Abstract
To date, no studies have investigated the association of the GWAS-identified SNPs with BC risk in Indian population. We investigated the association of 30 previously reported and replicated BC susceptibility SNPs in 1,204 cases and 1,212 controls from a hospital based case-control study conducted at the Tata Memorial Hospital, Mumbai. As a measure of total susceptibility burden, the polygenic risk score (PRS) for each individual was defined by the weighted sum of genotypes from 21 independent SNPs with weights derived from previously published estimates of association odds-ratios. Logistic regression models were used to assess risk associated with individual SNPs and overall PRS, and stratified by menopausal and receptor status. A total of 11 SNPs from eight genomic regions (FGFR2, 9q31.2, MAP3K, CCND1, ZM1Z1, RAD51L11, ESR1 and UST) showed statistically significant (p-value ≤ 0.05) evidence of association, either overall or when stratified by menopausal status or hormone receptor status. BC SNPs previously identified in Caucasian population showed evidence of replication in the Indian population mainly with respect to risk of postmenopausal and hormone receptor positive BC.
Similar content being viewed by others
Introduction
Breast cancer (BC) is a complex disease strongly influenced by genetic, environmental and lifestyle factors. Under the assumption that common disorders in humans are partly due to common low penetrant polymorphisms, a large number of genome wide association studies (GWAS) have been conducted during the last decade, primarily in Caucasian, European, Japanese and Chinese populations1,2,3. These studies along with projects such as HapMap and 1000G indicate that the prevalence of single nucleotide polymorphisms (SNPs) can vary in different populations, and in some cases, the risk estimates associated with BC also differ4,5,6,7.
BC rates have been increasing in India for the last decade. However, there are very few large-scale population based studies to identify risk factors related to lifestyle and/or genetics. Given that a high percentage of BC in India occurs in young and premenopausal women8, with a large proportion being triple negative tumours that typically have poorer prognosis9, it is important to understand the aetiology of BC in this population. We conducted a large scale case-control study at Tata Memorial Hospital (TMH), Mumbai, India, to identify lifestyle and genetic risk factors for BC. In this analysis, we focus on examining risk in established SNP loci for BC identified by GWAS studies in Caucasian and East Asian populations. To the best of our knowledge this is the first study in an Indian population to examine a large number of GWAS-identified BC risk loci in an adequately-powered population-based study.
Methods
We conducted a hospital based case-control study at TMH, Mumbai during the period of January 2009 to September 2013. Enrolled cases were females with primary BC seen at TMH, aged 20–69 years with a date of diagnosis not more than 6 months from date of interview. All BC cases enrolled in the study were histologically confirmed.
Female visitors aged 20–69 years, with no history of cancer, and who were accompanying cancer patients of any primary site, were eligible for inclusion as controls. Controls that were unrelated to BC patients were genotyped and included in the analysis. Controls were frequency-matched to cases by age and region of residence at enrolment. The study has been approved by TMH Institutional Review Board. Written informed consent was obtained from all study participants before enrolling them in the study. All methods were performed in accordance with the relevant guidelines and regulations.
Information collected on all subjects included data on residential status, reproductive history and anthropometric measurements. Women whose menstrual period had either stopped naturally, or due to oophorectomy, hysterectomy or any other reason for 12 months or more before the date of enrolment were classified as postmenopausal. The rest were treated as premenopausal. Estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) status for cases were obtained from hospital pathology records.
Apart from questionnaire data and anthropometric measurements a 10 ml blood sample was collected from each study participant and plasma and buffy coat were separated. Buffy coat samples were available for 1,214 cases (74.1% of all cases) and 1,293 controls (85.3% of all controls).
DNA Preparation and Assay Performance
Genomic DNA was extracted from buffy coat using the Qiagen QiAamp Blood DNA MidiKit and Macherey Nagel Nucleomag Blood kit. The concentration of each DNA sample was determined by Quant-iT PicoGreen assay. A total of 250 ng DNA was applied to SNP typing using Illumina’s GoldenGate Genotyping Custom SNP Panel assay (Illumina Inc., San Diego, CA)10. Illumina Golden Gate assay is reported to be highly accurate in humans with error rates in the order of 0.3–0.4%11. Genotyping was performed on 1,204 cases and 1,212 controls on 384 custom-selected SNPs. Plates were prepared containing randomly mixed cases and controls. Intraplate and interplate replicates (7% approx.) were included on all plates and in all batches.
Design of Custom SNP Panel
A customized panel of 384 SNPs was designed by including GWAS-identified BC risk loci, BC SNPs identified from candidate gene studies, SNPs previously reported to be associated with obesity related traits and other SNPs in obesity genes. This paper focuses on a subset of 31 BC GWAS SNPs identified in Caucasian and East Asian populations using the Human Genome Epidemiology (HuGE) Navigator and the National Institute of Health (NIH) GWAS Catalog12,13. We have also included 45 BC SNPs from candidate gene studies. Only 30 BC GWAS SNPs and 42 BC candidate SNPs were used for final analysis after quality assessment. For BC GWAS SNPs, only SNPs with p value < 5 × 10−8 were included in the analysis. Duplicate SNPs between the HuGE Navigator and NIH GWAS Catalog were removed. There were no overlapping SNPs between GWAS and candidate gene studies. The panel was designed in March 2011 and therefore SNPs identified after this time were not included.
Since results from candidate gene SNP studies have had a poor record of replication14, we have presented the results in Supplementary Table S3 and have not evaluated them in the main tables.
Quality Assessment
The reproducibility rate of replicate samples (n = 160) for all assays was >98%. Examination of negative controls indicated no inter-sample contamination. A designability rank score (0–1.0) was calculated for each SNP by Illumina for conversion of SNP into a successful GoldenGate Assay. All SNPs had a score of 1.0, indicating a high success rate. Following completion of the assay, data were cleaned using the Illumina Genome Studio software version 1.9.4. Automatic allele calling was performed using a GenCall (GC) threshold of 0.25. The software assigned three clusters on a graph based on the fluorescence obtained. Seventeen samples had a call rate <90% that were excluded and a total of 2,399 (1,194 cases and 1,205 control) samples were included in the final analysis. One SNP with MAF <1% and 3 SNPs with diffused clusters in our study population were excluded, yielding a list of 30 GWAS SNPs and 42 candidate SNPs (i.e. total 72 SNPs across 41 genes) for final analysis. All SNPs had call frequency above 95%. No deviation from hardy-weinberg equilibrium (HWE) (P < 0.001) was observed using the chi-square test, and all SNPs had Gen-train score value of 0.4 and above. All quality control dashboards provided in Genome Studio Software showed that the quality of the assays were satisfactory including allele specific extension, PCR uniformity, extension gap, first and second hybridization.
Out of 1,194 cases and 1,205 controls, information on menopausal status could be obtained on 1,193 cases (607 premenopausal and 586 postmenopausal) and 1,191 (650 premenopausal and 541 postmenopausal) controls respectively. When we stratified cases by hormone receptor status, there were 408 estrogen receptor positive (ER+)/progesterone receptor positive (PR+), 529 estrogen receptor negative (ER−)/progesterone receptor negative (PR−) irrespective of their HER2 status and 340 triple negative breast cancer (TNBC) cases.
Statistical Analysis
Unconditional logistic regression was used to estimate adjusted odds ratios (OR) and corresponding 95% confidence intervals (CI) between genotype and BC case-control status. The model was adjusted for age (continuous variable) and region of residence (North, South, East, West and Central India). An additive model of inheritance (continuous effect of increasing number of variant alleles - 0 versus 1 versus 2) was assumed and the genotypes were coded as 0 = wild type, 1 = heterozygous and 2 = homozygous variant. Further analyses were performed by menopausal and hormone receptor status. ORs for all SNPs were reported with respect to the risk allele as identified in previous GWAS. All statistical tests were two-sided, and a P value equal to or less than 0.05 was considered statistically significant.
To investigate the association between BC risk and total susceptibility burden defined by the combination of the SNPs, a polygenic risk score (PRS) was derived for each individual using the formula: PRS = β1x1 + β2x2 + … + βnxn. For any given SNP n, βn is the log-odds-ratio associated with risk allele reported in the literature from prior GWAS conducted in Caucasian population2 and xn is the number of risk alleles carried by an individual in our study. Only independent SNPs were included in the PRS analysis. If there were multiple SNPs in LD (r^2 > 0.2) from one region, the SNP with strongest association signal reported in previous GWAS was picked for our analysis, resulting in a total of 21 independent SNPs. Logistic regression models were used to estimate the odds ratios for BC by percentile of the PRS, with 25th percentile as the reference. Based on previously reported ORs of all the SNPs, their allele frequencies in the Indian population, and the sample, we estimated power of replication of each SNP in the current study.
The probability of observing a larger number of significant associations (P ≤ 0.05) than would be expected by chance is a function of the binomial distribution15. We conducted a global test for the hypothesis to evaluate whether the number of significant associations with BC (P ≤ 0.05) was greater than expected for the number of loci tested. All analyses were performed using the statistical software Stata version 12.016.
Results
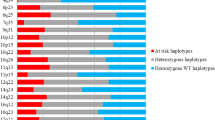
A total of 1,194 cases and 1,205 controls were included in the final analysis. Table 1 describes the distribution of cases and controls with respect to age, education, region of residence at enrolment, menopausal status, and family history of breast, ovary or endometrial cancer. The risk of TNBC increases two-fold in females with family history of breast, ovary or endometrial cancer (OR = 2.00; 95%CI = 1.01–3.97) (data not shown). Direction of effect were similar to previously reported GWAS for 22 of the 30 SNPs, but different for SNPs rs10069690 (TERT), rs13387042 (TNP1), rs1562430 (FAM84B), rs2180341 (RNF146), rs2981575 (FGFR2), rs3757318 (C6orf97), rs6504950 (STXBP4), rs999737 (RAD51L1) (Table 2). For the risk of overall BC, we confirm previously-reported associations between 5 GWAS-identified BC susceptibility SNPs, with an exception of rs2981575 which was associated with BC risk in our population but with reverse direction of effect (Table 2). Overall, out of the 30 GWAS BC SNPs analysed 22 SNPs showed effects in the same direction as that reported in previous GWAS. Applying the binomial test for enrichment indicated that the pattern was unlikely to be due to chance (p-value = 0.016). When cases were stratified by menopausal status, a number of BC GWAS-identified SNPs appeared to show stronger association in postmenopausal versus premenopausal women (Table 3). In particular, 7 SNPs achieved statistical significance (P ≤ 0.05) for association with postmenopausal BCs: FGFR2 (rs1219648, rs2981575, rs2981579 and rs2981582), MAP3K1 (rs889312), ESR1 (rs2046210) and 9q31.2 (rs865686) whereas only one SNP RAD51L1 (rs999737) showed association with premenopausal BCs. Details of all SNPs stratified on menopausal status are presented in Supplementary Table S1. Analyses performed on ER/PR status showed a total of 8 SNPs achieved statistical significance for association with ER+/PR+ BC. In addition, the SNP rs2046210 in ESR1 and rs9485372 (UST) showed statistically significant increased risk for BCs for ER−/PR− and TNBC but not for ER+/PR+ (Table 4). The minor alleles of rs1219648, rs2981579, rs2981582 (FGFR2), rs889312 (MAP3K1), rs614367 (CCND1) and rs704010 (ZMIZ1) increased the risk. The alleles T and C of rs2981575 (FGFR2) and rs999737 (RAD51L1) respectively decreased the risk of hormone receptor positive BC (Table 4). Details of all SNPs analysed by hormone receptor status are presented in Supplementary Table S2.
The distribution of the PRS was shifted upwards in cases when compared to controls. Among all the different outcomes considered, PRS showed strongest association with the risk of BC among postmenopausal cases. In particular, postmenopausal women in the highest quartile of the PRS score had an 83% increased BC risk when compared to women in the lowest quartile. Results were similar when study participants with family history of breast, ovary or endometrial cancer were excluded from analysis (Table 5).
We also attempted to replicate associations for 42 BC SNPs which were previously identified in candidate studies and observed significant association in 3 regions viz. rs2420946 (FGFR2), rs3218408 (XRCC2), rs1641535 and rs1641536 (ATP1B2) (Supplementary Table S3).
Discussion
We used 1,194 cases and 1,205 controls from a hospital based case-control study in India to report evidence of replication for susceptibility BC SNPs that have been previously identified primarily through GWAS conducted in Caucasian population. To our knowledge this is the first study to evaluate risk of BC and GWAS-identified SNPs in the Indian population. The study population is unique in that it is an unscreened population with no use of hormone replacement therapy and 52% premenopausal women. The average tumour size in our study cases was well over 2 cm17 and cases were not diagnosed mammographically.
Out of 30 GWAS-identified SNPs analysed, 11 SNPs from eight genomic regions (FGFR2, 9q31.2, MAP3K1, CCND1, ZM1Z1, RAD51L11, ESR1 and UST) showed statistically significant (P ≤ 0.05) evidence of association either overall or when stratified by menopausal status or when analysed separately by hormone receptor status BCs. The direction of effect was the same as the previously reported GWAS results for 22 of the 30 SNPs.
We also attempted to replicate associations for 42 SNPs reported to be associated with BC in candidate gene studies. We observed statistical significance in 4 SNPs (9.5%) indicating that association observed in candidate gene studies are more prone to false positive results.
Our current data support the conclusions of previous GWAS studies18,19 that FGFR2 (a tumour suppressor gene) polymorphisms (rs2981582 and rs2981575), first identified as susceptibility loci for BC in Caucasian population20,21, are associated with overall BC. The risk allele frequencies of FGFR2 SNPs were similar to those reported in previous GWAS, with the exception of rs2981575, which was much common in our population. rs865686 in 9q31.2 was significantly associated with BC, further the rare allele (G) frequency of rs865686 (MAF = 0.14) obtained in our study was comparable to that reported in a previous study in Asians (MAF = 0.09)22.
Consistent with results from Caucasian and East Asian populations, we found that both rs889312 (MAP3K1) and rs2046210 (close to ESR1) were associated with increased risk of overall BC in our Indian population. rs889312 lies in an LD block of approximately 280 kb which includes the MAP3K1 gene23. The MAP3K1 gene encodes a 196-kDa serine/threonine protein kinase that activates the extracellular signal regulated kinase (ERK), c-Jun NH2-terminal kinase (JNK) and nuclear factor-kB (NF-kB) pathways24. Downstream signal transduction genes regulate cell survival, differentiation, proliferation and apoptosis, and appear to be involved in tumour development and tumour progression25,26,27.
The SNP rs2046210 is located 180 kb upstream of the transcription initiation site of the first coding exon of the ESR1 gene. Considering the relative vicinity of rs2046210 to the ESR1 gene, it has been speculated that either the SNP itself, or causal variants in LD with it, might alter ESR1 gene expression, thus affecting susceptibility to BC. Functional genomic analyses and in vitro functional experiments conducted by Cai et al.28 provided no support for the potential involvement of the polymorphism itself in the regulation of ESR1. The function of this SNP therefore is still unclear; future fine-mapping of the BC susceptibility loci tagged by rs2046210 is warranted and the underlying biological mechanism of this polymorphism needs further investigation.
Results of our hormone receptor analyses successfully replicated previous GWAS reported loci for hormone receptor positive BCs in FGFR2 (rs1219648, rs2981575, rs2981579 and rs2981582)29; MAP3K1 (rs889312)30,31, CCND1 (rs614367) ZMIZ1 (rs704010)3, although the inverse direction of effect as compared to the previous GWAS report for FGFR2 (rs2981575) and RAD51L1 (rs999737) was unexpected. Our observed association of increased risk with respect to rs2046210 (ESR1) was consistent with previous studies suggesting that rs2046210 tended to increase BC risk in ER− tumour by a greater magnitude as compared to ER+ tumour32,33,34. Consistent with the literature, we also found SNP rs9485372 (UST) to be associated more with ER−/PR− and TNBC1.
Our analyses stratified on menopausal status showed 7 SNPs to be associated with postmenopausal BC, as opposed to only 1 SNP associated with premenopausal BC. SNPs in FGFR2 (rs2981582, rs2981575, rs1219648 and rs2981579), MAP3K1 (rs889312) and the 9q31.2 region (rs865686) were associated with BCs in postmenopausal women. Large scale GWAS studies have previously reported that the association of rs865686 is stronger in postmenopausal women2,22,35. Our observed associations of rs2046210 and BC were also consistent with prior literature suggesting that risk in postmenopausal BC cases was greater than risk in premenopausal BC cases4.
We observed evidence of replication of association for previously reported GWAS BC SNPs mostly among postmenopausal or ER+/PR+ BC patients. This is not surprising given that these SNPs were identified mainly in American/European populations comprised largely of postmenopausal/older women21,36,37. Among ER+/PR+ SNPs, most of SNPs (80%) were significant for postmenopausal women (data not shown). None of 19 SNPs which could not be replicated in present study (except rs2180341) had statistical power of 80% or more for replication (Supplementary Table S4), suggesting that larger sample sizes may be needed in order to detect an association. Nonetheless, the consistency analysis showed that the similarity in direction of effect for these 18 non-replicated SNPs was more than chance (P = 0.016). On the other hand, the null observation in our study for rs2180341 (an association initially observed in Ashkenazi Jewish GWAS population) even with 98% power for replication, is more likely to reflect a true difference in association across different ethnicities.
Our results of the 21 SNP PRS (including only the strongest SNP from LD >0.2 SNP groups) showed that postmenopausal women in the highest quartile of PRS had an OR of 1.83 (95% CI = 1.28–2.60) when compared to women in lowest quartile. An increase in risk of ER+/PR+ BCs was also observed, having OR = 1.36 (95% CI = 1.04–1.79) with per unit increase in PRS.
In conclusion, our study provides early evidence that the genetic architecture of postmenopausal and/or hormone receptor positive BC in the Indian population may be similar to that of Caucasian populations. More population-based studies in the Indian population are needed in order to identify additional BC susceptibility SNPs, especially for hormone receptor negative BCs.
Additional Information
How to cite this article: Nagrani, R. et al. Association of Genome-Wide Association Study (GWAS) Identified SNPs and Risk of Breast Cancer in an Indian Population. Sci. Rep. 7, 40963; doi: 10.1038/srep40963 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Long, J. et al. Genome-wide association study in east Asians identifies novel susceptibility loci for breast cancer. PLoS Genet. 8, e1002532 (2012).
Michailidou, K. et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 45, 353–361, 361–2 (2013).
Turnbull, C. et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 42, 504–507 (2010).
Zheng, W. et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 41, 324–328 (2009).
Couch, F. J. et al. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 9, e1003212 (2013).
Birney, E. & Soranzo, N. Human genomics: The end of the start for population sequencing. Nature 526, 52–53 (2015).
Consortium, I. H. 3 & others. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–58 (2010).
Ghiasvand, R., Adami, H.-O., Harirchi, I., Akrami, R. & Zendehdel, K. Higher incidence of premenopausal breast cancer in less developed countries; myth or truth? BMC Cancer 14, 343 (2014).
Gupta, S. Triple negative breast cancer: A continuing challenge. Indian J. Med. Paediatr. Oncol. Off. J. Indian Soc. Med. Paediatr. Oncol. 34, 1 (2013).
Steemers, F. J. & Gunderson, K. L. Illumina, Inc. Pharmacogenomics 6, 777–782 (2005).
Fan, J.-B. et al. Highly parallel SNP genotyping. In Cold Spring Harbor symposia on quantitative biology 68, 69–78 (Cold Spring Harbor Laboratory Press, 2003).
Yu, W. et al. GWAS Integrator: a bioinformatics tool to explore human genetic associations reported in published genome-wide association studies. Eur. J. Hum. Genet. EJHG 19, 1095–1099 (2011).
Hindorff, L. et al. A Catalog of Published Genome-Wide Association Studies. Available at: www.genome.gov/gwastudies. (Accessed: 10th February 2012).
Siontis, K. C., Patsopoulos, N. A. & Ioannidis, J. P. Replication of past candidate loci for common diseases and phenotypes in 100 genome-wide association studies. Eur. J. Hum. Genet. 18, 832–837 (2010).
Frayling, T. M. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science 316, 889–894 (2007).
StataCorp. Stata Statistical Software. (StataCorp LP, 2011).
Budrukkar, A. et al. Clinical outcomes of prospectively treated 140 women with early stage breast cancer using accelerated partial breast irradiation with 3 dimensional computerized tomography based brachytherapy. Radiother. Oncol. 115, 349–354 (2015).
Jia, C., Jia, C., Cai, Y., Ma, Y. & Fu, D. Quantitative assessment of the effect of FGFR2 gene polymorphism on the risk of breast cancer. Breast Cancer Res. Treat. 124, 521–528 (2010).
Zhang, J. et al. Current evidence on the relationship between three polymorphisms in the FGFR2 gene and breast cancer risk: a meta-analysis. Breast Cancer Res. Treat. 124, 419–424 (2010).
Easton, D. F. et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447, 1087–1093 (2007).
Hunter, D. J. et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat. Genet. 39, 870–874 (2007).
Warren, H. et al. 9q31.2-rs865686 as a Susceptibility Locus for Estrogen Receptor-Positive Breast Cancer: Evidence from the Breast Cancer Association Consortium. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 21, 1783–1791 (2012).
Jara, L. et al. Genetic variants in FGFR2 and MAP3K1 are associated with the risk of familial and early-onset breast cancer in a South-American population. Breast Cancer Res. Treat. 137, 559–569 (2013).
Mokhtari, D., Myers, J. W. & Welsh, N. MAPK kinase kinase-1 is essential for cytokine-induced c-Jun NH2-terminal kinase and nuclear factor-kappaB activation in human pancreatic islet cells. Diabetes 57, 1896–1904 (2008).
Dent, P. ERK plays the baddie (again). Cancer Biol. Ther. 14, 997–998 (2013).
Fan, Y., Mao, R. & Yang & J. NF-κB and STAT3 signaling pathways collaboratively link inflammation to cancer. Protein Cell 4, 176–185 (2013).
Seki, E., Brenner, D. A. & Karin, M. A liver full of JNK: signaling in regulation of cell function and disease pathogenesis, and clinical approaches. Gastroenterology 143, 307–320 (2012).
Cai, Q. et al. Replication and functional genomic analyses of the breast cancer susceptibility locus at 6q25. 1 generalize its importance in women of chinese, Japanese, and European ancestry. Cancer Res. 71, 1344–1355 (2011).
Tamaru, N. et al. Estrogen receptor-associated expression of keratinocyte growth factor and its possible role in the inhibition of apoptosis in human breast cancer. Lab. Invest. 84, 1460–1471 (2004).
Rebbeck, T. R. et al. Hormone-dependent effects of FGFR2 and MAP3K1 in breast cancer susceptibility in a population-based sample of post-menopausal African-American and European-American women. Carcinogenesis 30, 269–274 (2009).
Zheng, Q., Ye, J., Wu, H., Yu, Q. & Cao, J. Association between Mitogen-Activated Protein Kinase Kinase Kinase 1 Polymorphisms and Breast Cancer Susceptibility: A Meta-Analysis of 20 Case-Control Studies. PLoS ONE 9 (2014).
Yang, Z., Shen, J., Cao, Z. & Wang, B. Association between a novel polymorphism (rs2046210) of the 6q25.1 locus and breast cancer risk. Breast Cancer Res. Treat. 139, 267–275 (2013).
Zheng, W. et al. Common genetic determinants of breast-cancer risk in East Asian women: a collaborative study of 23 637 breast cancer cases and 25 579 controls. Hum. Mol. Genet. 22, 2539–2550 (2013).
Hein, R. et al. Comparison of 6q25 breast cancer hits from Asian and European Genome Wide Association Studies in the Breast Cancer Association Consortium (BCAC). PloS One 7, e42380 (2012).
Fletcher, O. et al. Novel Breast Cancer Susceptibility Locus at 9q31.2: Results of a Genome-Wide Association Study. J. Natl. Cancer Inst. 103, 425–435 (2011).
Thomas, G. et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat. Genet. 41, 579–584 (2009).
Long, J. et al. Evaluating genome-wide association study-identified breast cancer risk variants in African-American women. PloS One 8, e58350 (2013).
Acknowledgements
We would like to thank Ms. Janhavi Acharya, Ms. Swati Bandiwadekar, Ms. Shraddha Bonkar for data collection and Mr. Vaibhav Chikne, Ms. Sheetal Shedge for DNA extraction.
Author information
Authors and Affiliations
Contributions
R.D. study design; R.N., S.M., R.B., S.G. data acquisition; Pa.Bo., Pa.Br., M.A., R.N., S.M. conceived the experiment; R.N. and S.M. conducted the experiment; R.N., S.M., R.D., N.C., P.R. analysed and interpreted the results; R.N., R.D. manuscript preparation; R.N., P.R., N.C., M.A., R.D. manuscript editing. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Nagrani, R., Mhatre, S., Rajaraman, P. et al. Association of Genome-Wide Association Study (GWAS) Identified SNPs and Risk of Breast Cancer in an Indian Population. Sci Rep 7, 40963 (2017). https://doi.org/10.1038/srep40963
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep40963
- Springer Nature Limited
This article is cited by
-
Symposium report: breast cancer in India—trends, environmental exposures and clinical implications
Cancer Causes & Control (2021)
-
Low-penetrance susceptibility variants and postmenopausal oestrogen receptor positive breast cancer
Journal of Genetics (2020)