Abstract
Background
Spiroplasma citri BR3-3X and S. kunkelii CR2-3X cause serious diseases worldwide on citrus and maize species, respectively. S. citri BR3-3X harbors a plasmid, pBJS-Original (pBJS-O), that encodes the s piroplasma a dhesion r elated p rotein 1 (SARP1), a protein implicated in binding of the pathogen to cells of its leafhopper vector, Circulifer tenellus. The S. kunkelii CR2-3X plasmid, pSKU146, encodes a homolog of SARP1, Sk-ARP1. Due to the close phylogenetic relationship of the two pathogens, we hypothesized that the two plasmids are closely related as well.
Results
The nucleotide sequence of pBJS-O was determined and compared to the sequences of a plasmid from BR3-T (pBJS-T), which is a multiply passaged leafhopper transmissible derivative of BR3-3X, and to known plasmid sequences including that of pSKU146. In addition to arp1, the 13,374 bp pBJS-O sequence putatively contains nine genes, recognized as open reading frames (ORFs). Several pBJS-O ORFs have homologs on pSKU146. However, the sequences flanking soj-like genes on both plasmids were found to be more distant from one another than sequences in any other region. Further, unlike pSKU146, pBJS-O lacks the conserved oriT region characteristic of the IncP group of bacterial plasmids. We were unable to identify a region in pBJS-O resembling a known plasmid origin of transfer. In regions where sequence was available for the plasmid from both BR3-3X and BR3-T, the pBJS-T sequence had a 0.4 kb deletion relative to its progenitor, pBJS-O. Southern blot hybridization of extrachromosomal DNA from various S. citri strains and spiroplasma species to an arp-specific probe and a probe made from the entire plasmid DNA of BR3-3X revealed limited conservation of both sequences in the genus Spiroplasma. Finally, we also report the presence on the BR3-3X chromosome of arp2, an S. citri homolog of arp1 that encodes the predicted protein SARP2. The C-terminal domain of SARP2 is homologous to that of SARP1, but its N-terminal domain is distinct.
Conclusion
Our data suggest that pBJS is a novel S. citri plasmid that does not belong to any known plasmid incompatibility group. The differences between pBJS-O and pSKU146 suggest that one or more events of recombination have contributed to the divergence of the plasmids of the two sister Spiroplasma species; the plasmid from S. citri itself has diverged slightly during the derivation of S. citri BR3-T from BR3-3X. Our data also show that pBJS-O encodes the putative adhesin SARP1. The presence of traE and mob on pBJS-O suggests a role for the plasmid in spiroplasmal conjugation.
Similar content being viewed by others
Background
The phytopathogenic spiroplasmas and phytoplasmas, which cause serious diseases of economically important plant species worldwide [[1] and [2]], are wall-less prokaryotes phylogenetically related to Gram-positive eubacteria with low G+C content [3]. They are transmitted in nature by phloem-feeding insects, predominantly leafhoppers, in a propagative manner [4]. Even though there are close to forty recognized spiroplasma species, only three plant pathogenic spiroplasmas have been identified and characterized to date. S. citri [[5, 6] and [7]] is the causative agent of stubborn disease of citrus and brittle root disease of horseradish; S. kunkelii [[8, 9] and [10]] is the etiological agent of corn stunt; and S. phoeniceum [11] causes periwinkle yellows. Unlike phytoplasmas, spiroplasmas can be cultured in vitro. Therefore, the relationships between S. citri and its insect vectors, the beet leafhopper, Circulifer tenellus, and the related species, C. haematoceps [12], have been investigated extensively, serving as models for investigating the molecular aspects of mollicute-vector interactions.
Spiroplasma binding to insect host and non-host cells, both in tissue-culture and within the intact insect, has been reported [[13] and [14]]. The loss and restoration of the ability of S. citri to adhere to tissue-cultured C. tenellus cells was associated with degradation and restoration of P89 (designated SARP1), a spiroplasma membrane protein [14]. Due to the possible direct involvement of SARP1 in the spiroplasma-leafhopper interaction, it was hypothesized that SARP1 is an adhesin. Later, Berg et al [15] reported cloning and characterization of arp1, the gene encoding SARP1, from S. citri BR3-T. They also reported that mature SARP1 protein contains a novel domain at the N-terminus, called "sarpin", made of six repeats of 39–42 amino acids each.
S. citri harbors several extrachromosomal DNAs with unique restriction patterns [[16–18] and [19]]. S. citri lines, derived from a clone, and sister clones of the same lines showed differences in their extrachromosomal DNAs [20]. In addition to known plasmids, there are replicative forms (RFs) of several viruses and other uncharacterized circular extrachromosomal DNAs in S. citri [21].
Plasmids have also been noted in strains of S. kunkelii [22]. Recently, Davis and colleagues [23] reported the complete sequence of the S. kunkelii CR2-3X plasmid pSKU146, which encodes a homolog of SARP1, Sk-ARP1. In the present study, we isolated and characterized a related indigenous plasmid, designated pBJS-Original (pBJS-O), from S. citri BR3-3X. This is a report of the discovery, distribution and characterization of that plasmid. Among other genes, pBJS-O contains arp1. The significance of the discovery of pBJS-O in relation to our current understanding of the S. citri-leafhopper interactions and potential genetic manipulations in mollicutes is discussed. Implications for the evolution of both pBJS-O and pSKU146 are also presented.
Results
Detection and analysis of arp2
SARP1 has been characterized previously and the gene encoding it, arp1, has been cloned and sequenced [GenBank:AJ297706] from S. citri BR3-T [15]. In the process, an Rsa I restriction fragment was cloned and sequenced from BR3-T genomic DNA; the alignment of this fragment with AJ297706 revealed 92% similarity in the 3' 660 nucleotides of the former sequence. However in the 5' 55 bases of the total 715 bp, upstream from position 2370 in AJ297706, the new fragment was not similar to the known sequence. We designated this gene, which resembles but is not identical to arp1, as arp2 and its putative protein product as SARP2. As also noted by Bai et al. [22], the S. kunkelii CR2-3X genome contains two sequences similar to those of S. citri BR3-T arp genes. The predicted protein, Sk-ARP1 (for S. kunkelii adhesion related protein 1), encoded by the first sequence, Sk-arp1, contains seven rather than six sarpin repeats and has C-terminal domains resembling those of SARP1 [15]. The second sequence encodes a putative protein whose C-terminus is homologous to that of SARP1, but has an unrelated N-terminus. This protein is designated Sk-ARP2 (S. kunkelii adhesion related protein 2) and the corresponding gene is named Sk-arp2. SARP1 has sequence similarity with known adhesins. Fleury et al. [24] have shown that the predicted amino acid sequence of P40, a Mycoplasma agalactiae cytadhesin, is similar not only to that of SARP1 but also to the one of P50, an adhesin of M. hominis.
Isolation and distribution of Spiroplasma extrachromosomal DNA
We isolated extrachromosomal DNA from S. citri BR3-3X to test the hypothesis that this DNA contains an arp-like gene as in S. kunkelii. Restriction of the DNA with single enzymes, including Bgl II and Nde I, converted a DNA migrating with 9 kb into a fragment migrating close to 7 kb (Fig. 1). These results were consistent with the presence of a single major plasmid. We designated the plasmid pBJS-O. By nucleotide sequencing, we determined that the actual size of the plasmid was 13,374 bp and deposited the sequence in the EMBL Nucleotide Sequence Database [EMBL:AJ972409]
To test the conservation of pBJS-O in S. citri strains derived from S. citri BR3, plasmid preparations from S. citri BR3-3X and from BR3-G, BR3-T, BR3-M and BR3-P, lines derived from BR3-3X, were probed with a DNA fragment derived from arp1 (Fig. 2 and Table 1). All hybridized with the probe, producing two or more bands. To test the conservation of pBJS-O in other S. citri strains, other plant-associated spiroplasmas and the closest relative of S. citri, S. melliferum [25], the plasmids of S. kunkelii CR2-3X, S. melliferum, S. citri strains R8A2, ASP-1 and Beni Mellal, S. floricola and S. phoeniceum also were probed with the arp1-derived probe (Fig. 3A). Only S. kunkelii CR2-3X and S. melliferum reacted in the hybridization. However, when the same plasmids were probed with the whole pBJS-O plasmid as a probe (Fig. 3B), all the sample preparations, except those from S. citri Beni Mellal, S. floricola and S. phoeniceum, hybridized with the probe. All of the above Southern hybridization experiments revealed multiple reactive species in the plasmid preparations and the hybridization patterns of Eco RI-digested and undigested plasmid samples were very similar to each other. For comparison, the blots included Eco RI-digested chromosomal DNA of S. citri BR3-3X. A single hybridization signal distinct from those of plasmid preparations was observed (Fig. 2).
Southern blotting hybridization of (A) S. citri BR3-3X, BR3-G, BR3-T, and (B) S. citri BR3-M and BR3-P plasmid preparations to an arp1-derived probe. Eco RI-digested S. citri BR3-3X chromosomal DNA and Eco RI-digested and undigested plasmid preparations from BR3-3X, BR3-G, BR3-T, BR3-M and BR3-P are shown. D, digested with Eco RI; U, undigested. Hybridization in the marker lane is due to presence of short pBluescript vector sequences in the probe.
Southern blotting hybridization of undigested plasmid preparations from different S. citri strains and spiroplasma species to (A) an arp1-derived probe and (B) the whole pBJS-O probe. Plasmids from S. citri ASP-1, R8A2 and Beni Mellal, and from S. floricola, S. melliferum, S. phoeniceum and S. kunkelii CR2-3X were used. The blot shown in panel A was stripped and rehybridized using the whole pBJS-O probe, shown in panel B.
arp1 and arp2 locations in S. citri BR3-3X
The Southern blot hybridization results suggest that arp-related sequences are present on both a plasmid and the chromosome. arp1 and arp2 from BR3-T are nearly identical over a considerable portion of their nucleotide sequence. Hence, using a probe containing this conserved region should detect both genes. Nevertheless, arp1 and arp2 differ at several positions in those regions. To determine whether the BR3-3X plasmid and chromosomal sequences represented arp1 or arp2 genes, we determined parts of the sequences of BR3-3X plasmid and chromosomal DNAs by direct sequencing and by sequencing amplified PCR products. Comparison of the BR3-3X arp sequences with those of BR3-T revealed that the BR3-3X arp2 sequence had diverged more from the other three sequences than the latter had from each other (Figs. 4 and 5). At positions where the two BR3-T genes differed from one another, the chromosomal BR3-3X sequence had arp2 residues in 21 positions and arp1 residues in only 3 positions (Figs. 4A and 4B). Conversely, at arp1- and arp2-specific positions, the BR3-3X plasmid DNA had no arp2 residues and 28 arp1 residues. Further, at all 57 positions at which chromosomal and plasmid sequences differed, the BR3-3X plasmid and arp1 nucleotides were identical. Hence, we conclude that, in S. citri BR3-3X, the arp1 gene resides on a plasmid and that the arp2 gene most likely resides on the chromosome. The newly determined arp2 sequences from BR3-3X and BR3-T were deposited in the EMBL Nucleotide Sequence Database [EMBL:AM040506 and EMBL:AM040505, respectively].
Comparison of partial nucleotide sequences of S. citri BR3-3X chromosomal (BR3-3X_chr) and plasmid (BR3-3X_pl) sequences to those of available BR3-T arp1 and arp2 genes (BR3-T_arp1 and BR3-T_arp2, respectively). Only the regions containing polymorphic positions are shown. (A) region of arp1 positions 3572 to 3800 (AJ297706). (B) region of arp1 positions 4118 to 4177 and (C) region of arp1 positions 4658 to 4717). S. citri BR3-T arp2 is not available for the last sequence alignment. Gaps are denoted by dashed lines, whereas dots denote identical bases.
Complete pBJS-O sequencing and analysis
The 4273 bp sequence [GenBank:AJ297706] originally cloned and characterized from S. citri BR3-T [15] contains a partial ORF soj, followed by ORF2, P89 (arp1) and another partial ORF, ORF4. AJ297706 was used to design primers and initiate primer walking to determine the complete pBJS-O plasmid sequence and allow its characterization. During sequencing, a segment (from nucleotide 1–80) of the assembled sequence proved particularly difficult to sequence. It contained three of the six oligopurine/oligopyrimidine tracts of 12 or more bp in the entire plasmid sequence. That the sequence of the tracts was consistent with triple-helix formation suggests that this region of the plasmid may readily form triple-helical structures interfering with sequencing.
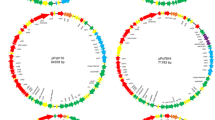
The total plasmid sequence is 13,374 bp in length and contains ten predicted ORFs (Fig. 6 and Table 2), of which orf2 (S. citri ORF2) has no homologs, and orf9 and orf10 appear to have distant relatives (E values 0.34 and 0.024, respectively). Of the ten, six putative pBJS-O-ORFs have homologs in pSKU146, the recently characterized S. kunkelii CR2-3X plasmid [16]: arp1 (adhesin protein; E value 0.0), orf4 (hypothetical protein pSKU146_11; E value 0.0), traE (conjugation ATPase; E value 0.0), orf6 (hypothetical protein pSKU146_13; E value 9 × 10-45), mob (mobilization protein; E value 0.0) and orf8 (hypothetical protein pSKU146_17; E value 1 × 10-103). Predicted products of traE and mob are similar to proteins involved in conjugative DNA transfer in other bacterial genera. In the regions where the plasmid sequence was available from both BR3-3X and BR3-T, pBJS-T (the plasmid from S. citri BR3-T) sequence had a 0.4 kb deletion relative to pBJS-O, bringing the orf4 gene close to arp1 and traE. In BR3-3X, however, arp1 and orf4 are separated by 281 bp. The nucleotide sequence variations between pBJS-O and pBJS-T were found to be clustered. Two regions of enhanced variation were observed over a 200 bp stretch in the ORF2-arp1 intergenic region (positions 2700 to 2900 in pBJS-O). In a comparable stretch from position 5262 to 5544 in the arp1-ORF4 intergenic region, a single stretch of dissimilarity was found.
Algorithm TMHMM v. 2.0 was used to predict the locations of transmembrane helices and intervening loops in the putative products of traE (Fig. 7), mob and orf4. Although the TraE polypeptide was predicted to contain three transmembrane helices, the third helix was predicted at a lower probability than were the other two. Assuming the presence of three transmembrane helices, the protein was predicted to have the N-terminal region (about 10% of the length of the polypeptide) in the cytosol and almost all of the rest of the protein extracellular.
Predicted locations of transmembrane helices and intervening loops in the putative protein encoded by ORF5 (traE) of pBJS-O. The sequential amino acid positions in the primary sequence of the polypeptide are on the X-axis, while the probability score of each residue for being in a transmembrane helix is on the Y-axis in red. The blue and pink curves denote the probability of each amino acid in the sequence to be cytosolic or extracellular, respectively. In the schematic representation of the protein domains at the top, blue lines show the cytosolic portions, purple ones denote the extracellular portions and the thick horizontal bars denote the predicted transmembrane portions of the polypeptide, respectively.
Plasmid pSKU146 from S. kunkelii CR2-3X encodes the S. kunkelii homolog of SARP1, Sk-ARP1. In addition to sk-arp1, pSKU146 contains 17 ORFs. The pSKU146-ORFs having counterparts on pBJS-O were listed above. However, although both plasmids contain genes encoding the ParA-like protein, Soj, sequences surrounding those genes are more distant from one another than are sequences in any other regions. Further, unlike pSKU146, pBJS-O lacks the conserved oriT region characteristic of the IncP group of bacterial plasmids. Also, we were unable to identify a region in pBJS-O resembling a known plasmid origin of transfer.
Discussion
In the present study we report isolation, distribution and structural characterization of pBJS-O, an indigenous S. citri BR3-3X plasmid. We also present evidence that pBJS-O harbors arp1, the gene encoding SARP1, and describe the presence on the BR3-3X chromosome of arp2, an S. citri homolog of arp1. Finally, the sequences of pBJS-O, pBJS-T and the S. kunkelii CR2-3X plasmid, pSKU146, in relation to plasmid evolution are discussed.
Conservation of arp and pBJS-O sequences in Spiroplasma
In Southern hybridizations, the similarity in the hybridization patterns of Eco RI digested versus undigested pBJS-O preparations, despite the presence of two GAATTC recognition sequences, may be due to an adenine methylation system in S. citri. Restriction site modification in S. citri has been reported elsewhere. Rascoe et al. [26] detected multiple bands of S. citri extrachromosomal DNA by Southern blotting, which they attributed to incomplete restriction due to variable restriction site modification in the DNA, and Ye et al. [27] reported protection of an Eco RI site in the S. citri 16S rDNA. Moreover, differential methylation of restriction sites in the RF of the spiroplasma virus, SVTS2, allowed Sha et al. [28] to clone the full-length DNA.
S. citri BR3-3X showed probe-reactive sequences in both the chromosomal and extrachromosomal DNA fractions. However, that the patterns of hybridization of the two fractions differed significantly from each other demonstrates that the two fractions of BR3-3X DNA were not appreciably cross-contaminated. Sequence analyses of DNA from the two fractions showed that, in BR3-3X, arp1 resides on pBJS-O and arp2 on the chromosome. Hybridization of S. citri ASP-1 and R8A2 plasmid preparations with the pBJS-O probe (Fig. 3B), but not with the arp1 probe (Fig. 3A), indicates that each of these two strains contained a plasmid related to pBJS-O, which differed from pBJS-O in lacking arp1. Although S. citri ASP-1 and R8A2 were originally derived from the same parent strain, both have undergone extensive cultivation in vitro since their first isolation, which may have contributed to the differences between their plasmids and pBJS-O. Moreover, the differences in the maintenance regimes of the various spiroplasmas tested may have contributed to the evolution of their plasmids. In this paper we could not correlate pBJS-O and pBJS-O like sequences with either transmissibility or phytopathogenicity of the spiroplasmas tested. However, it is still hypothesized that SARP1 is involved in S. citri transmission by the insect vector.
Frequent chromosomal rearrangements such as inversions and deletions, leading to genome instability, have been reported in spiroplasmas, such as in the lines derived from S. citri strain BR3 [[29] and [30]]. In the present study, we detected a 0.4 kbp deletion in pBJS-T relative to pBJS-O. Unlike BR3-3X, which was stored frozen, S. citri BR3-T was maintained for several years in turnip plants via transmission by the natural insect vector C. tenellus, possibly leading to the sequence differences between pBJS-T and pBJS-O. A recombinational chromosomal rearrangement is indicated by the 5'-sequence differences between arp1 and arp2 reported above.
Recombination likely also played a role in the generation of pBJS-O like plasmids. The gene organization on pBJS-O is similar to that of the recently characterized IncP-like S. kunkelii CR2-3X plasmid, pSKU146. Yet, the two plasmids have substantially different sequences in the region including the soj-like gene in both plasmids and the IncP oriT sequence in pSKU146. Highly similar sequences in the remainder of the two plasmids suggest that recombination events have occurred during the generation of one or both plasmids.
Phage sequences have been implicated in many recombination events in spiroplasmas. Only a short region with similarity to a phage gene was found in pBJS-O. However, the observation of strong stops to sequencing reactions in the region of nucleotides 1 to 80 is reminiscent of a strong stop encountered during the sequencing of the SVTS2 phage [31]. This strong stop was attributed to potential secondary structure putatively involved in phage packaging. It is, thus, possible that pBJS-O has some phage-like properties.
pBJS-O genes
As mentioned above, ORF3, encoding SARP1, and adjacent ORFs [GenBank:AJ297706], had been cloned and characterized from S. citri BR3-T [15]. ORF3 was flanked downstream by a partial ORF (ORF4) having no known homologs. Upstream, ORF3 was flanked by ORF2, encoding a hypothetical protein with no similarity to any known protein, and ORF1, a partial ORF encoding a putative homolog of a ParA-like protein, Soj, which oscillates from pole to pole [32] and is important for chromosome partitioning in Bacillus subtilis [33]. In this study, the putative protein product of orf4 was predicted to contain eight transmembrane helices. Due to a 0.4 kb deletion in the derivation of pBJS-T, orf4 is possibly a part of the same transcription unit as arp1 and traE in this strain. In BR3-3X, arp1 and orf4 are separated by 281 bp, suggesting that they are transcribed separately. Consistent with different translational constraints on this region in BR3-T and BR3-3X, this region contains a large proportion of the differences between the lines. The translation start site of traE was predicted to be ten nucleotides upstream of the orf4 translation stop site.
Consistent with the observation of Bai et al. [22], putative products of the other pBJS-O ORFs, traE [34] and mob [35], are homologous to proteins that are components of the bacterial type IV secretion system involved in conjugative DNA transfer. Members of the TraE family of proteins are thought to form pili that, in addition to conjugation, are involved in processes like virus infection and biofilm formation. Bai et al. [22] reported the presence of three conserved transmembrane helices in four TraE homologs that they characterized from S. kunkelii M2. Ozbek et al. [36], in their transmission electron micrographs, reported the presence of structures resembling fimbriae and pili in S. kunkelii and Bai et al. [22] considered whether the structures may be involved in conjugation. Bové [37] reported that rod-shaped spiroplasma viruses, approximately 230–280 by 10–15 μm in size, can also be surface-associated. Because they can attach perpendicularly to the host membrane at their tips [[38] and [39]], they might resemble the structures reported as pili/fimbriae. In the putative TraE homolog reported here, unlike its S. kunkelii counterpart, two transmembrane helices were predicted at high probability and a third one at moderate probability. Should the third not actually be a transmembrane helix, the ATP binding site would be located intracellularly rather than extracellularly.
pBJS-O gene organization and evolution
Unlike pSKU146, pBJS-O was found to lack the conserved oriT region characteristic of the IncP group of plasmids. We were also unable to identify a region in pBJS-O resembling any other known plasmid origins of transfer, suggesting that pBJS-O belongs to a hitherto unidentified group of plasmids. Horizontal transfer of a promiscuous plasmid, such as an IncP plasmid, between phylogenetically related and unrelated bacteria would help the hosts quickly adapt to different niches [40]. It is possible that an IncP-like plasmid was acquired by the ancestor of S. citri and S. kunkelii. The plasmid may have co-evolved with the host chromosomes after the divergence of the two species, leading to the emergence of pBJS-O and pSKU146, respectively, and to the adaptation of the pathogens to phylogenetically distinct leafhopper vectors and plant hosts.
Future directions
Molecular genetic tools such as cloning and transposon-mediated mutagenesis are available for the study of mollicutes [41]. Cloned genes were expressed in S. citri GII-3 using artificial plasmids based on the S. citri chromosomal oriC [[42, 43] and [44]]. However, those plasmids tend to integrate into the S. citri chromosome. When pCJ32, a derivative of the oriC plasmid pBOT1, containing an internal fragment of the gene scm1 (a motility-related S. citri gene), was transformed into S. citri GII-3 cells it successfully integrated into the host chromosome by homologous recombination and disrupted scm1, resulting in non-motile S. citri GII-3 mutants [45]. However, attempts to use pBOT1 in S. citri BR3-3X have been unsuccessful (F. Ye, unpublished data), possibly due to the incompatibility of the plasmid with the host. The indigenous S. citri BR-3X plasmid, pBJS-O, will help us develop a better vector for genetic manipulation not only in S. citri BR3-3X but also in other spiroplasmas.
Conclusion
We have shown that the S. citri BR3-3X plasmid, pBJS-O, encodes the putative adhesin SARP1. This is the first report of an S. citri plasmid encoding a putative adhesin. We have further shown that the arp1-like gene, arp2, resides on the BR3-3X chromosome. The indigenous S. citri BR3-3X plasmid, pBJS-O, will be useful for the development of a better vector for genetic manipulation not only in S. citri BR3-3X but also in other spiroplasmas. Our data also suggest that pBJS-O is a novel S. citri plasmid that does not belong to any known plasmid incompatibility group. The differences between pBJS-O and pSKU146 suggest that recombination has contributed to the divergence of the two plasmids.
Methods
Spiroplasmas
S. citri BR3 was isolated from horseradish plants with brittle root disease [7]. S. citri BR3-T, derived from the triply cloned parental isolate (BR3-3X) by repeated transmission in turnips via its insect vector C. tenellus, is insect-transmissible. BR3-M, derived by passage in liquid medium 43 times, is also a transmissible line. The lines BR3-G (maintained in periwinkle plants by graft transmission) and BR3-P (passed in liquid medium more than 130 times) are insect non-transmissible [46]. S. citri R8A2, isolated from citrus in Morocco [47], and its non-helical derivative ASP-1 (both obtained from R.E. Davis, USDA/ARS, Beltsville, MD), are non-transmissible. Also provided by R.E. Davis were S. citri Beni Mellal, originally isolated from C. hematoceps collected in Morocco; S. melliferum TS2, isolated from honeybees and S. floricola 23-6, isolated from a flower surface [48]. S. phoeniceum P40, a gift from G. Gasparich (Towson University, Towson, MD), was originally isolated from periwinkle in Syria [11]. S. kunkelii CR2-3X was isolated by one of us (J. Fletcher) from stunt-diseased corn collected in Costa Rica [49]. All spiroplasmas, except S. kunkelii CR2-3X, were grown in LD8 broth medium [50] at 31°C. The latter was grown in LD8A3 broth medium [51] at 28°C.
Purification of chromosomal and extrachromosomal ds DNAs from spiroplasmas
For Southern blot hybridization and PCR, extrachromosomal double-stranded (ds) DNA of spiroplasma strains was isolated using the QIAprep Spin Miniprep and the QIAGEN Plasmid Mini Kits (Qiagen, Santa Clarita, CA), following the manufacturer's protocols. For primer walking, S. citri BR3-3X extrachromosomal DNA was isolated using a previously published procedure [28]. The isolation of chromosomal DNA from S. citri BR3-3X cells was performed according to Murray and Thompson [52] and using 1.4 M NaCl, 2.5% cetyltrimethylammonium bromide [CTAB], 100 mM Tris-HCl, pH 8.0, and 20 mM EDTA in the extraction buffer.
PCR and sequencing using S. citri BR3-3X plasmid and chromosomal DNAs
To amplify the 3'- and flanking regions of arp genes from S. citri BR3-3X plasmid and chromosomal DNAs, two oligonucleotides were designed, forward (#7686) 5'-AACACTATTTTCACTGCGG-3', from the S. citri BR3-T arp1 sequence (GenBank accession number AJ297706), and reverse (#7960) 5'-TTTTCCATTGTTTTTGTCTCC-3', from the sequence homologous to ORF4 from the plasmid pSKU146 (pSKU146_11; accession number NC_006400). The PCR was carried out in a DNA thermal cycler (MJ Research, Waltham, MA) performing 35 cycles, each of 30 sec at 94°C, 1 min at 42°C and 3 min at 72°C. Reactions were performed separately in a volume of 50 μl containing 2.5 Units Taq polymerase (Promega), 0.20 μM primers, 200 μM of each dNTP, 1.5 mM MgCl2, and 100–150 ng BR3-3X plasmid and ~ 3.5 μg chromosomal DNA. The amplicons were sequenced using ~ 100 ng of each of the PCR products, 10 μM of the same primers used in the PCR in separate reactions by the ABI PRISM BigDye Terminator Cycle Sequencing method (version 1.0, Applied Biosystems, Foster City, CA) with an ABI PRISM 3700 Automated DNA Analyzer (Perkin Elmer Biosystems, Foster City, CA).
Southern blotting
Extrachromosomal DNA of each spiroplasma strain was digested with Eco RI (Life Technologies, Inc.) for 4 h at 37°C. The fragments were separated by electrophoresis on a 0.75% (w/v) agarose gel in 1× TAE running buffer and transferred to Hybond-N+ nylon membranes (Amersham Biosciences, Uppsala, Sweden) according to standard procedures. The blots were subsequently hybridized to Dig-11-UTP-labeled arp1-derived and whole-plasmid probes, labeled using a DIG DNA Labeling Kit (Roche Molecular Biochemicals, Indianapolis, IN), following the manufacturer's instructions. The arp1-derived probe was obtained by PCR, using clone pP89B (an Rsa I fragment of S. citri BR3-T genomic DNA; [15]) as template, and primer pair T7 and #7483 (5'-TTTAACATCAACCGAACCC-3'). The probe comprised 657 bp of a DNA segment from S. citri BR3-T (AJ297706; positions 2315–2989) and 72 bp derived from the cloning vector (pBluescript). PCR was carried out in a DNA thermal cycler performing 34 cycles, each of 30 sec at 94°C, 30 sec at 54°C and 1 min at 72°C. Reactions were performed in a volume of 50 μl containing 1 Unit Taq polymerase, 0.25 μM primers, 250 μM of each dNTP, 50 – 100 ng template DNA, and 2.5 mM MgCl2. Hybridizations were performed at 55°C in Church buffer (0.5 M sodium phosphate buffer, pH 7.2, 7% SDS, and 1 mM EDTA) overnight followed by four washes, each of 20 min, at 55°C in washing buffer (40 mM sodium phosphate buffer, pH 7.2, containing 0.1% SDS). Detection of the DIG-labeled probes was performed using a DIG Luminescent Detection Kit (Roche) following the manufacturer's protocol.
Complete nucleotide sequencing of pBJS-O
The sequence AJ297706 was used to design primers to initiate primer walking to completely sequence and characterize the unknown portion of pBJS-O. The sequencing reactions were performed using ~ 1.2 μg of pBJS-O DNA and 40 μM of primers with the ABI PRISM BigDye Terminator Cycle Sequencing method and the ABI PRISM 3700 Automated DNA Analyzer, as mentioned above. The total 134 sequence reads with an average length of 600 bases gave us about 6× coverage of the entire plasmid sequence. The fragments were assembled from the trace files using the software package PipeOnline 2.0 [53]. Physical gaps in the sequence were closed by PCR and cloning of the products into vector pGEM-T (Promega). The clones were sequenced using primers T7 and SP6. The consensus sequence of the final assembly was annotated using the BLASTX search program [54] and the ORF Finder tool at NCBI, in which a minimum length of 100 bases was used for the nucleotide sequence of a putative ORF. The nucleotide and amino acid sequence analysis tools offered by the Biology Workbench at the San Diego Supercomputer Center, such as ClustalW and PHYLIP for generating the unrooted phylogenetic tree of the S. citri arp sequences, were used to further analyze the plasmid and the polypeptide sequences. BLASTN and BLASTP searches were carried out to find out relationships with the closest homologs. S. kunkelii CR2-3X genome sequence data were accessed and BLAST searches were performed at the Spiroplasma Genome Sequencing Project Web site mentioned above.
Note added in proof
During review of this manuscript sequences of plasmids from a different strain of S. citri were released [GenBank:AJ969069, GenBank:AJ969070, GenBank:AJ969071, GenBank:AJ969072, GenBank:AJ969073, GenBank:AJ969074].
References
McCoy RE, Caudwell A, Chang CJ, Chen TA, Chiykowski LN, Cousin MT, Dale JL, deLeeuw GTN, Golino DA, Hackett KJ, Kirkpatrick BC, Marwitz R, Petzold H, Sinha RC, Sugiura M, Whitcomb RF, Yang IL, Zhu BM, Seemûller E: Plant diseases associated with mycoplasma-like organisms. The Mycoplasmas. Edited by: Whitcomb RF, Tully JG. 1989, New York: Academic Press, 5: 545-640.
Garnier M, Foissac X, Gaurivaud P, Laigret F, Renaudin J, Saillard C, Bové JM: Mycoplasmas, plants, insect vectors: a matrimonial triangle. C R Acad Sci Paris. 2001, 324: 923-928.
Woese CR: Bacterial evolution. Microbiol Rev. 1987, 51: 221-271.
Whitcomb RF, Tully JG, Bové JM, Saglio P: Spiroplasmas and acholeplasmas: multiplication in insects. Science. 1973, 182: 1251-1253.
Saglio P, Lhospital M, Lafleche D, Dupont G, Bové JM, Tully JG, Freundt EA: Spiroplasma citri gen. and sp. n.: a Mycoplasma-like Organism Associated with "Stubborn" Disease of Citrus. Int J Syst Bacteriol. 1973, 23: 191-204.
Calavan EC, Bové JM: Ecology of Spiroplasma citri. The Mycoplasmas. Edited by: Whitcomb RF, Tully JG. 1989, New York: Academic Press, 5: 425-485.
Fletcher J, Schultz GA, Davis RE, Eastman CE, Goodman RE: Brittle root disease of horseradish: Evidence for an etiological role of Spiroplasma citri. Phytopathology. 1981, 71: 1073-1080.
Chen TA, Liao CH: Corn stunt spiroplasma: isolation, cultivation, and proof of pathogenicity. Science. 1975, 188: 1015-1017.
Williamson DL, Whitcomb RF: Plant mycoplasmas: a cultivable spiroplasma causes corn stunt disease. Science. 1975, 188: 1018-1020.
Whitcomb RF, Chen TA, Williamson DL, Liao C, Tully JG, Bové JM, Mouches C, Rose DL, Coan ME, Clark TB: Spiroplasma kunkelii sp. nov.: Characterization of the Etiological Agent of Corn Stunt Disease. Int J Syst Bact. 1986, 36: 170-178.
Saillard C, Vignault JC, Gadeau A, Carle P, Garnier M, Fos A, Bové JM, Tully JG, Whitcomb RF: Discovery of a new plant-pathogenic spiroplasma. Isr J Med Sci. 1984, 20: 1013-1015.
Bové JM, Renaudin J, Saillard C, Foissac X, Garnier M: Spiroplasma citri, a plant pathogenic mollicute: relationships with its two hosts, the plant and the leafhopper vector. Annu Rev Phytopathol. 2003, 41: 483-500. 10.1146/annurev.phyto.41.052102.104034.
Steiner T, McGarrity GJ, Bové JM, Phillips DM, Garnier M: Insect cell cultures in the study of attachment and pathogenicity of spiroplasmas and mycoplasmas. Ann Microbiol (Paris). 1984, 135A: 47-53.
Yu J, Wayadande AC, Fletcher J: Spiroplasma citri surface protein P89 implicated in adhesion to cells of the vector Circulifer tenellus. Phytopathology. 2000, 90: 716-722.
Berg M, Melcher U, Fletcher J: Characterization of Spiroplasma citri adhesion related protein SARP1, which contains a domain of a novel family designated sarpin. Gene. 2001, 275: 57-64. 10.1016/S0378-1119(01)00655-2.
Barber CE, Archer DB, Daniels MJ: Molecular biology of spiroplasma plasmids. Yale J Biol Med. 1983, 56: 777-781.
Gasparich GE, Hackett KJ, Clark EA, Renaudin J, Whitcomb RF: Occurrence of extrachromosomal deoxyribonucleic acids in spiroplasmas associated with plants, insects, and ticks. Plasmid. 1993, 29: 81-93. 10.1006/plas.1993.1011.
Mouches C, Barroso G, Gadeau A, Bové JM: Characterization of two cryptic plasmids from Spiroplasma citri and occurrence of their DNA sequences among various spiroplasmas. Ann Microbiol Paris. 1984, 135A: 17-24.
Archer DB, Best J, Barber C: Isolation and Restriction Mapping of a Spiroplasma Plasmid. J Gen Microbiol. 1981, 126: 511-514.
Fletcher J, Shaw ME, Baker GR, Dugan KJ, Ye F, Sha Y, Zuck PD, Myers GD: Molecular characterization of Spiroplasma citri BR3 lines that differ in transmissibility by the leafhopper Circulifer tenellus. Can J Microbiol. 1996, 42: 124-131.
Melcher U, Fletcher J: Genetic variation in Spiroplasma citri. Eur J Plant Pathol. 1999, 105: 519-533. 10.1023/A:1008757716803.
Bai X, Fazzolari T, Hogenhout SA: Identification and characterization of traE genes of Spiroplasma kunkelii. Gene. 2004, 336: 81-91. 10.1016/j.gene.2004.03.022.
Davis RE, Dally EL, Jomantiene R, Zhao Y, Roe B, Lin SP, Shao J: Cryptic plasmid pSKU146 from the wall-less plant pathogen Spiroplasma kunkelii encodes an adhesin and components of a type IV translocation-related conjugation system. Plasmid. 2005, 53: 179-190. 10.1016/j.plasmid.2004.09.002.
Fleury B, Bergonier D, Berthelot X, Peterhans E, Frey J, Vilei EM: Characterization of P40, a cytadhesin of Mycoplasma agalactiae. Infect Immun. 2002, 70: 5612-5621. 10.1128/IAI.70.10.5612-5621.2002.
Gasparich GE: Spiroplasmas: evolution, adaptation and diversity. Front Biosci. 2002, 7: d619-640.
Rascoe J, Melcher U, Fletcher J: Evaluation of variations in gene presence and expression among lines of Spiroplasma citri using arbitrarily-primed PCR of cDNA. Phytopathology. 1996, 86: S96-
Ye F, Laigret F, Carle P, Bové JM: Chromosomal heterogeneity among various strains of Spiroplasma citri. Int J Syst Bact. 1995, 45: 729-734.
Sha Y, Melcher U, Davis RE, Fletcher J: Resistance of Spiroplasma citri lines to the virus SVTS2 is associated with integration of viral DNA sequences into host chromosomal and extrachromosomal DNA. Appl Environ Microbiol. 1995, 61: 3950-3959.
Melcher U, Sha Y, Ye F, Fletcher J: Mechanisms of spiroplasma genome variation associated with SpV1-like viral DNA inferred from sequence comparisons. Microb Comp Genomics. 1999, 4: 29-46.
Ye F, Melcher U, Rascoe JE, Fletcher J: Extensive chromosome aberrations in Spiroplasma citri strain BR3. Biochem Genet. 1996, 34: 269-286.
Sha Y, Melcher U, Davis RE, Fletcher J: Common elements of spiroplasma plectroviruses revealed by nucleotide sequence of SVTS2. Virus Genes. 2000, 20: 47-56. 10.1023/A:1008108106916.
Quisel JD, Lin DC, Grossman AD: Control of development by altered localization of a transcription factor in B. subtilis. Mol Cell. 1999, 4: 665-672. 10.1016/S1097-2765(00)80377-9.
Ireton K, Gunther IV NW, Grossman AD: spoOJ is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994, 176: 5320-5329.
Winans SC, Walker GC: Conjugal transfer system of the N incompatibility plasmid pKM101. J Bacteriol. 1985, 161: 402-410.
Clewell D, Helinski D: Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an open circular DNA form. Proc Natl Acad Sci. 1969, 62: 1159-1166.
Ozbek E, Miller SA, Meulia T, Hogenhout SA: Infection and replication sites of Spiroplasma kunkelii (Class: Mollicutes) in midgut and Malpighian tubules of the leafhopper Dalbulus maidis. J Invert Pathol. 2003, 82: 167-175. 10.1016/S0022-2011(03)00031-4.
Bové JM: Wall-less prokaryotes of plants. Ann Rev Phytopathol. 1984, 22: 361-396. 10.1146/annurev.py.22.090184.002045.
Townsend R, Markham PG, Plaskitt KA: Multiplication and morphology of Spiroplasma citri in the leafhopper Euscelidius variegatus. Ann Appl Biol. 1977, 87: 307-313.
Sha Y-S: Molecular characterization of spiroplasma viruses and the mechanism of resistance of Spiroplasma citri lines to infection by the virus SVTS2. PhD thesis. 1993, Oklahoma State University, Entomology and Plant Pathology Department
Nojiri H, Shintani M, Omori T: Divergence of mobile genetic elements involved in the distribution of xenobiotic-catabolic capacity. Appl Microbiol Biotechnol. 2004, 64: 154-174. 10.1007/s00253-003-1509-y.
Renaudin J: Extrachromosomal elements and gene transfer. Molecular Biology and Pathogenicity of Mycoplasmas. Edited by: Razin S, Herrmann R. 2002, New York: Kluwer Academic Publishers/Plenum Press, 347-370.
Renaudin J, Bové JM: Plasmid and viral vectors for gene cloning and expression in Spiroplasma citri. Molecular and Diagnostic Procedures in Mycoplasmology. Edited by: Razin S, Tully JG. 1995, New York: Academic Press, 167-178.
Renaudin J, Marais A, Verdin E, Duret S, Foissac X, Laigret F, Bové JM: Integrative and free Spiroplasma citri oriC plasmids: expression of the Spiroplasma phoeniceum spiralin in Spiroplasma citri. J Bacteriol. 1995, 177: 2800-2877.
Ye F, Renaudin J, Bové JM, Laigret F: Cloning and sequencing of the replication origin (oriC) of the Spiroplasma citri chromosome and construction of autonomously replicating artificial plasmids. Curr Microbiol. 1994, 29: 23-29. 10.1007/BF01570187.
Duret S, Danet JL, Garnier M, Renaudin J: Gene disruption through homologous recombination in Spiroplasma citri: an scm1-disrupted motility mutant is pathogenic. J Bacteriol. 1999, 181: 7449-7456.
Wayadande AC, Fletcher J: Transmission of Spiroplasma citri lines and their ability to cross gut and salivary gland barriers within the leafhopper vector Circulifer tenellus. Phytopathology. 1995, 85: 1256-1259.
Saglio P, Lafleche D, Bonissol C, Bové JM: Culture in vitro des mycoplasmes associés au stubborn des agrumes et leur observation au microscope électronique. C R Acad Sci Paris. 1971, 272: 1387-1390.
Davis RE, Lee IM, Basciano LK: Spiroplasmas: serological grouping of strains associated with plants and insects. Can J Microbiol. 1979, 25: 861-866.
Zhao Y, Hammond RW, Jomantiene R, Dally EL, Lee IM, Jia H, Wu H, Lin S, Zhang P, Kenton S, Najar FZ, Hua A, Roe BA, Fletcher J, Davis RE: Gene content and organization of an 85-kbp DNA segment from the genome of the phytopathogenic mollicute Spiroplasma kunkelii. Molec Genet Genom. 2003, 269: 592-602. 10.1007/s00438-003-0878-3.
Chen TA, Davis RE: Cultivation of spiroplasmas. The Mycoplasmas. Edited by: Whitcomb RF, Tully JG. 1979, New York: Academic Press, 65-79.
Lee I-M, Davis RE: Serum-free media for cultivation of spiroplasmas. Can J Microbiol. 1989, 35: 1092-1099.
Murray MG, Thompson WF: Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8: 4321-4325.
Ayoubi P, Jin X, Leite S, Liu X, Martajaja J, Abduraham A, Wan Q, Yan W, Misawa E, Prade RA: PipeOnline 2.0: automated EST processing and functional data sorting. Nucleic Acids Res. 2002, 30: 4761-4769. 10.1093/nar/gkf585.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ: Basic local alignment search tool. J Mol Biol. 1990, 215: 403-410. 10.1006/jmbi.1990.9999.
S. kunkelii Genome Project. [http://www.genome.ou.edu/spiro.html]
ORF Finder. [http://www.ncbi.nlm.nih.gov/gorf/gorf.html]
Biology Workbench. [http://workbench.sdsc.edu/]
Acknowledgements
We are indebted to Dr. Robert E. Davis of USDA/ARS, Beltsville, MD for personally communicating his work on the plasmid related to pBJS-O from S. kunkelii CR2-3X and sharing with us some of the unpublished data. Staff members of the Recombinant DNA/Protein Resource Facility at OSU are thanked for oligonucleotide synthesis, DNA sequencing, and valuable technical assistance and advice. Dr. Samir Gunjan is thanked for providing technical help in cloning pBJS-O PCR products while filling the gaps in the sequence. The people involved in the S. kunkelii CR2-3X genome sequencing project [B.A. Roe, S.P. Lin, H.G. Jia, H.M. Wu, D. Kupfer, and R.E. Davis] are thanked for making the sequence data publicly available.
We are grateful to Drs. Moses N. Vijaykumar, Richard C. Essenberg and Astri Wayadande for critically reviewing the manuscript. This work was supported by grants from the United States Department of Agriculture, the Robert J. Sirny Professorship to UM, and the Oklahoma Agricultural Experiment Station, whose Director has approved the manuscript for publication.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
BDJ performed isolation, distribution and sequence characterization of pBJS-O. JR carried out pBJS-O sequencing and assisted BDJ in primer design and sequence assembly. MB performed S. citri BR3-T arp2 gene cloning and sequencing, and also assisted BDJ in pBJS-O distribution experiments. BDJ, MB, UM and JF planned the research, BDJ and UM wrote the manuscript and MB and JF reviewed it.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Joshi, B.D., Berg, M., Rogers, J. et al. Sequence comparisons of plasmids pBJS-O of Spiroplasma citri and pSKU146 of S. kunkelii: implications for plasmid evolution. BMC Genomics 6, 175 (2005). https://doi.org/10.1186/1471-2164-6-175
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2164-6-175