Abstract
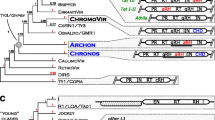
At present, only fragmentary data about the biodiversity and structural features of L1 retrotransposons of angiosperms related to individual elements from few genomes are available. There is no clear cladistic classification of transposable elements of this family. Structural data on elements from particular groups of angiosperm L1 retrotransposons are also scarce. For these reasons, a comprehensive structural and phylogenetic analysis of L1 retrotransposons in angiosperms has been undertaken. We have compiled information on 19 genomes of angiosperm species that was available in databases and discerned three clades of L1 elements on the base of the phylogeny of the conserved reverse transcriptase region and on their structural organization. It is demonstrated that the emergence of new protein types that form the ribonucleoprotein particles of the retrotransposons and the acquisition of RNH protein-encoding regions by several elements were crucial steps in the formation of new L1 retrotransposon types.
Similar content being viewed by others
References
Anisimova, M. and Gascuel, O., Approximate Likelihood-Ratio Test for Branches: A Fast, Accurate, and Powerful Alternative, Syst. Biol., 2006, vol. 55, pp. 539–552.
Chaw, S.M., Chang, C.C., Chen, H.L., and Li, W.H., Dating the Monocot-Dicot Divergence and the Origin of Core Eudicots using Whole Chloroplast Genomes, J. Mol. Evol., 2004, vol. 58, pp. 424–441.
Eddy, S.R., Profile Hidden Markov Models, Bioinformatics, 1998, vol. 14, pp. 755–763.
Edgar, R.C., MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput, Nucleic Acids Res., 2004, vol. 32, pp. 1792–1797.
Guindon, S., Dufayard, J.F., Lefort, V., et al., New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0, Syst. Biol., 2010, vol. 59, pp. 307–321.
Heitkam, T. and Schmidt, T., BNR—a LINE Family from Beta vulgaris—Contains a RRM Domain in Open Reading Frame 1 and Defines a L1 Sub-Clade Present in Diverse Plant Genomes, Plant J., 2009, vol. 59, pp. 872–882.
Higashiyama, T., Noutoshi, Y., Fujie, M., and Yamada, T., Zepp, a LINE-Like Retrotransposon Accumulated in the Chlorella Telomeric Region, EMBO J., 1997, vol. 16, pp. 3715–3723.
Jurka, J., Kapitonov, V.V., Pavlicek, A., et al., Repbase Update, a Database of Eukaryotic Repetitive Elements, Cytogenet. Genome Res., 2005, vol. 110, pp. 462–467.
Khazina, E. and Weichenrieder, O., Non-LTR Retrotransposons Encode Noncanonical RRM Domains in Their First Open Reading Frame, Proc. Natl. Acad. Sci. U.S.A., 2009, vol. 106, pp. 731–736.
Kojima, K.K. and Fujiwara, H., Cross-Genome Screening of Novel Sequence-Specific Non-LTR Retrotransposons: Various Multicopy RNA Genes and Microsatellites Are Selected as Targets, Mol. Biol. Evol., 2004, vol. 21, pp. 207–217.
Kojima, K.K. and Fujiwara, H., An Extra Ordinary Retrotransposon Family Encoding Dual Endonucleases, Genome Res., 2005, vol. 15, pp. 1106–1117.
Kolosha, V.O. and Martin, S.L., In vitro Properties of the First ORF Protein from Mouse LINE-1 Support Its Role in Ribonucleoprotein Particle Formation during Retrotransposition, Proc. Natl. Acad. Sci. USA, 1997, vol. 94, pp. 10155–10160.
Lander, E.S., Linton, L.M., Birren, B., et al., Initial Sequencing and Analysis of the Human Genome, Nature, 2001, vol. 409, pp. 860–921.
Leeton, P.R. and Smyth, D.R., An Abundant LINE-Like Element Amplified in the Genome of Lilium speciosum, Mol. Gen. Genet., 1993, vol. 237, pp. 97–104.
Ma, J. and Bennetzen, J.L., Rapid Recent Growth and Divergence of Rice Nuclear Genomes, Proc. Natl. Acad. Sci. USA, 2004, vol. 101, pp. 12404–12410.
Malik, H.S., Ribonuclease H Evolution in Retrotransposable Elements, Cytogenet. Genome Res., 2005, vol. 110, pp. 392–401.
Marchler-Bauer, A., Lu, S., Anderson, J.B., et al., CDD: a Conserved Domain Database for the Functional Annotation of Proteins, Nucleic Acids Res., 2011, vol. 39, pp. 225–229.
Maris, C., Dominguez, C., and Allain, F.H., The RNA Recognition Motif, a Plastic RNA-Binding Platform to Regulate Post-Transcriptional Gene Expression, FEBSJ, 2005, vol. 272, pp. 2118–2131.
Martin, S.L., The ORF1 Protein Encoded by LINE-1: Structure and Function during L1 Retrotransposition, J. Biomed. Biotechnol, 2006.
Matsui, T., Tanaka, T., Endoh, H., et al., The RNA Recognition Mechanism of Human Immunodeficiency Virus (HIV) Type 2 NCp8 Is Different from that of HIV-1 NCp7, Biochemistry, 2009, vol. 48, pp. 4314–4323.
Noma, K., Ohtsubo, E., and Ohtsubo, H., Non-LTR Retrotransposons (LINEs) as Ubiquitous Components of Plant Genomes, Mol. Gen. Genet., 1999, vol. 261, pp. 71–79.
Noma, K., Ohtsubo, H., and Ohtsubo, E., ATLN Elements, LINEs from Arabidopsis thaliana: Identification and Characterization, DNA Res., 2000, vol. 7, pp. 291–303.
Novikova, O., Fet, V., and Blinov, A., Non-LTR Retrotransposons in Fungi, Funct. Integr. Genomics, 2008, vol. 9, pp. 27–42.
Rho, M., Tang H. MGE Scannon-LTR: Computational Identification and Classification of Autonomous Non-LTR Retrotransposons in Eukaryotic Genomes, Nucleic Acids Res., 2009, vol. 37, p. e143.
Sakamoto, K., Ohmido, N., Fukui, K., et al., Site-Specific Accumulation of a LINE-Like Retrotransposon in a Sex Chromosome of the Dioecious Plant Cannabis sativa, Plant. Mol. Biol., 2000, vol. 44, pp. 723–732.
Schwarz-Sommer, Z., Leclercq, L., Gobel, E., and Saedler, H., Cin4, An Insert Altering the Structure of the A1 Gene in Zea mays, Exhibits Properties of Nonviral Retrotransposons, EMBO J., 1987, vol. 6, pp. 3873–3880.
Sding, J., Protein Homology Detection by HMM-HMM Comparison, Bioinformatics, 2005, vol. 21, pp. 951–960.
Sergeeva, E.M. and Salina, E.A., Mobile Elements and the Evolution of the Plant Genome, Vavilov. Zh. Genet. Selekts., 2011, vol. 15, no. 2, pp. 382–398.
Sormacheva, N.D. and Blinov, A.G., LTR Retrotransposons of Plants, Vavilov. Zh. Genet. Selekts., 2011, vol. 15, no. 2, pp. 351–381.
Tamura, K., Peterson, D., Peterson, N., et al., MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods, Mol. Biol. Evol., 2011. http://www.kumarlab.net/publications
Turcotte, K., Srinivasan, S., and Bureau, T., Survey of Transposable Elements from Rice Genomic Sequences, Plant J., 2001, vol. 25, pp. 169–179.
Vershinin, A.V., Druka, A., Alkhimova, A.G., et al., LINEs and gypsy-Like Retrotransposons in Hordeum Species, Plant. Mol. Biol., 2002, vol. 49, pp. 1–14.
Wenke, T., Holtgrwe, D., Horn, A.V., et al., An Abundant and Heavily Truncated Non-LTR Retrotransposon (LINE) Family in Beta vulgaris, Plant. Mol. Biol., 2009, vol. 71, pp. 585–597.
Wright, D.A., Ke, N., Smalle, J., et al., Multiple Non-LTR Retrotransposons in the Genome of Arabidopsis thaliana, Genetics, 1996, vol. 142, pp. 569–578.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © G.A. Smyshlyaev, A.G. Blinov, 2011, published in Vavilovskii Zhurnal Genetiki i Selektsii, 2011, Vol. 15, No. 3, pp. 563–571.
Rights and permissions
About this article
Cite this article
Smyshlyaev, G.A., Blinov, A.G. Evolution and biodiversity of L1 retrotransposons in angiosperm genomes. Russ J Genet Appl Res 2, 72–78 (2012). https://doi.org/10.1134/S2079059712010133
Published:
Issue Date:
DOI: https://doi.org/10.1134/S2079059712010133