Abstract
Transposable elements (TEs) comprise a significant part of eukaryotic genomes being a major source of genome instability and mutagenesis. Cellular defense systems suppress the TE expansion at all stages of their life cycle. Piwi proteins and Piwi-interacting RNAs (piRNAs) are key elements of the anti-transposon defense system, which control TE activity in metazoan gonads preventing inheritable transpositions and developmental defects. In this review, we discuss various regulatory mechanisms by which small RNAs combat TE activity. However, active transposons persist, suggesting these powerful anti-transposon defense mechanisms have a limited capacity. A growing body of evidence suggests that increased TE activity coincides with genome reprogramming and telomere lengthening in different species. In the Drosophila fruit fly, whose telomeres consist only of retrotransposons, a piRNA-mediated mechanism is required for telomere maintenance and their length control. Therefore, the efficacy of protective mechanisms must be finely balanced in order not only to suppress the activity of transposons, but also to maintain the proper length and stability of telomeres. Structural and functional relationship between the telomere homeostasis and LINE1 retrotransposon in human cells indicates a close link between selfish TEs and the vital structure of the genome, telomere. This relationship, which permits the retention of active TEs in the genome, is reportedly a legacy of the retrotransposon origin of telomeres. The maintenance of telomeres and the execution of other crucial roles that TEs acquired during the process of their domestication in the genome serve as a type of payment for such a “service”.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Currently, a bulk of data has been accumulated to demonstrate a close relationship between the key cellular processes and the regulation of the activity of transposable elements (TEs). This relationship enables survival of TEs in the genomes despite strong defense mechanisms limiting their activity. What mechanism underlies this global genomic trade-off? TEs serve as a rich source of evolutionary changes in the genome: they offer enhancers, promoters, exons, splicing sites, architectural elements and participate in the key mechanisms of development and immune response [1-4]. TEs contribute to the maintenance of essential chromosome structures such as centromeres and ribosomal RNA gene loci [5-8]. In this review, we focus on the origin and maintenance of telomeres, one of the most compelling cases of the role of retrotransposons in genome evolution.
Telomeres – the ends of linear chromosomes – have captivated attention of many scientists for more than 50 years. In 1971-1973, Aleksey M. Olovnikov published his brilliant prediction regarding the “Achilles’ heel of the double helix,” or chromosome end under-replication [9, 10]. He also suggested existence of a specialized DNA polymerase, which compensates for the shortening of telomeric DNA. Much later this enzyme, now known as telomerase, was discovered. It turned out that this was a reverse transcriptase, which acts in a complex with the RNA template [11]. The ends of the primary linear chromosomes are thought to be protected by the attachments of retroelements. From this point of view, the telomerase ribonucleoprotein complex can be considered as a specialized retroelement that evolved to protect the chromosome ends [12]. In fact, phylogenetic analysis of the reverse transcriptase of retrotransposons and telomerase revealed that they originated from a common ancient retroelement enzyme [13-15]. Telomerase maintains telomeres in most organisms, but there are other ways to elongate chromosome ends. There are many modern species of animals whose telomeres are supported by the retrotransposon attachments. These are many species of insects and, in particular, the Drosophilidae family. It is believed that Drosophila has lost telomerase, and the specialized telomeric retrotransposons are used to maintain telomeres. The telomeres of the silkworm Bombyx mori are of mixed type, and they are maintained by the specific telomeric retroelements and low-activity telomerase. Under certain conditions, retroelements are also attached to the human telomeres and to telomeres of other species using telomerase [16, 17]. In these cases, the double strand break at the chromosome end is used as a convenient target for TE retrotranspositions. However, mosquitoes do not have telomerase or telomeric retroelements, and the telomeres lengthening appeared to be mediated by the recombination of short satellite-like repeats [18, 19].
Telomeres maintained by TE transpositions differ in the nature of telomeric repeats from telomeres maintained by telomerase, but they serve as a reminder of the retrotransposon origin of telomeres. The study of the regulation of Drosophila telomeres, which consist of retrotransposons, reveals a surprising similarity, if not identity, between the mechanisms that control telomere maintenance and TE activity. This similarity leads us to the conclusion that there is a close functional link between retrotransposons and telomeres in the genome, which, according to recent evidence, is found not only in Drosophila but also in mammals.
WHY IS TELOMERASE LOST IN MANY INSECT AND DROSOPHILA SPECIES?
Telomerase has been lost in many plants and animals in the process of evolution. Instead, telomeres are extended by other mechanisms in these cases. In Diptera the telomerase gene was lost about 270 million years ago [20]. Members of the Diptera group are one of the most numerous and prosperous species of animals, despite their lack of telomerase. The gene coding for telomerase has not been detected in the genome of Drosophila, and telomere elongation occurs due to transpositions of specialized TEs. The most thoroughly studied are the telomeric retroelements of Drosophila melanogaster. They are represented by three families of LINE retrotransposons (Long Interspersed Nuclear Elements) – HeT-A, TART, and TAHRE [21, 22]. At the same time, Bombyx mori has low-activity telomerase, with telomeric niches actively filled with specialized telomere retrotransposons SART and TRAS [23]. At present, the evolutionary pressures underlying the apparent reverse evolution and rejection of telomerase remain unknown.
Alexey M. Olovnikov was always interested in exceptions to the rules as he endeavored to explain various mysterious phenomena of nature. In this chapter, we would like to cite his interesting ideas concerning the loss of telomerase in Drosophila, which he expressed in personal correspondence: “It is known that the chromosomes of the salivary gland cells in D. melanogaster larva are able to undergo many of endoreplication rounds. In addition, there is a somatic synapse of homologous chromosomes. Therefore, the lateral conjugation of sister chromatids, tightly connected together along their entire length, should necessarily create a mechanical barrier for the formation of a telomerase telomere. Such telomere should have a three-dimensional telomere loop. In the formation of a three-dimensional telomeric complex, hundreds of conjugated chromatid ends, tightly united in a single polytenic bunch, would create compelling steric obstacles to each other in the formation of their 3D telomeric complex. Therefore, polytenization forced Drosophila to abandon telomerase. In contrast, the G-quadruplex formation, which protects the telomeric retrotransposon end on each chromatid, does not require the telomeric loop and is therefore easily compatible with lateral chromatid conjugation. Presumably, polytenization was the main reason for choosing an alternative method of protecting Drosophila telomeres. As it is well known, chromosome polytenization in salivary gland cells in Drosophila larva is necessary for production of large amounts of glue before pupation. It is likely that organisms that need increased gene copy number and have telomerase telomeres do not use dense chromatid packaging. For example, in ciliates that have a polyploid macronucleus and telomerase, chromosomes are fragmented. In theory, the following alternative is also acceptable: if telomeres, unlike the rest of the polytenized chromatids, are not side-conjugated (and, therefore, free from the previously mentioned steric hindrance), then this expands the possibilities of using the telomerase method of telomere protection. Therefore, there may be species in which polytene chromosomes and telomerase-like proteins are used at some developmental stages, but the ends of the chromosomes are not paired. Such termini have been cytogenetically observed, for example, in specialized polytene cells and at the ends of meiotic pachytene chromosomes of the legume plant Vigna unguiculata [24, 25]. A tendency to split some chromosomal ends into oligotene bundles, can be seen in the polytene chromosomes of certain species [26]” (from the letter of A. M. Olovnikov to A. I. Kalmykova, September 2017).
Indeed, recently it was reported that telomeric retrotransposons tend to form G-quadruplexes (secondary structures formed by guanine-rich DNA sequences) not only in the Drosophila species, but also in other species [27]. Such structures can protect the ends of linear chromosomes in the absence of a telomeric loop typical for the telomeres maintained by telomerase. Existence of alternative ways of telomere maintenance gives a unique opportunity to explore emergence of the functional analogues in nature. Researching Drosophila telomeres allowed us to take a fresh look at the protective mechanisms of TE control and their roles in telomere function. The most striking example is the involvement of small Piwi-interacting RNA (piRNA) and the piRNA pathway in the control of Drosophila telomere length in the germline.
piRNA PATHWAY: SOURCES AND TARGETS OF piRNAs
TEs were found in all studied species and are represented by several classes and numerous families [28]. TEs make up half of the human genome and 20% of the D. melanogaster genome [29] and are the primary source of mutations that can cause both cancer and severe developmental disorders [30, 31]. Various mechanisms are used to limit the activity of TEs in somatic cells and prevent inherited transpositions in the germline. Mechanisms which work at the level of transcription of TEs are key to prevent the initial stage of their reproduction. Transcriptional silencing is achieved by two main mechanisms leading to the formation of an inactive chromatin structure. The first and most conservative mechanism of chromatin compaction is associated with changes in histone modifications, in particular with methylation of lysine 9 of histone 3 (H3K9me), which next leads to the binding and spread of the heterochromatin protein 1 (HP1) [32]. Another powerful mechanism of repression of transcription is DNA methylation by cytosine-5′-methyltransferases [33]. In the genome, both DNA methylation and histone modifications target TEs to prevent their transcription. The main question is how are these general mechanisms recruited to the TEs? The KRAB-ZFP (KRAB-containing zinc finger proteins) family proteins play an important role in the recognition and methylation of DNA and histones at endogenous retroviruses in somatic cells of vertebrates [34, 35].
For most plants and animals, RNA interference is the most conservative and nearly universal mechanism for distinguishing between “self” and “non-self”. The pathways involving Argonaute family proteins and associated short RNAs are termed nucleic acid immunity because these mechanisms may recognize and eliminate foreign nucleic acids belonging to viruses, TEs, or transgenes based on nucleic acid sequence. siRNA (small interfering RNA) of 21 nucleotides in length and RNA interference protect somatic cells from viruses. The proteins of the Piwi subfamily of the Argonaute family and the associated piRNAs with a length of 24-30 nucleotides provide protection against TEs and viruses in the animal gonads [36, 37]. The distinctive feature of this system is that the Piwi–piRNA complex is able to induce TE chromatin modifications in a sequence-specific manner, i.e., to cause transcriptional silencing. Piwi nuclear proteins in complex with piRNAs induce the formation of heterochromatin, using the complementarity of piRNA and nascent mRNA of TEs. This is a multi-stage process that requires the interaction of several linker and accessory proteins, post-translational modifications that result in assembly, conformational changes, and eventually stabilization of the Piwi-piRNA-driven chromatin protein complex, and, finally, recruitment of universal heterochromatin factors to TEs [36, 38]. Such tight regulation is essential to turn off the TEs and prevent erroneous repression of cellular genes.
piRNA-mediated silencing is a multi-stage process. The main steps of this process are the formation of long single-strand RNA precursors in the nucleus, their processing into mature piRNAs in the cytoplasm, and piRNA-mediated silencing, which can occur both in the nucleus (transcriptional silencing) and in the cytoplasm (post-transcriptional silencing).
To perform their functions, mature piRNAs should complementarily interact with the transcripts of TE, i.e., they should be antisense to the mRNA of TEs. Indeed, a significant portion of piRNAs in the gonads corresponds to TEs, but are not generated from the mRNA of TEs. The origin of such antisense TE RNAs in the genome is not quite clear but may involve the following mechanisms. piRNAs are thought to be formed from long single-strand endogenous RNAs known as piRNAs precursors (pre-piRNAs) [39, 40]. Our knowledge on this issue is mainly obtained from the D. melanogaster oogenesis model. Studies of the genomic origin of piRNAs in Drosophila have led to the conclusion that the sources of piRNAs and their targets are located at different genomic loci (Fig. 1a). This assumption was based on the small RNA sequencing data that revealed the regions in pericentromeric heterochromatin with high density of the single-mapped (in other words, unique) piRNAs. Such loci were termed piRNA clusters [39]. These extended regions of the genome, up to 200 kb in size, enriched in damaged TE copies, encode unusually long read-through transcripts that are processed into mature piRNAs that target euchromatic active TEs. This is a remarkable illustration of how the genome regions that were previously thought of as “junk” DNA are really being used functionally. It should be mentioned that this scenario has not been proved to be universal.
Where do piRNA precursors come from? a) Heterochromatic pericentromeric piRNA clusters are the sources of anti-transposon piRNAs that target the TE active copies. The mapping of small RNAs to the chromosome is schematically shown with the activity of uni- and dual-strand piRNA clusters depicted below. The Piwi–piRNA complex recognizes and silences TE active copies (black triangle). b) TE active copies form de novo piRNA clusters. piRNA signature of the TE-associated piRNA clusters is schematically shown. sRNAseq – small RNA sequencing. c) Transgenic model demonstrates how active TEs in euchromatin could become piRNA clusters. Piwi protein in complex with endogenous piRNA to I-element recognizes RNA in the complementary transgenic locus, promoting generation of transcripts from both genomic strands, which are then processed into mature piRNAs.
In Drosophila, there are two types of piRNA clusters, uni-strand and dual-strand, which are transcribed in one or two directions, respectively. Both types of piRNA clusters produce predominantly antisense piRNA relative to TEs. The reason why they are antisense is easier to understand considering the uni-strand piRNA clusters, in which all TE remnants are in an inverted position relative to the pre-piRNA transcription direction. The best-known uni-strand piRNA cluster is the Drosophila flamenco locus, which operates in ovarian follicular cells. It contains many inactive TE copies on the minus genomic strand relative to the direction of transcription, therefore the piRNAs processed from long precursor transcripts are complementary to the transposon mRNAs and guide their transcriptional silencing [39, 41]. flamenco-like loci generate piRNAs against endogenous retroviruses related to the gypsy family. This intricate scenario nevertheless is recurring in evolution. flamenco-like uni-strand piRNA clusters have been found in mosquitoes and other Drosophila species [42-44]. Moreover, during mouse spermatogenesis, two flamenco-like uni-strand piRNA clusters produce anti-transposon piRNAs [45]. The second type of heterochromatic piRNA clusters – dual-strand clusters – are considered as principal players in the anti-transposon defense in the Drosophila germline. These loci contain chaotically oriented TE copies disrupted by other TE insertions. Dual-strand piRNA clusters are bidirectionally transcribed resulting in the generation of long non-coding piRNA precursors from both genomic strands [46]. Long piRNA precursors are exported to the cytoplasm where they are processed into mature piRNAs. piRNA processing is a conserved well-established mechanism in different species. The outer mitochondrial membrane and perinuclear compartment serve as platform for compartmentalization of piRNA generation and maturation. piRNAs are cleaved as a result of activity of various specialized ribonucleases. Cytoplasmic Piwi subfamily proteins perform piRNA amplification using sense and antisense TE transcripts and piRNA precursors from clusters, which leads to the formation of both sense and antisense piRNAs. This mechanism, known as “ping-pong,” is found in the gonads of all the species studied, and leads not only to cleavage of TE mRNA (post-transcriptional silencing), but also to production of new piRNAs. Recent comprehensive reviews describe in details the processes of piRNA formation and amplification, as well as the piRNA processing machinery [36, 37, 47].
Dual-strand piRNA clusters are thought to represent a repository of information in the genome about prior TE invasions, as well as a kind of trap for TEs, because their insertions in the piRNA cluster would lead to the production of piRNAs and the suppression of the activity of cognate copies in the genome [48]. While dual-strand piRNA clusters were shown to be species-specific for Drosophila and a few arthropod species, these types of transposon-specific piRNA sources have not been identified in other investigated species, including mammals. The existence of a conservative mechanism for the synthesis of piRNA precursors that are antisense to TEs is still a matter of debate. For example, sense and antisense piRNAs are produced from individual copies of evolutionarily young active retrotransposons in mammals, despite the presence of a significant number of damaged TE copies in the genome [45].
In the piRNA pathway mechanism for transposon silencing, active transposons are expected to be the primary targets of piRNAs. A more thorough examination of small RNA libraries derived from D. melanogaster ovaries revealed that not only heterochromatic piRNA clusters, but also full-length euchromatic TEs generate short RNAs, both piRNAs and siRNAs. This shows that at the sites of recent TE insertions, local dual-strand piRNA clusters occur [49], which is similar to the scenario of piRNA generation from the active copies of LINE1 in mammals [45]. A distinctive feature of the TE-associated piRNA clusters is a “piRNA signature”. It is an asymmetric profile of small RNA distribution upstream and downstream of TEs resulting from the read-through transcription of piRNA precursors into the TE flanking genomic regions (Fig. 1b). This signature was successfully used for the prediction of non-annotated TE insertions. Moreover, the expansion of small RNA production outside TEs can suppress the expression of the neighboring genes [49]. Thus, active TEs serve not only as targets for the piRNA system, which promotes chromatin compaction, but also as a source of small RNAs, as they can produce de novo si- and piRNAs. The Drosophila telomeric retrotransposons, which are arrayed as tandem repeats in the telomere, are also the target and source of piRNAs, which allow feedback regulation of the telomeric retrotransposons expression, as addressed in more depth in the following section.
The mechanism underlying the formation of new TE-associated piRNA clusters, particularly the mechanism of activation of bidirectional transcription, is not totally understood. Antisense promoters are known only for a few TEs and are rather an exception. For example, the LINE1 human retrotransposon antisense promoter and the transcripts derived from it are involved in siRNA-mediated LINE1 transposition suppression [50, 51]. When the Piwi–piRNA complex recognizes complementary transcripts, an uncommon type of transcription, convergent transcription, is thought to be triggered at genomic loci harboring active copies of TEs. This process depends on the genomic environment of the TE insertion: endogenous convergent transcription at the TE insertion locus promotes the formation of a new piRNA cluster [52]. Transcripts generated from both genomic strands are then processed into mature pi/siRNAs. Such a scenario has an obvious biological significance, as it would result in amplification of protective small RNAs against most dangerous, transcriptionally active TEs. A transgenic model containing a DNA fragment targeted by endogenous piRNAs elucidated some details of the de novo piRNA cluster formation [53-55]. It was shown that transgenic constructs containing a fragment of I-element can become new piRNA clusters. The formation of piRNA clusters de novo is accompanied by the appearance of weak transcription from the minus strand and the generation of sense and antisense pi/siRNAs from the entire transgene and from genomic sequences at a distance of 1 to 10 kb from the transgene [56, 57] (Fig. 1c).
A scenario in which an active TE is a source of piRNAs should be evolutionarily beneficial and have selective advantages. It is unclear why extended heterochromatic piRNA clusters, which are maintained by a unique transcription mechanism, have survived throughout evolution in Drosophila and other arthropods. The removal of the most extended pericentromeric piRNA clusters in D. melanogaster were found to not lead to TE derepression [58]. This suggests that TE silencing in the stable laboratory line is not primarily mediated by heterochromatic piRNA clusters. This function is most likely carried out by TE-associated piRNA clusters, which are generated via the assistance of piRNAs acquired from the mother with the oocyte cytoplasm [59].
Differences in TE silencing strategies between species may be related to different habitats. Horizontal TE transfer has an impact on natural arthropod populations, including the re-invasion of previously lost TEs in the genome [60, 61]. The memory of earlier TE invasions, which is kept in the form of TE fragments in piRNA clusters, can protect the population from TE re-infection due to the existence of complementary piRNAs and the activation of piRNA-silencing [39, 52, 57, 59, 62].
The idea that active TE copies serve as the primary targets of the piRNA pathway is emphasized by the recently discovered mechanism of a co-transcriptional degradation of TE transcripts [63]. Despite the establishment of heterochromatin in loci with active copies of TEs, they can still be transcribed in the germline. The excess of the TE transcripts in the Drosophila germline is removed through the activity of the nuclear Ccr4–Not complex, which has deadenylase activity [64]. Ccr4–Not is recruited to the transposon transcripts co-transcriptionally by the Piwi-piRNA nuclear complex. Most likely, the polyA tail at the 3′-end of the TE mRNA is eliminated due to deadenylase activity of the Ccr4–Not complex. The nuclear RNA quality control system can identify these transcripts as aberrant and subject them to exonuclease cleavage. Noteworthy, the targets of nuclear Ccr4–Not are mainly active full-length TEs and telomeric retrotransposons [63].
To summarize, piRNAs can trigger a number of important processes aimed at suppressing TE activity (Fig. 2). In the nucleus, piRNAs induce transcriptional silencing leading to heterochromatin assembly at the TE loci [65-67]. The piRNA system also functions at the co-transcriptional level, causing nucleases to degrade TE transcripts at transcription sites [63]. In the cytoplasm, the piRNA system cleaves TE transcripts (post-transcriptional silencing), resulting in piRNA amplification and their subsequent inheritance through the oocyte cytoplasm [39, 59, 68-71]. Importantly, piRNAs can initiate de novo piRNA cluster establishment at full-length euchromatic TE insertions leading to the expression of antisense TE transcripts and a burst of piRNA production against most active TEs [49]. Furthermore, the fast evolution of proteins participating in the piRNA pathway allows this system to be highly adaptable to new targets [72-74]. Despite the high capacity of piRNAs and other anti-transposon defense systems, selfish TEs successfully bypass them, continuing their propagation, resulting in deleterious mutations, diseases, and developmental disorders. A paradoxical situation has emerged in which the suppression of TE activity requires its activity, and as a result, the piRNA system operates. Apparently, the far from perfect efficacy of these and other defense systems allows TEs to multiply within the tolerable limits, producing material for genome evolution and positive selection, while also allowing some critical systems, such as telomeres, to function. The latter phenomenon will be discussed below.
The piRNA silencing system is a multi-level mechanism. In Drosophila, piRNAs can induce the following processes: a) transcriptional silencing and chromatin compaction; b) co-transcriptional degradation of nascent RNA in the nucleus via assistance of the deadenylase nuclear complex Ccr4–Not; c) post-transcriptional RNA degradation and amplification of piRNAs in the cytoplasm; d) de novo piRNA production at active TEs due to activation of antisense transcription; e) epigenetic transgenerational memory (transfer of maternal piRNAs in complex with Piwi proteins to offspring through the germ plasm of the oocyte).
ROLE OF piRNAs IN REGULATION OF Drosophila TELOMERES
In Drosophila, disruption of the piRNA system causes not only TE activation but also excessive telomere extension due to the increased frequency of retrotransposon attachments to chromosome ends [75]. This occurs because the germline pool of piRNAs complementary to telomeric retrotransposons is reduced. The piRNA pathway processes all TE transcripts in the germline, producing piRNA without differentiating between telomeric and parasitic retrotransposons. Disruption of the piRNA pathway leads to a decrease in piRNA abundance, which is accompanied by the accumulation of telomeric TE transcripts. These transcripts serve as intermediates for the extension of telomeres. Drosophila telomeres are maintained by the retrotranspositions of specialized telomeric TEs that are not found in other genomic loci. Telomeric retroelements are both the sources and the targets of piRNAs, making them a unique example of a self-regulated TE-associated piRNA cluster. Bidirectional promoters of Drosophila telomeric retrotransposons organized in head-to-tail arrays ensure the generation of both sense and antisense transcripts, which are then used to produce telomeric piRNAs [76-79]. Drosophila is not the only example of such regulation. Piwi proteins are also involved in the regulation of SART and TRAS telomeric retrotransposon transpositions in the silkworms [80].
The heterochromatin state is a characteristic of telomeres. Heterochromatin factors are recruited to telomeric retrotransposons via the assistance of the piRNA system in the Drosophila germline [81]. When the piRNA system is disrupted, the levels of the key heterochromatin protein HP1 and the histone modification H3K9me3 decrease at telomeres. The shift of telomeric clusters from the nuclear periphery, along with a global change in telomeric chromatin structure, is found in the germline of Drosophila piRNA mutants [81]. Furthermore, the most abundant D. melanogaster telomeric repeat, HeT-A, is co-transcriptionally regulated in the nucleus by recruiting the Ccr4–Not deadenylase complex [63]. Confocal microscopy revealed the presence of structures nearby telomeres that contained the Ccr4–Not protein complex, the Piwi protein, and RNA nuclear export factors [63]. Interestingly, non-canonical deadenylases Ccr4 and Caf1, which are localized in Cajal nuclear bodies, are involved in the biogenesis of human telomerase RNA component [82]. Thus, the piRNA system and Ccr4–Not deadenylase are implicated in both the reduction of activity of selfish TEs and in the Drosophila telomere regulation.
Telomere length in Drosophila is negatively regulated by the piRNA system; the more actively it functions, the less frequently telomere lengthening takes place. In order to maintain the frequency of attachment of telomeric retroelements to the ends of chromosomes required for normal development, a complicated feedback mechanism controls the level of telomeric transcripts, telomeric piRNAs, and the state of telomeric chromatin. The fact that the piRNA-mediated mechanism should effectively suppress TE activity complicates telomere length control even more. Indeed, piRNA system disruption causes not only an increase in the frequency of telomeric attachments, but also TE activation and sterility [75]. The opposite situation, i.e., activation of the piRNA system and excessive telomere shortening, was observed in one of the natural Drosophila strains. Differences in the extent of anti-transposon protection among natural Drosophila populations can be explained by the high variability of proteins involved in the piRNA pathway and variable efficacy of primary piRNA processing [83]. Flies, which had the most efficient piRNA processing, lacked full-length copies of the HeT-A retrotransposon, the main structural element of telomeres. This strain had reduced fertility and viability. Therefore, enhancement of piRNA-mediated protection leading to strong repression of transposons, can also cause telomere shortening. This result implies that limits exist for improvements in the efficiency of the piRNA defense system because it needs to achieve a balance between protecting the genome against transposon propagation and allowing adequate telomere maintenance (Fig. 3).
WHEN AND WHY ARE TRANSPOSABLE ELEMENTS ACTIVATED DURING DEVELOPMENT?
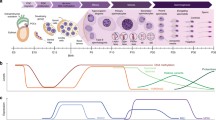
The stages of development during which TEs are activated could point to the mechanisms of TE control (Fig. 4, a-c). Surprisingly, short-term transposon activation is observed during early stages of gametogenesis in a variety of species [84]. There is a brief developmental window at the early stages of D. melanogaster oogenesis when Piwi levels decrease and TEs mobilize [85]. The TE transcripts produced at this stage are processed into piRNAs by the cytoplasmic piRNA amplification machinery. When Piwi expression is restored these piRNAs mediate transcriptional silencing of TEs at later stages of oogenesis [86]. When the Piwi protein is absent during the early stages of oogenesis, transcriptional activation of telomeric retroelements (HeT-A and TART) is detected, which leads to the formation of telomere elongation intermediates [66, 87, 88]. In D. melanogaster, such intermediates are spherical ribonucleoprotein (RNP) particles composed of the retrotransposon HeT-A-encoded Gag protein, packed with HeT-A RNA, and capable of being targeted to telomeres [89, 90]. For the first time, such HeT-A spheres were found in actively proliferating larval brain cells, and their appearance on telomeres coincided with telomere replication [89]. The main structural component of Drosophila telomeres is the non-autonomous retroelement HeT-A, lacking reverse transcriptase. Reverse transcriptase is provided by other telomeric retroelements – TART and/or TAHRE. TART reverse transcriptase was also discovered in the HeT-A spheres in neuroblasts [91]. Most likely, HeT-A spheres are necessary for the retrotransposition of telomeric elements to the chromosome ends to elongate Drosophila telomeres. Transcription of Drosophila telomeric retrotransposons HeT-A and TART is normally detected in the ovarian germ cysts, and when the piRNA system is disrupted, spherical particles containing RNA and the Gag protein encoded by HeT-A arise [87]. Telomere elongation occurs at the same stages at which TEs are activated. This is most likely due to a decrease in piRNA protection and the accumulation of telomeric element transcripts, as well as global heterochromatin decompaction and greater accessibility of chromosomal ends in mitotically active gamete progenitor cells. Indeed, disruption of various heterochromatin factors such as HP1, histone demethylase Lsd1 and its cofactor Ova, methyltransferases dSetdB1 and Su(var)3-9 lead to both TE activation and telomeric transcript accumulation [92-98]. Mutation of Su(var)2-5 gene encoding HP1, causes telomere elongation [98]. A similar scenario has been observed in other animals. In mammals, a wave of de novo DNA methylation is typical for prenatal male germline development. As a result, expression of young active subfamilies of retroelement LINE1 occurs in primordial germ cells [99]. At later stages of spermatogenesis, emergence of Piwi subfamily protein, MILI2 in mouse, triggers piRNA production, recruitment of DNA methylase and establishment of piRNA-mediated de novo DNA methylation at LINE1 sequences [100].
Developmental stages of transposon activation. a) When Piwi protein levels fall during Drosophila oogenesis, TEs and telomeric retrotransposons are activated in the germ cysts. At this step, piRNAs are produced, which cause transcriptional silencing at the later stages of oogenesis. Telomere elongation is anticipated to occur at the same stage due to retrotranspositions of telomeric retroelements to the chromosome ends. b) TE activation is associated with global demethylation in mouse primordial germ cells during spermatogenesis. Then, piRNA-mediated de novo DNA methylation of LINE1 active copies is established. c) Transposon mobilization and telomere elongation are observed during first zygotic divisions in mammalian embryogenesis. d) The inhibition of LINE1 by piRNAs may have an indirect effect on LINE1 telomeric functions in mammalian germ cells (hypothesis).
In the mammalian development, two waves of demethylation and remethylation of DNA occur – in primordial germ cells and in the early embryogenesis [101]. Global demethylation of the genome in the primordial germ cells during prenatal development is believed to be required for epigenome reprogramming and the establishment de novo DNA methylation patterns [102, 103]. DNA demethylation is also required for telomere extension in embryonic stem cells which is associated with increased levels of transcription of many retrotransposons [104-106]. The transcription factor Zscan4 (Zinc finger and SCAN domain containing 4) plays a critical role in maintaining the DNA demethylation state and heterochromatin derepression in embryonic stem cells and 2-cell embryos [107]. Zscan4 increases the expression of genes involved in homologous recombination, which stimulates the recombination mechanism of telomere lengthening in embryonic stem cells, 2-cell embryos, and ALT (alternative lengthening of telomeres) tumor cells that use recombination for telomere lengthening [105, 106, 108]. In preimplantation embryos, the activation of LINE1 retrotransposons and endogenous retroviruses is also observed [109-112]. Interestingly, LINE1 inhibition leads to impaired expression of pluripotency factors, including Zscan4, and blocks telomere elongation in embryonic stem cells [113]. In turn, LINE1 RNA acts as a transcriptional repressor of the Dux gene, which is required for embryonic cells to exit the 2C state and continue development [114]. It was also shown that early differentiation gene promoters contain regulatory regions of endogenous retroviruses and are regulated by retroviral proteins [115]. Thus, transposon activation occurs during brief periods of development associated with genome reprogramming and telomere elongation. Furthermore, these processes are linked by a sophisticated regulatory network, the proper balance of which is required for normal development.
Natural aging and premature aging syndromes both exhibit chromatin decompaction and activation of expression of TEs and telomeric repeats, resulting in DNA damage and cell death [116, 117], which emphasizes the commonality of epigenetic regulation of telomeres and TEs at different developmental stages.
INVOLVEMENT OF LINE1 IN THE MAINTENANCE OF MAMMALIAN TELOMERES
Similarities between TE control and telomere regulation mechanisms are evident in Drosophila where telomeres are elongated by the retrotranspositions of retroelements. However, in the vast majority of species, telomeres are maintained by the activity of a specialized reverse transcriptase – telomerase – the components of which are encoded by cellular genes. New evidence demonstrates that the retrotransposons are involved in the functioning of telomeres in mammalian cells. In the absence of the components of the protective telomere complex shelterin, the LINE1 retrotransposon can attach to the telomere in human cells via reverse transcription and become a structural part of telomeric DNA [17]. In addition, LINE1 is also able to play a role in telomere function. LINE1 knockdown in cancer cells resulted in a reduced expression of shelterin proteins, decreased telomerase activity and telomere shortening [118]. In line with this observation, inhibition of LINE1 activity in the mouse 2-cell embryos blocked telomere elongation and genome reprogramming [113]. Furthermore, LINE1 RNPs were found directly at telomeric ends in human cancer cells and mouse 2-cell embryos, where they interacted with telomeric repeat-containing RNA (TERRA) [113, 119]. LINE1 appears to be involved in telomere biogenesis, however it is unknown how LINE1 is targeted there or which telomere components it recognizes. Given the role of LINE1 in telomere function, it is tempting to speculate about a potential relationship between the piRNA system that regulates LINE1 expression and telomeres in telomerase-using mammals (Fig. 4d).
All together, these findings indicate that the systems regulating telomere maintenance and transposon control in the mammalian genome are connected with respect to temporal regulation and function. Research on this relationship will contribute to a better understanding of the mechanisms of telomere regulation in development, aging, and cancer cells.
CONCLUSION
The interaction between the host genome and TEs is commonly referred to as a genomic conflict, and the processes behind this conflict are complicated and debatable [120]. When the molecular mechanisms of TE control were not yet known, analysis of TE population dynamics revealed a balance between TE spread rate and the mechanisms that limit this process. As a result, the genome has a rather stable amount of TE copies [121]. Excessive reduction of TE activity and their copy number does not appear to provide a selective benefit to the host, as recent methods of genomic data analysis of natural Drosophila populations indicate. For example, the amount of piRNA does not correlate with TE transposition activity, therefore, the piRNA system is not optimally adapted to protect the genome from TEs [122]. The continuous improvement of the adaptable piRNA system is probably limited by its participation in key regulatory functions, which creates a conflict between TEs and the genome. Incomplete suppression of TEs allows important cellular functions to be performed by the domesticated TEs and consequently ensures the survival of selfish TEs in the genome. What is the mechanism of this global genomic compromise? Part of the solution, we believe, can be found in the retrotransposon origin of telomeres and telomerase. Transposon activity is associated with genome reprogramming, erasing of epigenetic marks and telomere elongation. A dual role of the piRNA system in telomere protection and transposon silencing in Drosophila resulted from the nature of Drosophila telomeres which are made up of retrotransposons per se. However, telomerase can also be considered as a specialized retroelement that retained its functional relationship with genomic retrotransposons. Thus, we conclude that protective systems can only partially suppress the activity of selfish TEs since they must be balanced to carry out essential tasks, like those related to telomere maintenance. However, this is a blessing in disguise: an increasing body of evidence indicates that domesticated TEs make a significant contribution to the diversity of regulatory mechanisms that ultimately confer evolutionary benefits to the host genome.
References
Fueyo, R., Judd, J., Feschotte, C., and Wysocka, J. (2022) Roles of transposable elements in the regulation of mammalian transcription, Nat. Rev. Mol. Cell Biol., 23, 481-497, https://doi.org/10.1038/s41580-022-00457-y.
Modzelewski, A. J., Gan Chong, J., Wang, T., and He, L. (2022) Mammalian genome innovation through transposon domestication, Nat. Cell Biol., 24, 1332-1340, https://doi.org/10.1038/s41556-022-00970-4.
Almojil, D., Bourgeois, Y., Falis, M., Hariyani, I., Wilcox, J., and Boissinot, S. (2021) The structural, functional and evolutionary impact of transposable elements in eukaryotes, Genes (Basel), 12, 918, https://doi.org/10.3390/genes12060918.
Nishihara, H. (2020) Transposable elements as genetic accelerators of evolution: contribution to genome size, gene regulatory network rewiring and morphological innovation, Genes Genet. Syst., 94, 269-281, https://doi.org/10.1266/ggs.19-00029.
Hartley, G., and O’Neill, R. J. (2019) Centromere repeats: hidden gems of the genome, Genes (Basel), 10, 223, https://doi.org/10.3390/genes10030223.
Chang, C. H., Chavan, A., Palladino, J., Wei, X., Martins, N. M. C., Santinello, B., Chen, C. C., Erceg, J., Beliveau, B. J., Wu, C. T., Larracuente, A. M., and Mellone, B. G. (2019) Islands of retroelements are major components of Drosophila centromeres, PLoS Biol., 17, e3000241, https://doi.org/10.1371/journal.pbio.3000241.
Chueh, A. C., Northrop, E. L., Brettingham-Moore, K. H., Choo, K. H., and Wong, L. H. (2009) LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin, PLoS Genet., 5, e1000354, https://doi.org/10.1371/journal.pgen.1000354.
Nelson, J. O., Slicko, A., and Yamashita, Y. M. (2023) The retrotransposon R2 maintains Drosophila ribosomal DNA repeats, Proc. Natl. Acad. Sci. USA, 120, e2221613120, https://doi.org/10.1073/pnas.2221613120.
Olovnikov, A. M. (1971) Principle of marginotomy in template synthesis of polynucleotides [in Russian], Dokl. Akad. Nauk SSSR, 201, 1496-1499.
Olovnikov, A. M. (1973) A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon, J. Theor. Biol., 41, 181-190, https://doi.org/10.1016/0022-5193(73)90198-7.
Blackburn, E. H. (1992) Telomerases, Annu. Rev. Biochem., 61, 113-129, https://doi.org/10.1146/annurev.bi.61.070192.000553.
Garavis, M., Gonzalez, C., and Villasante, A. (2013) On the origin of the eukaryotic chromosome: the role of noncanonical DNA structures in telomere evolution, Genome Biol. Evol., 5, 1142-1150, https://doi.org/10.1093/gbe/evt079.
Gladyshev, E. A., and Arkhipova, I. R. (2007) Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes, Proc. Natl. Acad. Sci. USA, 104, 9352-9357, https://doi.org/10.1073/pnas.0702741104.
Nakamura, T. M., and Cech, T. R. (1998) Reversing time: origin of telomerase, Cell, 92, 587-590, https://doi.org/10.1016/s0092-8674(00)81123-x.
Eickbush, T. H. (1997) Telomerase and retrotransposons: which came first? Science, 277, 911-912, https://doi.org/10.1126/science.277.5328.911.
Kordyukova, M., Olovnikov, I., and Kalmykova, A. (2018) Transposon control mechanisms in telomere biology, Curr. Opin. Genet. Dev., 49, 56-62, https://doi.org/10.1016/j.gde.2018.03.002.
Morrish, T. A., Garcia-Perez, J. L., Stamato, T. D., Taccioli, G. E., Sekiguchi, J., and Moran, J. V. (2007) Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres, Nature, 446, 208-212, https://doi.org/10.1038/nature05560.
Roth, C. W., Kobeski, F., Walter, M. F., and Biessmann, H. (1997) Chromosome end elongation by recombination in the mosquito Anopheles gambiae, Mol. Cell. Biol., 17, 5176-5183, https://doi.org/10.1128/MCB.17.9.5176.
Compton, A., Liang, J., Chen, C., Lukyanchikova, V., Qi, Y., Potters, M., Settlage, R., Miller, D., Deschamps, S., Mao, C., Llaca, V., Sharakhov, I. V., and Tu, Z. (2020) The beginning of the end: a chromosomal assembly of the new world malaria mosquito ends with a novel telomere, G3 (Bethesda), 10, 3811-3819, https://doi.org/10.1534/g3.120.401654.
Mason, J. M., Randall, T. A., and Capkova Frydrychova, R. (2016) Telomerase lost? Chromosoma, 125, 65-73, https://doi.org/10.1007/s00412-015-0528-7.
Pardue, M. L., and DeBaryshe, P. G. (2008) Drosophila telomeres: a variation on the telomerase theme, Fly, 2, 101-110, https://doi.org/10.4161/fly.6393.
Casacuberta, E. (2017) Drosophila: retrotransposons making up telomeres, Viruses, 9, 192, https://doi.org/10.3390/v9070192.
Fujiwara, H., Osanai, M., Matsumoto, T., and Kojima, K. K. (2005) Telomere-specific non-LTR retrotransposons and telomere maintenance in the silkworm, Bombyx mori, Chromosome Res., 13, 455-467, https://doi.org/10.1007/s10577-005-0990-9.
Guerra, M., Kenton, A., and Bennett, M. D. (1996) rDNA sites in mitotic and polytene chromosomes of Vigna unguiculata (L.) Walp. and Phaseolus coccineus L. revealed by in situ hybridization, Ann. Botany, 78, 157-161, https://doi.org/10.1006/anbo.1996.0108.
Iwata-Otsubo, A., Lin, J. Y., Gill, N., and Jackson, S. A. (2016) Highly distinct chromosomal structures in cowpea (Vigna unguiculata), as revealed by molecular cytogenetic analysis, Chromosome Res., 24, 197-216, https://doi.org/10.1007/s10577-015-9515-3.
Zhimulev, I. F. (1996) Morphology and structure of polytene chromosomes, Adv. Genet., 34, 1-497, https://doi.org/10.1016/s0065-2660(08)60533-7.
Jedlicka, P., Tokan, V., Kejnovska, I., Hobza, R., and Kejnovsky, E. (2023) Telomeric retrotransposons show propensity to form G-quadruplexes in various eukaryotic species, Mob. DNA, 14, 3, https://doi.org/10.1186/s13100-023-00291-9.
Wells, J. N., and Feschotte, C. (2020) A field guide to eukaryotic transposable elements, Annu. Rev. Genet., 54, 539-561, https://doi.org/10.1146/annurev-genet-040620-022145.
Merel, V., Boulesteix, M., Fablet, M., and Vieira, C. (2020) Transposable elements in Drosophila, Mob. DNA, 11, 23, https://doi.org/10.1186/s13100-020-00213-z.
Anwar, S. L., Wulaningsih, W., and Lehmann, U. (2017) Transposable elements in human cancer: causes and consequences of deregulation, Int. J. Mol. Sci., 18, 974, https://doi.org/10.3390/ijms18050974.
Huang, C. R., Burns, K. H., and Boeke, J. D. (2012) Active transposition in genomes, Annu. Rev. Genet., 46, 651-675, https://doi.org/10.1146/annurev-genet-110711-155616.
Lomberk, G., Wallrath, L., and Urrutia, R. (2006) The heterochromatin protein 1 family, Genome Biol., 7, 228, https://doi.org/10.1186/gb-2006-7-7-228.
Lyko, F. (2018) The DNA methyltransferase family: a versatile toolkit for epigenetic regulation, Nat. Rev. Genet., 19, 81-92, https://doi.org/10.1038/nrg.2017.80.
Ecco, G., Cassano, M., Kauzlaric, A., Duc, J., Coluccio, A., Offner, S., Imbeault, M., Rowe, H. M., Turelli, P., and Trono, D. (2016) Transposable elements and their KRAB-ZFP controllers regulate gene expression in adult tissues, Dev. Cell, 36, 611-623, https://doi.org/10.1016/j.devcel.2016.02.024.
Yang, P., Wang, Y., and Macfarlan, T. S. (2017) The role of KRAB-ZFPs in transposable element repression and mammalian evolution, Trends Genet., 33, 871-881, https://doi.org/10.1016/j.tig.2017.08.006.
Czech, B., Munafo, M., Ciabrelli, F., Eastwood, E. L., Fabry, M. H., Kneuss, E., and Hannon, G. J. (2018) piRNA-guided genome defense: from biogenesis to silencing, Annu. Rev. Genet., 52, 131-157, https://doi.org/10.1146/annurev-genet-120417-031441.
Ozata, D. M., Gainetdinov, I., Zoch, A., O’Carroll, D., and Zamore, P. D. (2019) PIWI-interacting RNAs: small RNAs with big functions, Nat. Rev. Genet., 20, 89-108, https://doi.org/10.1038/s41576-018-0073-3.
Andreev, V. I., Yu, C., Wang, J., Schnabl, J., Tirian, L., Gehre, M., Handler, D., Duchek, P., Novatchkova, M., Baumgartner, L., Meixner, K., Sienski, G., Patel, D. J., and Brennecke, J. (2022) Panoramix SUMOylation on chromatin connects the piRNA pathway to the cellular heterochromatin machinery, Nat. Struct. Mol. Biol., 29, 130-142, https://doi.org/10.1038/s41594-022-00721-x.
Brennecke, J., Aravin, A. A., Stark, A., Dus, M., Kellis, M., Sachidanandam, R., and Hannon, G. J. (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila, Cell, 128, 1089-1103, https://doi.org/10.1016/j.cell.2007.01.043.
Aravin, A., Gaidatzis, D., Pfeffer, S., Lagos-Quintana, M., Landgraf, P., Iovino, N., Morris, P., Brownstein, M. J., Kuramochi-Miyagawa, S., Nakano, T., Chien, M., Russo, J. J., Ju, J., Sheridan, R., Sander, C., Zavolan, M., and Tuschl, T. (2006) A novel class of small RNAs bind to MILI protein in mouse testes, Nature, 442, 203-207, https://doi.org/10.1038/nature04916.
Sarot, E., Payen-Groschene, G., Bucheton, A., and Pelisson, A. (2004) Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene, Genetics, 166, 1313-1321, https://doi.org/10.1534/genetics.166.3.1313.
Aguiar, E., de Almeida, J. P. P., Queiroz, L. R., Oliveira, L. S., Olmo, R. P., de Faria, I., Imler, J. L., Gruber, A., Matthews, B. J., and Marques, J. T. (2020) A single unidirectional piRNA cluster similar to the flamenco locus is the major source of EVE-derived transcription and small RNAs in Aedes aegypti mosquitoes, RNA, 26, 581-594, https://doi.org/10.1261/rna.073965.119.
Rozhkov, N. V., Zelentsova, E. S., Shostak, N. G., and Evgen’ev, M. B. (2011) Expression of Drosophila virilis retroelements and role of small RNAs in their intrastrain transposition, PLoS One, 6, e21883, https://doi.org/10.1371/journal.pone.0021883.
Van Lopik, J., Alizada, A., Trapotsi, M. A., Hannon, G. J., Bornelöv, S., and Czech Nicholson, B. (2023) Unistrand piRNA clusters are an evolutionarily conserved mechanism to suppress endogenous retroviruses across the Drosophila genus, Nat. Commun., 14, 7337, https://doi.org/10.1038/s41467-023-42787-1.
Aravin, A. A., Sachidanandam, R., Bourc’his, D., Schaefer, C., Pezic, D., Toth, K. F., Bestor, T., and Hannon, G. J. (2008) A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice, Mol. Cell, 31, 785-799, https://doi.org/10.1016/j.molcel.2008.09.003.
Andersen, P. R., Tirian, L., Vunjak, M., and Brennecke, J. (2017) A heterochromatin-dependent transcription machinery drives piRNA expression, Nature, 549, 54-59, https://doi.org/10.1038/nature23482.
Sato, K., and Siomi, M. C. (2020) The piRNA pathway in Drosophila ovarian germ and somatic cells, Proc. Jpn. Acad. Ser. B Phys. Biol. Sci., 96, 32-42, https://doi.org/10.2183/pjab.96.003.
Khurana, J. S., Wang, J., Xu, J., Koppetsch, B. S., Thomson, T. C., Nowosielska, A., Li, C., Zamore, P. D., Weng, Z., and Theurkauf, W. E. (2011) Adaptation to P element transposon invasion in Drosophila melanogaster, Cell, 147, 1551-1563, https://doi.org/10.1016/j.cell.2011.11.042.
Shpiz, S., Ryazansky, S., Olovnikov, I., Abramov, Y., and Kalmykova, A. (2014) Euchromatic transposon insertions trigger production of novel Pi- and endo-siRNAs at the target sites in the drosophila germline, PLoS Genet., 10, e1004138, https://doi.org/10.1371/journal.pgen.1004138.
Speek, M. (2001) Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes, Mol. Cell. Biol., 21, 1973-1985, https://doi.org/10.1128/MCB.21.6.1973-1985.2001.
Yang, N., and Kazazian, H. H., Jr. (2006) L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells, Nat. Struct. Mol. Biol., 13, 763-771, https://doi.org/10.1038/nsmb1141.
Komarov, P. A., Sokolova, O., Akulenko, N., Brasset, E., Jensen, S., and Kalmykova, A. (2020) Epigenetic requirements for triggering heterochromatinization and Piwi-interacting RNA production from transgenes in the Drosophila germline, Cells, 9, 922, https://doi.org/10.3390/cells9040922.
De Vanssay, A., Bouge, A. L., Boivin, A., Hermant, C., Teysset, L., Delmarre, V., Antoniewski, C., and Ronsseray, S. (2012) Paramutation in Drosophila linked to emergence of a piRNA-producing locus, Nature, 490, 112-115, https://doi.org/10.1038/nature11416.
Josse, T., Teysset, L., Todeschini, A. L., Sidor, C. M., Anxolabehere, D., and Ronsseray, S. (2007) Telomeric trans-silencing: an epigenetic repression combining RNA silencing and heterochromatin formation, PLoS Genet., 3, 1633-1643, https://doi.org/10.1371/journal.pgen.0030158.
Muerdter, F., Olovnikov, I., Molaro, A., Rozhkov, N. V., Czech, B., Gordon, A., Hannon, G. J., and Aravin, A. A. (2012) Production of artificial piRNAs in flies and mice, RNA, 18, 42-52, https://doi.org/10.1261/rna.029769.111.
Akulenko, N., Ryazansky, S., Morgunova, V., Komarov, P. A., Olovnikov, I., Vaury, C., Jensen, S., and Kalmykova, A. (2018) Transcriptional and chromatin changes accompanying de novo formation of transgenic piRNA clusters, RNA, 24, 574-584, https://doi.org/10.1261/rna.062851.117.
Olovnikov, I., Ryazansky, S., Shpiz, S., Lavrov, S., Abramov, Y., Vaury, C., Jensen, S., and Kalmykova, A. (2013) De novo piRNA cluster formation in the Drosophila germ line triggered by transgenes containing a transcribed transposon fragment, Nucleic Acids Res., 41, 5757-5768, https://doi.org/10.1093/nar/gkt310.
Gebert, D., Neubert, L. K., Lloyd, C., Gui, J., Lehmann, R., and Teixeira, F. K. (2021) Large Drosophila germline piRNA clusters are evolutionarily labile and dispensable for transposon regulation, Mol. Cell, 81, 3965-3978, https://doi.org/10.1016/j.molcel.2021.07.011.
Brennecke, J., Malone, C. D., Aravin, A. A., Sachidanandam, R., Stark, A., and Hannon, G. J. (2008) An epigenetic role for maternally inherited piRNAs in transposon silencing, Science, 322, 1387-1392, https://doi.org/10.1126/science.1165171.
Blumenstiel, J. P. (2019) Birth, school, work, death, and resurrection: the life stages and dynamics of transposable element proliferation, Genes (Basel), 10, 336, https://doi.org/10.3390/genes10050336.
Wallau, G. L., Vieira, C., and Loreto, E. L. S. (2018) Genetic exchange in eukaryotes through horizontal transfer: connected by the mobilome, Mob. DNA, 9, 6, https://doi.org/10.1186/s13100-018-0112-9.
Jensen, S., Gassama, M. P., and Heidmann, T. (1999) Taming of transposable elements by homology-dependent gene silencing, Nat. Genet., 21, 209-212, https://doi.org/10.1038/5997.
Kordyukova, M., Sokolova, O., Morgunova, V., Ryazansky, S., Akulenko, N., Glukhov, S., and Kalmykova, A. (2020) Nuclear Ccr4-Not mediates the degradation of telomeric and transposon transcripts at chromatin in the Drosophila germline, Nucleic Acids Res., 48, 141-156, https://doi.org/10.1093/nar/gkz1072.
Collart, M. A., and Panasenko, O. O. (2012) The Ccr4 – not complex, Gene, 492, 42-53, https://doi.org/10.1016/j.gene.2011.09.033.
Rozhkov, N. V., Hammell, M., and Hannon, G. J. (2013) Multiple roles for Piwi in silencing Drosophila transposons, Genes Dev., 27, 400-412, https://doi.org/10.1101/gad.209767.112.
Shpiz, S., Olovnikov, I., Sergeeva, A., Lavrov, S., Abramov, Y., Savitsky, M., and Kalmykova, A. (2011) Mechanism of the piRNA-mediated silencing of Drosophila telomeric retrotransposons, Nucleic Acids Res., 39, 8703-8711, https://doi.org/10.1093/nar/gkr552.
Sienski, G., Donertas, D., and Brennecke, J. (2012) Transcriptional silencing of transposons by piwi and maelstrom and its impact on chromatin state and gene expression, Cell, 151, 964-980, https://doi.org/10.1016/j.cell.2012.10.040.
Akkouche, A., Mugat, B., Barckmann, B., Varela-Chavez, C., Li, B., Raffel, R., Pelisson, A., and Chambeyron, S. (2017) Piwi is required during Drosophila embryogenesis to license dual-strand piRNA clusters for transposon repression in adult ovaries, Mol. Cell, 66, 411-419, https://doi.org/10.1016/j.molcel.2017.03.017.
Gunawardane, L. S., Saito, K., Nishida, K. M., Miyoshi, K., Kawamura, Y., Nagami, T., Siomi, H., and Siomi, M. C. (2007) A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila, Science, 315, 1587-1590, https://doi.org/10.1126/science.1140494.
Han, B. W., Wang, W., Li, C., Weng, Z., and Zamore, P. D. (2015) Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production, Science, 348, 817-821, https://doi.org/10.1126/science.aaa1264.
Mohn, F., Handler, D., and Brennecke, J. (2015) Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis, Science, 348, 812-817, https://doi.org/10.1126/science.aaa1039.
Lewis, S. H., Salmela, H., and Obbard, D. J. (2016) Duplication and diversification of dipteran argonaute genes, and the evolutionary divergence of Piwi and aubergine, Genome Biol. Evol., 8, 507-518, https://doi.org/10.1093/gbe/evw018.
Parhad, S. S., Tu, S., Weng, Z., and Theurkauf, W. E. (2017) Adaptive evolution leads to cross-species incompatibility in the piRNA transposon silencing machinery, Dev. Cell, 43, 60-70 e65, https://doi.org/10.1016/j.devcel.2017.08.012.
Vermaak, D., Henikoff, S., and Malik, H. S. (2005) Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila, PLoS Genet., 1, 96-108, https://doi.org/10.1371/journal.pgen.0010009.
Savitsky, M., Kwon, D., Georgiev, P., Kalmykova, A., and Gvozdev, V. (2006) Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline, Genes Dev., 20, 345-354, https://doi.org/10.1101/gad.370206.
Danilevskaya, O. N., Traverse, K. L., Hogan, N. C., DeBaryshe, P. G., and Pardue, M. L. (1999) The two Drosophila telomeric transposable elements have very different patterns of transcription, Mol. Cell. Biol., 19, 873-881, https://doi.org/10.1128/MCB.19.1.873.
Maxwell, P. H., Belote, J. M., and Levis, R. W. (2006) Identification of multiple transcription initiation, polyadenylation, and splice sites in the Drosophila melanogaster TART family of telomeric retrotransposons, Nucleic Acids Res., 34, 5498-5507.
Radion, E., Ryazansky, S., Akulenko, N., Rozovsky, Y., Kwon, D., Morgunova, V., Olovnikov, I., and Kalmykova, A. (2017) Telomeric retrotransposon HeT-A contains a bidirectional promoter that initiates divergent transcription of piRNA precursors in Drosophila germline, J. Mol. Biol., 429, 3280-3289, https://doi.org/10.1016/j.jmb.2016.12.002.
Shpiz, S., Kwon, D., Rozovsky, Y., and Kalmykova, A. (2009) rasiRNA pathway controls antisense expression of Drosophila telomeric retrotransposons in the nucleus, Nucleic Acids Res., 37, 268-278, https://doi.org/10.1093/nar/gkn960.
Tatsuke, T., Sakashita, K., Masaki, Y., Lee, J. M., Kawaguchi, Y., and Kusakabe, T. (2010) The telomere-specific non-LTR retrotransposons SART1 and TRAS1 are suppressed by Piwi subfamily proteins in the silkworm, Bombyx mori, Cell. Mol. Biol. Lett., 15, 118-133, https://doi.org/10.2478/s11658-009-0038-9.
Radion, E., Morgunova, V., Ryazansky, S., Akulenko, N., Lavrov, S., Abramov, Y., Komarov, P. A., Glukhov, S. I., Olovnikov, I., and Kalmykova, A. (2018) Key role of piRNAs in telomeric chromatin maintenance and telomere nuclear positioning in Drosophila germline, Epigenetics Chromatin, 11, 40, https://doi.org/10.1186/s13072-018-0210-4.
Wagner, E., Clement, S. L., and Lykke-Andersen, J. (2007) An unconventional human Ccr4-Caf1 deadenylase complex in nuclear cajal bodies, Mol. Cell. Biol., 27, 1686-1695, https://doi.org/10.1128/MCB.01483-06.
Ryazansky, S., Radion, E., Mironova, A., Akulenko, N., Abramov, Y., Morgunova, V., Kordyukova, M. Y., Olovnikov, I., and Kalmykova, A. (2017) Natural variation of piRNA expression affects immunity to transposable elements, PLoS Genet., 13, e1006731, https://doi.org/10.1371/journal.pgen.1006731.
Maupetit-Mehouas, S., and Vaury, C. (2020) Transposon reactivation in the germline may be useful for both transposons and their host genomes, Cells, 9, 1172, https://doi.org/10.3390/cells9051172.
Dufourt, J., Dennis, C., Boivin, A., Gueguen, N., Theron, E., Goriaux, C., Pouchin, P., Ronsseray, S., Brasset, E., and Vaury, C. (2014) Spatio-temporal requirements for transposable element piRNA-mediated silencing during Drosophila oogenesis, Nucleic Acids Res., 42, 2512-2524, https://doi.org/10.1093/nar/gkt1184.
Theron, E., Maupetit-Mehouas, S., Pouchin, P., Baudet, L., Brasset, E., and Vaury, C. (2018) The interplay between the Argonaute proteins Piwi and Aub within Drosophila germarium is critical for oogenesis, piRNA biogenesis and TE silencing, Nucleic acids Res., 46, 10052-10065, https://doi.org/10.1093/nar/gky695.
Kordyukova, M., Morgunova, V., Olovnikov, I., Komarov, P. A., Mironova, A., Olenkina, O. M., and Kalmykova, A. (2018) Subcellular localization and Egl-mediated transport of telomeric retrotransposon HeT-A ribonucleoprotein particles in the Drosophila germline and early embryogenesis, PLoS One, 13, e0201787, https://doi.org/10.1371/journal.pone.0201787.
Sokolova, O., Morgunova, V., Sizova, T. V., Komarov, P. A., Olenkina, O. M., Babaev, D. S., Mikhaleva, E. A., Kwon, D. A., Erokhin, M., and Kalmykova, A. (2023) The insulator BEAF32 controls the spatial-temporal expression profile of the telomeric retrotransposon TART in the Drosophila germline, Development, 150, dev201678, https://doi.org/10.1242/dev.201678.
Zhang, L., Beaucher, M., Cheng, Y., and Rong, Y. S. (2014) Coordination of transposon expression with DNA replication in the targeting of telomeric retrotransposons in Drosophila, EMBO J., 33, 1148-1158, https://doi.org/10.1002/embj.201386940.
Rashkova, S., Karam, S. E., Kellum, R., and Pardue, M. L. (2002) Gag proteins of the two Drosophila telomeric retrotransposons are targeted to chromosome ends, J. Cell Biol., 159, 397-402, https://doi.org/10.1083/jcb.200205039.
Lopez-Panades, E., Gavis, E. R., and Casacuberta, E. (2015) Specific localization of the Drosophila telomere transposon proteins and RNAs, give insight in their behavior, control and telomere biology in this organism, PLoS One, 10, e0128573, https://doi.org/10.1371/journal.pone.0128573.
Lepesant, J. M. J., Iampietro, C., Galeota, E., Auge, B., Aguirrenbengoa, M., Merce, C., Chaubet, C., Rocher, V., Haenlin, M., Waltzer, L., Pelizzola, M., and Di Stefano, L. (2020) A dual role of dLsd1 in oogenesis: regulating developmental genes and repressing transposons, Nucleic Acids Res., 48, 1206-1224, https://doi.org/10.1093/nar/gkz1142.
Yang, F., Quan, Z., Huang, H., He, M., Liu, X., Cai, T., and Xi, R. (2019) Ovaries absent links dLsd1 to HP1a for local H3K4 demethylation required for heterochromatic gene silencing, Elife, 8, e40806, https://doi.org/10.7554/eLife.40806.
Sienski, G., Batki, J., Senti, K. A., Donertas, D., Tirian, L., Meixner, K., and Brennecke, J. (2015) Silencio/CG9754 connects the Piwi-piRNA complex to the cellular heterochromatin machinery, Genes Dev., 29, 2258-2271, https://doi.org/10.1101/gad.271908.115.
Penke, T. J., McKay, D. J., Strahl, B. D., Matera, A. G., and Duronio, R. J. (2016) Direct interrogation of the role of H3K9 in metazoan heterochromatin function, Genes Dev., 30, 1866-1880, https://doi.org/10.1101/gad.286278.116.
Teo, R. Y. W., Anand, A., Sridhar, V., Okamura, K., and Kai, T. (2018) Heterochromatin protein 1a functions for piRNA biogenesis predominantly from pericentric and telomeric regions in Drosophila, Nat. Commun., 9, 1735, https://doi.org/10.1038/s41467-018-03908-3.
Wang, S. H., and Elgin, S. C. (2011) Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line, Proc. Natl. Acad. Sci. USA, 108, 21164-21169, https://doi.org/10.1073/pnas.1107892109.
Savitsky, M., Kravchuk, O., Melnikova, L., and Georgiev, P. (2002) Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster, Mol. Cell. Biol., 22, 3204-3218, https://doi.org/10.1128/MCB.22.9.3204-3218.2002.
Molaro, A., Falciatori, I., Hodges, E., Aravin, A. A., Marran, K., Rafii, S., McCombie, W. R., Smith, A. D., and Hannon, G. J. (2014) Two waves of de novo methylation during mouse germ cell development, Genes Dev., 28, 1544-1549, https://doi.org/10.1101/gad.244350.114.
Zoch, A., Auchynnikava, T., Berrens, R. V., Kabayama, Y., Schopp, T., Heep, M., Vasiliauskaite, L., Perez-Rico, Y. A., Cook, A. G., Shkumatava, A., Rappsilber, J., Allshire, R. C., and O’Carroll, D. (2020) SPOCD1 is an essential executor of piRNA-directed de novo DNA methylation, Nature, 584, 635-639, https://doi.org/10.1038/s41586-020-2557-5.
Zeng, Y., and Chen, T. (2019) DNA methylation reprogramming during mammalian development, Genes (Basel), 10, 257, https://doi.org/10.3390/genes10040257.
Shirane, K., Kurimoto, K., Yabuta, Y., Yamaji, M., Satoh, J., Ito, S., Watanabe, A., Hayashi, K., Saitou, M., and Sasaki, H. (2016) Global landscape and regulatory principles of DNA methylation reprogramming for germ cell specification by mouse pluripotent stem cells, Dev. Cell, 39, 87-103, https://doi.org/10.1016/j.devcel.2016.08.008.
Kohlrausch, F. B., Berteli, T. S., Wang, F., Navarro, P. A., and Keefe, D. L. (2022) Control of LINE-1 expression maintains genome integrity in germline and early embryo development, Reprod. Sci., 29, 328-340, https://doi.org/10.1007/s43032-021-00461-1.
Akiyama, T., Xin, L., Oda, M., Sharov, A. A., Amano, M., Piao, Y., Cadet, J. S., Dudekula, D. B., Qian, Y., Wang, W., Ko, S. B., and Ko, M. S. (2015) Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells, DNA Res., 22, 307-318, https://doi.org/10.1093/dnares/dsv013.
Dan, J., Rousseau, P., Hardikar, S., Veland, N., Wong, J., Autexier, C., and Chen, T. (2017) Zscan4 inhibits maintenance DNA methylation to facilitate telomere elongation in mouse embryonic stem cells, Cell Rep., 20, 1936-1949, https://doi.org/10.1016/j.celrep.2017.07.070.
Zalzman, M., Falco, G., Sharova, L. V., Nishiyama, A., Thomas, M., Lee, S. L., Stagg, C. A., Hoang, H. G., Yang, H. T., Indig, F. E., Wersto, R. P., and Ko, M. S. (2010) Zscan4 regulates telomere elongation and genomic stability in ES cells, Nature, 464, 858-863, https://doi.org/10.1038/nature08882.
Thool, M., Sundaravadivelu, P. K., Sudhagar, S., and Thummer, R. P. (2022) A comprehensive review on the role of ZSCAN4 in embryonic development, stem cells, and cancer, Stem Cell Rev. Rep., 18, 2740-2756, https://doi.org/10.1007/s12015-022-10412-1.
Dan, J., Zhou, Z., Wang, F., Wang, H., Guo, R., Keefe, D. L., and Liu, L. (2022) Zscan4 contributes to telomere maintenance in telomerase-deficient late generation mouse escs and human ALT cancer cells, Cells, 11, 456, https://doi.org/10.3390/cells11030456.
Peaston, A. E., Evsikov, A. V., Graber, J. H., de Vries, W. N., Holbrook, A. E., Solter, D., and Knowles, B. B. (2004) Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos, Dev Cell., 7, 597-606, https://doi.org/10.1016/j.devcel.2004.09.004.
Kigami, D., Minami, N., Takayama, H., and Imai, H. (2003) MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos, Biol. Reprod., 68, 651-654, https://doi.org/10.1095/biolreprod.102.007906.
Fadloun, A., Le Gras, S., Jost, B., Ziegler-Birling, C., Takahashi, H., Gorab, E., Carninci, P., and Torres-Padilla, M. E. (2013) Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA, Nat. Struct. Mol. Biol., 20, 332-338, https://doi.org/10.1038/nsmb.2495.
Eckersley-Maslin, M. A., Svensson, V., Krueger, C., Stubbs, T. M., Giehr, P., Krueger, F., Miragaia, R. J., Kyriakopoulos, C., Berrens, R. V., Milagre, I., Walter, J., Teichmann, S. A., and Reik, W. (2016) MERVL/Zscan4 network activation results in transient genome-wide DNA demethylation of mESCs, Cell Rep., 17, 179-192, https://doi.org/10.1016/j.celrep.2016.08.087.
Wang, F., Chamani, I. J., Luo, D., Chan, K., Navarro, P. A., and Keefe, D. L. (2021) Inhibition of LINE-1 retrotransposition represses telomere reprogramming during mouse 2-cell embryo development, J. Assist Reprod. Genet., 38, 3145-3153, https://doi.org/10.1007/s10815-021-02331-w.
Percharde, M., Lin, C. J., Yin, Y., Guan, J., Peixoto, G. A., Bulut-Karslioglu, A., Biechele, S., Huang, B., Shen, X., and Ramalho-Santos, M. (2018) A LINE1-nucleolin partnership regulates early development and ESC identity, Cell, 174, 391-405, https://doi.org/10.1016/j.cell.2018.05.043.
Macfarlan, T. S., Gifford, W. D., Driscoll, S., Lettieri, K., Rowe, H. M., Bonanomi, D., Firth, A., Singer, O., Trono, D., and Pfaff, S. L. (2012) Embryonic stem cell potency fluctuates with endogenous retrovirus activity, Nature, 487, 57-63, https://doi.org/10.1038/nature11244.
Ghosh, S., and Zhou, Z. (2014) Genetics of aging, progeria and lamin disorders, Curr. Opin. Genet. Dev., 26, 41-46, https://doi.org/10.1016/j.gde.2014.05.003.
Gorbunova, V., Seluanov, A., Mita, P., McKerrow, W., Fenyo, D., Boeke, J. D., Linker, S. B., Gage, F. H., Kreiling, J. A., Petrashen, A. P., Woodham, T. A., Taylor, J. R., Helfand, S. L., and Sedivy, J. M. (2021) The role of retrotransposable elements in ageing and age-associated diseases, Nature, 596, 43-53, https://doi.org/10.1038/s41586-021-03542-y.
Aschacher, T., Wolf, B., Enzmann, F., Kienzl, P., Messner, B., Sampl, S., Svoboda, M., Mechtcheriakova, D., Holzmann, K., and Bergmann, M. (2016) LINE-1 induces hTERT and ensures telomere maintenance in tumour cell lines, Oncogene, 35, 94-104, https://doi.org/10.1038/onc.2015.65.
Aschacher, T., Wolf, B., Aschacher, O., Enzmann, F., Laszlo, V., Messner, B., Turkcan, A., Weis, S., Spiegl-Kreinecker, S., Holzmann, K., Laufer, G., Ehrlich, M., and Bergmann, M. (2020) Long interspersed element-1 ribonucleoprotein particles protect telomeric ends in alternative lengthening of telomeres dependent cells, Neoplasia, 22, 61-75, https://doi.org/10.1016/j.neo.2019.11.002.
Cosby, R. L., Chang, N. C., and Feschotte, C. (2019) Host-transposon interactions: conflict, cooperation, and cooption, Genes Dev., 33, 1098-1116, https://doi.org/10.1101/gad.327312.119.
Charlesworth, B., and Langley, C. H. (1989) The population genetics of Drosophila transposable elements, Annu. Rev. Genet., 23, 251-287, https://doi.org/10.1146/annurev.ge.23.120189.001343.
Kelleher, E. S., and Barbash, D. A. (2013) Analysis of piRNA-mediated silencing of active TEs in Drosophila melanogaster suggests limits on the evolution of host genome defense, Mol. Biol. Evol., 30, 1816-1829, https://doi.org/10.1093/molbev/mst081.
Funding
This work was financially supported by the Russian Science Foundation (project no. 23-24-00025).
Author information
Authors and Affiliations
Contributions
A.I.K. conception and writing the manuscript; O.A.S. writing and editing the manuscript.
Corresponding author
Ethics declarations
The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
Open access. This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kalmykova, A.I., Sokolova, O.A. Retrotransposons and Telomeres. Biochemistry Moscow 88, 1739–1753 (2023). https://doi.org/10.1134/S0006297923110068
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297923110068