui Z, Lai Q, Dong C, Shao Z: Biodiversity of polycyclic aromatic hydrocarbon-degrading bacteria from deep sea sediments of the Middle Atlantic Ridge. Environ Microbiol. 2008, 10: 2138-2149. 10.1111/j.1462-2920.2008.01637.x.
Zhou MY, Chen XL, Zhao HL, Dang HY, Luan WX, Zhang XY: Diversity of both the cultivable protease-producing bacteria and their extracellular proteases in the sediments of the South China Sea. Microb Ecol. 2009, 58: 582-590. 10.1007/s00248-009-9506-z.
Qin GK, Zhu L, Chen XL, Wang PG, Zhang YZ: Structural characterization and ecological roles of a novel exopolysaccharide from the deep-sea psychrotolerant bacterium Pseudoalteromonas sp. SM9913. Microbiology. 2007, 153: 1566-1572. 10.1099/mic.0.2006/003327-0.
Cusano AM, Parrilli E, Marino G, Tutino ML: A novel genetic system for recombinant protein secretion in the Antarctic Pseudoalteromonas haloplanktis TAC125. Microb Cell Fact. 2006, 14: 5-40.
Papa R, Rippa V, Sannia G, Marino G, Duilio A: An efficient cold inducible expression system developed in Pseudoalteromonas haloplanktis TAC125. J Biotechnol. 2007, 127: 199-210. 10.1016/j.jbiotec.2006.07.003.
Parrilli E, De Vizio D, Cirulli C, Tutino ML: Development of an improved Pseudoalteromonas haloplanktis TAC125 strain for recombinant protein secretion at low temperature. Microb Cell Fact. 2008, 7: 2- 10.1186/1475-2859-7-2.
Siani L, Papa R, Di Donato A, Sannia G: Recombinant expression of Toluene o-Xylene Monooxygenase (ToMO) from Pseudomonas stutzeri OX1 in the marine Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. J Biotechnol. 2006, 126: 334-341. 10.1016/j.jbiotec.2006.04.027.
Chen XL, Xie BB, Lu JT, He HL, Zhang YZ: A novel type of subtilase from the psychrotolerant bacterium Pseudoalteromonas sp. SM9913: catalytic and structural properties of deseasin MCP-01. Microbiology. 2007, 153: 2116-2125. 10.1099/mic.0.2007/006056-0.
Chen XL, Xie BB, Bian F, Zhao GY, Zhao HL, He HL, Zhou BC, Zhang YZ: Ecological function of myroilysin, a novel bacterial M12 metalloprotease with elastinolytic activity and a synergistic role in collagen hydrolysis, in biodegradation of deep-sea high-molecular-weight organic nitrogen. Appl Environ Microbiol. 2009, 75: 1838-1844. 10.1128/AEM.02285-08.
Zhao GY, Chen XL, Zhao HL, Xie BB, Zhou BC, Zhang YZ: Hydrolysis of insoluble collagen by deseasin MCP-01 from deep-sea Pseudoalteromonas sp. SM9913. J Biol Chem. 2008, 283: 36100-36107. 10.1074/jbc.M804438200.
Yan BQ, Chen XL, Hou XY, He HL, Zhou BC, Zhang YZ: Molecular analysis of the gene encoding a cold-adapted halophilic subtilase from deep-sea psychrotolerant bacterium Pseudoalteromonas sp. SM9913: cloning, expression, characterization and function analysis of the C-terminal PPC domains. Extremophiles. 2009, 13: 725-733. 10.1007/s00792-009-0263-1.
Xie BB, Bian F, Chen XL, He HL, Guo J, Gao X, Zeng YX, Chen B, Zhou BC, Zhang YZ: Cold adaptation of zinc metalloproteases in the thermolysin family from deep sea and arctic sea ice bacteria revealed by catalytic and structural properties and molecular dynamics: new insights into relationship between conformational flexibility and hydrogen bonding. J Biol Chem. 2009, 284: 9257-9269. 10.1074/jbc.M808421200.
Wang YK, Zhao GY, Li Y, Chen XL, Xie BB, Su HN, Lv YH, He HL, Liu H, Hu J, Zhou BC, Zhang YZ: Mechanistic insight into the function of the C-terminal PKD domain of the collagenolytic serine protease deseasin MCP-01 from deep sea Pseudoalteromonas sp. SM9913: binding of the PKD domain to collagen results in collagen swelling but does not unwind the collagen triple helix. J Biol Chem. 2010, 285: 14285-14291. 10.1074/jbc.M109.087023.
Sambrook J, Fritsch EF, Maniatis T: Molecular cloning: a laboratory manual. 1989, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2,
Lukashin AV, Borodovsky M: GeneMark. hmm: new solutions for gene finding. Nucleic Acids Res. 1998, 26: 1107-1115. 10.1093/nar/26.4.1107.
Tatusova TA, Madden TL: Blast2 sequences-a new tool for comparing proteins and nucleotide sequences. FEMS Microbiol Lett. 1999, 174: 247-250. 10.1111/j.1574-6968.1999.tb13575.x.
Qin Z, Shen M, Cohen SN: Identification and characterization of a pSLA2 plasmid locus required for linear DNA replication and circular plasmid stable inheritance in Streptomyces lividans. J Bacteriol. 2003, 185: 6575-6582. 10.1128/JB.185.22.6575-6582.2003.
Kurusu Y, Yoshimura S, Tanaka M, Nakamura T, Maruyama A, Higashihara T: Genetic transformation system for a psychrotrophic deep-sea bacterium: isolation and characterization of a psychrotrophic plasmid. Mar Biotechnol (NY). 2001, 3: 96-99. 10.1007/s101260000067. 10.1007/s101260000067.
van de Guchte M, van der Vossen JM, Kok J, Venema G: Construction of a Lactococcal Expression Vector: Expression of Hen Egg White Lysozyme in Lactococcus lactis subsp. Lactis. Appl Environ Microbiol. 1989, 55: 224-228.
Bhat TM, Wa KM: Methods for measuring cellulases activities. Methods Enzymol. 1988, 160: 87-144.
Miller GL: Use of dinitrosalicylic acid reagent for determination of reducing sugars. Anal Chem. 1959, 31: 426-428. 10.1021/ac60147a030. 10.1021/ac60147a030.
Tutino ML, Duilio A, Parrilli R, Remaut E, Sannia G, Marino G: A novel replication element from an Antarctic plasmid as a tool for the expression of proteins at low temperature. Extremophiles. 2001, 5: 257-264. 10.1007/s007920100203.
Cieslinski H, Werbowy K, Kur J, Turkiewicz M: Molecular characterization of a cryptic plasmid from the psychrotrophic antarctic bacterium Pseudoalteromonas sp. 643A. Plasmid. 2008, 60: 154-158. 10.1016/j.plasmid.2008.06.002.
Rohmer L, Kjemtrup S, Marchesini P, Dangl JL: Nucleotide sequence, functional characterization and evolution of pFKN, a virulence plasmid in Pseudomonas syringae pathovar maculicola. Mol Microbiol. 2003, 47: 1545-1562. 10.1046/j.1365-2958.2003.03402.x.
Zhao Y, Ma Z, Sundin GW: Comparative genomic analysis of the pPT23A plasmid family of Pseudomonas syringae. J Bacteriol. 2005, 187: 2113-2126. 10.1128/JB.187.6.2113-2126.2005.
Ma Z, Smith JJ, Zhao Y, Jackson RW, Arnold DL, Murillo J, Sundin GW: Phylogenetic analysis of the pPT23A plasmid family of Pseudomonas syringae. Appl Environ Microbiol. 2007, 73: 1287-1295. 10.1128/AEM.01923-06.
Ilyina TV, Koonin EV: Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992, 20: 3279-3285. 10.1093/nar/20.13.3279.
Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Zhang D, Bryant SH: CDD: a conserved domain database for protein classification. Nucleic Acids Res. 2005, 33: 192-196. 10.1093/nar/gni191. 10.1093/nar/gni191.
Giraldo R, Diaz-Orejas R: Similarities between the DNA replication initiators of Gram-negative bacteria plasmids (RepA) and eukaryotes (Orc4p)/archaea (Cdc6p). Proc Natl Acad Sci USA. 2001, 98: 4938-4943. 10.1073/pnas.081079298.
Iyer LM, Koonin EV, Leipe DD, Aravind L: Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 2005, 33: 3875-3896. 10.1093/nar/gki702.
Hubert C, Katarzyna W, Józef K, Marianna T: Molecular characterization of a cryptic plasmid from the psychrotrophic antarctic bacterium Pseudoalteromonas sp. 643A. Plasmid. 2008, 60: 154-158. 10.1016/j.plasmid.2008.06.002.
del Solar G, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R: Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998, 62: 434-464.
Feller G, Thiry M, Arpigny JL, Gerday C: Cloning and expression in Escherichia coli of three lipase-encoding genes from the psychrotrophic antarctic strain Moraxella TA144. Gene (Amst). 1991, 102: 111-115.
Duilio A, Tutino LM, Marino G: Recombinant protein production in Antarctic Gram-negative bacteria. Methods Mol Biol. 2004, 267: 225-237.
Miyake R, Kawamoto J, Wei YL, Kitagawa M, Kato I, Kurihara T, Esaki N: Construction of a low-temperature protein expression system using a cold adapted bacterium Shewanella sp. Strain Ac10, as a host. Appl Environ Microbiol. 2007, 73: 4849-4856. 10.1128/AEM.00824-07.
Acknowledgements
The work was supported by National Natural Science Foundation of China (31025001, 40876072), Hi-Tech Research and Development program of China (2006AA09Z414, SQ2010AA0900521004), Natural Science Foundation of Shandong Province, China (JQ200910, ZR2009DZ002), Special Fund of China for Marine-scientific Research in the Public Interest (201005032-6), Foundation for Young Excellent Scientists in Shandong Province, China (2007BS07007).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
XC and YZ designed the project; YY and BC provided the strains; DZ, ZY, PL and ZW performed the research; DZ. ZY, XC, MS, BZ and YZ analyzed the data; DZ wrote the paper; YZ and XC critically reviewed the paper. All authors approved the final manuscript.
Electronic supplementary material
12934_2011_522_MOESM1_ESM.PNG
Additional file 1: Figure S1: Agrose gel electrophoresis analysis of total genominc DNA from BSi20429. Genomic DNA, including the chromosome and plasmid, was isolated and then diluted by 10 and 100 times and electrophoresed in a 0.7% agarose gel. M, Trans15K DNA ladder (TransGen Biotech); Genomic DNA in lane 1 to lane3 were non-diluted, 10-time diluted and 100-time diluted. The open circular (OC) and covalently closed circular (CCC) forms of pSM429 are indicated. (PNG 92 KB)
12934_2011_522_MOESM2_ESM.PNG
Additional file 2: Figure S2: Agrose gel electrophoresis of plasmid pWD. Lanes 1, 2 and 3 show the plasmid pWD isolated from transformed SM20429; Lane 4 shows the original plasmid pWD isolated from transformed E. coli. (PNG 45 KB)
12934_2011_522_MOESM3_ESM.PDF
Additional file 3: Characteristics of the cellulase Cel308 from the psychrophilic bacterium Pseudoalteromonas BSw20308. Purification, characterization and gene cloning of the cold-adapted cellulase Cel308 from the psychrophilic bacterium Pseudoalteromonas sp. BSw 20308. It contains Figure S3 and Figure S4. (PDF 94 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Zhao, DL., Yu, ZC., Li, PY. et al. Characterization of a cryptic plasmid pSM429 and its application for heterologous expression in psychrophilic Pseudoalteromonas. Microb Cell Fact 10, 30 (2011). https://doi.org/10.1186/1475-2859-10-30
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2859-10-30