Summary
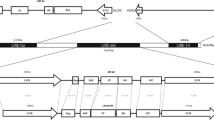
The genomes of Lilium species are very large, containing 30–40 million kilobase pairs of DNA. An abundant fragment of 3.5 kb was released by BamHI digestion of genomic DNA of Lilium speciosum. Analysis of 20 genomic clones containing sequences homologous to the fragment showed it to be part of a 4.45 kb dispersed repeat, which was named del2. Sequence analysis of one full element and regions of four others revealed del2 to be a non-LTR (long terminal repeat) retrotransposon. It is flanked by short direct repeats of from 4 to 13 bp and a run of adenines occurs at one end (the proposed 3′ end), 63 by downstream from a polyadenylation signal. A possible RNA polymerase II promoter similar to that found in Drosophila I and F group elements is present internally 30 by downstream from the 5′ end. Two degenerate open reading frames (ORFs) are present, the 5′ ORF containing a gag-related cysteine motif, and the 3′ ORF containing a different cysteine motif also found in most non-LTR retrotransposons. The 3′ ORF also has regions with homology to reverse transcriptase sequences, which are most similar to those in Cin4 of maize, the Ll LINE elements of humans and mice and the R2 ribosomal DNA inserts of insects. The majority of del2 elements occur as the full 4.45 kb element. They account for an estimated 4 % of the L. speciosum genome and are present in approximately 250 000 copies. del2-related sequences were also detected in 12 other monocot species. del2 is the most abundant non-LTR retrotransposon identified so far and reveals that LINE-like elements have been greatly amplified in some plant genomes just as they have in mammals.
Similar content being viewed by others
References
Besansky NJ (1990) A retrotransposable element from the mosquito Anopheles gambiae. Mol Cell Biol 10:863–871
Bucheton A (1990) I transposable elements and I-R hybrid dysgenesis in Drosophila. Trends Genet 6:16–21
Burke WD, Calalang CC, Eickbush TH (1987) The site-specific ribosomal insertion element type II of Bombyx mori (R2Bm) contains the coding sequence for a reverse transcriptase-like enzyme. Mol Cell Biol 7:2221–2230
Burton FH, Loeb DD, Voliva CF, Martin SL, Edgell MH, Hutchinson CA (1986) Conservation throughout Mammalia and extensive protein encoding capacity of the highly repeated DNA long interspersed sequence one. J Mol Biol 187:291–304
Covey SN (1986) Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauli-flower mosaic virus. Nucleic Acids Res 14:623–633
Di Nocera PP (1988) Close relationship between non-viral retroposons in Drosophila melanogaster. Nucleic Acids Res 16:4041–4052
Di Nocera PP, Casari G (1987) Related polypeptides are encoded by Drosophila F elements, I factors, and mammalian L1 sequences. Proc Natl Acad Sci USA 84:5843–5847
Evans JP, Palmiter RD (1991) Retrotransposition of a mouse L1 element. Proc Natl Acad Sci USA 88:8792–8795
Fanning T, Singer M (1987) The LINE-1 DNA sequences in four mammalian orders predict proteins that conserve homologies in retrovirus proteins. Nucleic Acids Res 15:2251–2260
Fawcett DH, Lister CK, Kellett E, Finnegan DJ (1986) Transposable elements controlling I-R hybrid dysgenesis in D. melanogaster are similar to mammalian LINEs. Cell 47:1007–1015
Finnegan DJ (1989) The I factor and I-R hybrid dysgenesis in Drosophila melanogaster. In: Berg DE, Howe MM (eds) Mobile DNA. American Society for Microbiology, Washington DC, pp 503–517
Friedman BE, Bouchard RA, Stern H (1982) DNA sequences repaired at pachytene exhibit strong homology among distantly related plants. Chromosoma 87:409–424
Garrett JE, Knutzon DS, Carroll D (1989) Composite transposable element in the Xenopus laevis genome. Mol Cell Biol 9:3018–3027
Hutchinson CA, Hardies SC, Loeb DD, Shehee WR, Edgell MH (1989) LINEs and related retrotransposons: long interspersed repeated sequences in the eucaryotic genome. In: Berg DE, Howe MM (eds) Mobile DNA. American Society for Microbiology, Washington DC, pp 593–617
Ivanov VA, Melnikov AA, Siunov AV, Fodor II, Ilyin YV (1991) Authentic reverse transcriptase is encoded by jockey mobile Drosophila element related to mammalian LINEs. EMBO J 10:2489–2495
Jakubczak JL, Xiong Y, Eickbush TH (1990) Type I (R1) and type II (R2) ribosomal DNA insertions of Drosophila melanogaster are retrotransposable elements closely related to those of Bombyx mori. J Mol Biol 212:37–52
Jakubczak JL, Burke WD, Eickbush TH (1991) Retrotransposable elements R1 and R2 interrupt the rRNA genes of most insects. Proc Natl Acad Sci USA 88:3295–3299
Jensen S, Heidmann T (1991) An indicator gene for detection of germline retrotransposition in transgenic Drosophila demonstrates RNA-mediated transposition of the LINE I element. EMBO J 10:1927–1937
Joseph JL, Sentry JW, Smyth DR (1990) Interspecies distribution of abundant DNA sequences in Lilium. J Mol Evol 30:146–154
Kimmel BE, Ole-Moiyol OK, Young JR (1987) Ingi, a 5.2-kb dispersed sequence element from Trypanosoma brucei that carries half a smaller mobile element at either end and has homology with mammalian LINES. Mol Cell Biol 7:1465–1475
Mathias SL, Scott AF, Kazazian HH, Boeke JD, Gabriel A (1991) Reverse transcriptase encoded by a human transposable element. Science 254:1808–1810
Minchiotti G, Di Nocera PP (1991) Convergent transcription initiates from oppositely oriented promoters within the 5′ end regions of Drosophila melanogaster F elements. Mol Cell Biol 11:5171–5180
Mizrokhi LJ, Georgieva SG, Ilyin YV (1988) jockey, a mobile Drosophila element similar to mammalian LINEs, is transcribed from the internal promoter by RNA polymerase II. Cell 54:685–691
Murphy NB, Pays A, Tebabi P, Coquelet H, Guyaux M, Steinert M, Pays E (1987) Trypanosoma brucei repeated element with unusual structural and transcriptional properties. J Mol Biol 195:855–871
O'Hare K, Alley MRK, Cullingford TE, Driver A, Sanderson MJ (1991) DNA sequence of the Doe retroposon in the white-1 mutant of Drosophila melanogaster and of secondary insertions in the phenotypically altered derivatives white-honey and white-eosin. Mol Gen Genet 225:17–24
Pélisson A, Finnegan DJ, Bucheton A (1991) Evidence for retrotransposition of the I factor, a LINE element of Drosophila melanogaster. Proc Natl Acad Sci USA 88:4907–4910
Priimägi AF, Mizrokhi LJ, Ilyin YV (1988) The Drosophila mobile element jockey belongs to LINEs and contains coding sequences homologus to some retroviral proteins. Gene 70:253–262
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: A laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Schwarz-Sommer Z, Leclercq L, Göbel E, Saedler H (1987) Cin4, an insert altering the structure of the A1 gene in Zea mays, exhibits properties of nonviral retrotransposons. EMBO J 6:3873–3880
Sentry JW, Smyth DR (1985) A family of repeated sequences dispersed through the genome of Lilium henryi. Chromosoma 92:149–155
Sentry JW, Smyth DR (1989) An element with long terminal repeats and its variant arrangements in the genome of Lilium henryi. Mol Gen Genet 215:349–354
Smyth DR, Kalitsis P, Joseph JL, Sentry JW (1989) Plant retrotransposon from Lilium henryi is related to Ty3 of yeast and the gypsy group of Drosophila. Proc Natl Acad Sci USA 86:5015–5019
Swergold GD (1990) Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol 10:6718–6729
Xiong Y, Eickbush TH (1988) The site-specific ribosomal DNA insertion element R1 Bm belongs to a class of non-long-terminalrepeat retrotransposons. Mol Cell Biol 8:114–123
Xiong Y, Eickbush TH (1990) Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9:3353–3362
Author information
Authors and Affiliations
Additional information
Communicated by D. Finnegan
Rights and permissions
About this article
Cite this article
Leeton, P.R.J., Smyth, D.R. An abundant LINE-like element amplified in the genome of Lilium speciosum . Molec. Gen. Genet. 237, 97–104 (1993). https://doi.org/10.1007/BF00282789
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00282789