Abstract
Assessments of biodiversity and ecosystem status can benefit from DNA metabarcoding as a means to streamline sample processing and specimen identification. Moreover, processing the fixation medium instead of the precious material introduces straightforward protocols that allow subsequent focus on certain organisms detected among the preserved specimens. In this study, we present a proof of concept via the analysis of freshwater invertebrate samples from the Tatra Mountain lakes (Slovakia). Besides highlighting a match between the lake-specific environmental conditions and the results of our fixative DNA metabarcoding, we observed an option to fine-tune the fixation time: to prefer two weeks over a day or a month. This effect emerged from the presence/absence of individual taxa rather than from coarse per-sample records of taxonomic richness, demonstrating that metabarcoding studies—and efforts to optimize their protocols—can use the robust metrics to explore even subtle trends. We also provide evidence that fixative DNA might better capture large freshwater species than terrestrial or meiofauna.
Similar content being viewed by others
Introduction
Benthic invertebrates play an essential role within their ecosystems and in freshwater biomonitoring1. However, the state of their diversity, which depends on the availability of suitable habitats, is declining due to numerous anthropogenic impacts2. These affect even remote ecosystems, such as mountain lakes3,4, which usually harbor unique and relatively isolated invertebrate communities5,6 but are difficult to access for sampling. The arising global need for efficient, reliable, and detailed biodiversity assessments has been addressed inter alia by the adoption of DNA metabarcoding, a method based on high-throughput sequencing of species-specific genetic markers amplified from various types of environmental samples7,8,9,10. Identification of species this way requires adequate reference barcode databases, which are currently comprehensive in many areas and taxa, but gaps still need to be filled11. Certain limitations are always in place, e.g., those flowing from the choice of sampling protocol or PCR primer specificity, so a single analysis cannot capture each organism in the ecosystem8,9,10. Nevertheless, DNA metabarcoding has already enhanced bioassessments and ecological studies, often detecting even taxa that are overlooked by conventional approaches12,13,14,15.
Invertebrate specimens gathered in whole-community “bulk” samples for metabarcoding are commonly sorted and homogenized prior to tissue-based DNA extraction16. Alternatively, it was proposed to omit sorting and specimen destruction by simply extracting DNA from sample fixative (usually ethanol)17. Satisfying congruence with more conventional approaches encouraged further use of fixative DNA17,18,19,20,21. In addition, it was shown that the recovery of DNA from storage ethanol can be improved through the choice of extraction method, longer storage duration (up to 14 days)20, shaking or freezing18. Providing more time for the specimens to shed their DNA into the fixative is an attractively simple means of improvement, but optimal interval has not been stated yet. Martins et al.20 only calculated an unconfirmed optimum in richness between 7 and 14th day of storage. Due to the lack of samples within and beyond that interval, the authors did not rule out continuation of the increasing trend with time.
As for DNA degradation, it is commonly avoided by storing samples cooled (+ 4 or − 20 °C) and replacing the fixative upon return from the field13,22,23,24. However, the original fixative may be of particular interest as a better source of DNA eluted from specimen surfaces, regurgitated stomachs, detritus, etc.18. The quality of DNA from the original fixative was sufficient throughout the period observed by Martins et al.20, and also in tunicate metabarcoding after 3 and 6 months, where it became problematic only at 12 months of storage25, which is generally beyond practical consideration. Furthermore, DNA quality in samples fixed with > 90% ethanol should not suffer from the slight, approximately 5% decrease in the concentration of the original fixative observed during long-term storage26. One experiment even concluded that the success of bulk-sample metabarcoding was independent of storage time up to 1 year, regardless of cooling27. On the other hand, fixative as a source of DNA might be more sensitive to storage conditions24 than the bulk samples.
Despite the optimistic conclusions of the abovementioned fixative metabarcoding studies, they also point to weak detection of organisms with attributes such as small size or extensive sclerotization18,28, which was especially troublesome for terrestrial or marine samples29,30,31,32. Perhaps for this reason, the taxonomical spectrum recovered from Malaise-trap fixative DNA by Chimeno et al.33 did not reflect the differences in community composition among the examined habitats, which was only achieved through the corresponding bulk samples. It is therefore important to acknowledge recent advances in the scalability of bulk sample processing34, in nondestructive DNA extraction for terrestrial arthropods31,35, and in the specificity toward invertebrates when metabarcoding is applied to environmental DNA (eDNA) from water36. Nevertheless, the fixative-based approach could still be perceived as a promising, voucher-preserving complement or compromise between bulk and water, at least for freshwater fauna.
This study was established within a broader, long-term assessment of fauna inhabiting the Tatra Mountain lakes (Slovakia, C Europe). Their lake water composition varies in wide ranges, mainly reflecting the soil and vegetation cover of their catchments4,37. This natural vegetational gradient from forested catchments, through different proportions of alpine meadows and scree fields to bare rock areas, provides a pronounced gradient of nutrient availability in individual lakes in a small region sharing the same acidification history and climate development4. The resulting sharp decline in the concentrations of dissolved organic carbon (DOC), organic nitrogen (orgN), and total phosphorus (TP) in lake water with decreasing amount of catchment soils and vegetation thus implies possible between-lake differences in food web structures and fauna, detectable with DNA methods. These pristine ecosystems are also known for the occurrence of endangered species and glacial relicts38,39,40. Thus, we chose fixative DNA as a tool with the potential to screen bulk samples nondestructively, motivated by the prospects of simpler metabarcoding protocols and more efficient workflows for individual DNA barcoding via enhanced preselection of specimens. Predicting which taxa in which samples contain new genetic lineages could facilitate subsequent targeted barcoding of samples relevant for filling the gaps in reference databases and studying the population status of species in these sensitive and threatened but hardly accessible ecosystems.
Here, we focus on fixative DNA metabarcoding of benthic samples taken from 15 remote mountain lakes, testing the effects of storage duration and taxon’s ecological attributes on taxonomic richness and individual detection probabilities. We also address the aforementioned doubts about the applicability of fixative DNA by presenting how the community composition reflects lake environmental conditions. We hypothesize that (a) longer fixation time positively affects the outcomes of fixative DNA metabarcoding (14 or 28 days vs. 1 day of storage), (b) small-bodied or terrestrial organisms are detected less likely, and (c) contrasting characteristics of the lakes and their catchments are manifested by dissimilarities in the community composition of the fauna determined from the fixative DNA.
Methods
Study area
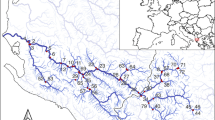
The Tatra Mountains are the highest alpine system within the Carpathian chain (up to 2655 m a.s.l.), located on the Slovak-Polish border and Baltic-Black Sea watershed (Fig. 1). Up to 260 lakes and ponds of glacial origin (locally called “pleso” in Slovak or “staw” in Polish) have been reported, predominantly from the alpine zone above 1800 m a.s.l., more than half of them being seasonal or small water bodies (< 0.01 ha)37,41. Their physicochemical characteristics and response to acidic deposition have been well documented in recent decades4,37,42. For the present study, we chose a subset of 15 lakes along a wide vegetation gradient from forest to rocky alpine catchments, including two lakes recovering from strong acidification and two dystrophic lakes (Fig. 1, Table 1, Supplementary Table S1). The bedrock in the catchment area of these lakes consists mainly of granodiorite in the central massif, and also contains gneiss and mica schist in the western part (lakes RO01, RO03, JM01 and JM06, Fig. 1). The soils are acidic and are dominated by humus-rich podzols in the forest zone, by shallow, undeveloped podzols, leptosols and regosols in the alpine meadows, and by sparse till soils beneath the surface stones in the scree fields37. This zonation is generally reflected by the substrate types in lake littoral (gravel, sand, organic sediment or even forest soil and vegetation).
Location of the 15 sampled lakes in the Tatra Mountains, with a broader European background (bottom left, the Carpathian chain highlighted). Made with QGIS 2.18 qgis.org and modified from SANtosito (Wikimedia Commons, https://commons.wikimedia.org/wiki/File:Tatra_Mountains_location_map.svg, accessed 4th Dec 2023, distributed under CC BY-SA 4.0 https://creativecommons.org/licenses/by-sa/4.0/ and the same license applies to Figure 1).
Sampling and DNA extraction
Samples of littoral benthos were taken from the 15 lakes in September 2020 by the kicking technique43 using nets with 0.3 mm mesh size. All accessible types of littoral habitats and substrates were sampled without any intention to normalize their variability, aiming for 10 min of sampling effort. On the site, the sampled material was decanted, transferred into a 1 L bottle, and fixed with 96% ethanol (aiming for a constant substrate:fixative ratio, filling to twice the substrate’s height in the bottle). The bottles were cooled to − 5 °C after returning to the field station and subsequently stored at − 20 °C. Duplicates of 35 mL of the fixative were taken from each of the 15 benthic samples upon thorough shaking and settling on days 1, 14, and 28 after visiting the respective lakes. These 90 subsamples were stored at − 20 °C until further processing, where another level of replication led to the final amount of 180 samples.
After shaking and settling, 1 mL of the subsamples was preserved for gas chromatography to assess the ethanol concentration (Supplementary Methods). Subsequently, 30 mL of the fixative was pressed through 0.45 μm filters (Whatman ME25, 25 mm diameter), which were immediately torn into small pieces and transferred into 1.5 mL tubes for drying overnight at room temperature. DNA extraction was performed the next day according to the protocol of Leese et al.36, which includes lysis with Proteinase K and TNES buffer, protein precipitation with NaCl, and DNA precipitation with ethanol. Concentration of DNA in the 90 extracts was measured fluorometrically (QuantusTM Fluorometer, QuantiFluor® ONE dsDNA System, Promega).
Library preparation and sequencing
A 418 bp fragment of the mitochondrial cytochrome c oxidase subunit 1 (COI) gene was targeted in a two-step PCR (polymerase chain reaction)44. In PCR1, the fragment was amplified from the DNA extracts (1 μL) using BF3/BR2 primers45 (final concentration 0.4 μM each), AccuStart™ II PCR ToughMix, and GelTrack Loading Dye (1×, Quantabio), filled to a total volume of 10 μL with nuclease-free water (Sigma-Aldrich). The thermal cycling program started at 94 °C for 5 min and proceeded with 25 cycles of 94 °C for 30 s, 50 °C for 30 s, 70 °C for 50 s, and a final elongation at 70 °C for 10 min. Similar conditions were used for PCR2, but the number of cycles was decreased to 18, and the elongation was adjusted to 65 °C for 2 min per cycle. Also, PCR1 products (1 μL) served as the DNA template for PCR2, and each well acquired a unique combination of tagged BF3/BR2 fusion primers44 instead of the basic primers.
All DNA extracts were diluted ten times before being added to PCR1, which overcame the PCR inhibition (common among the samples) without impairing the appearance of gel bands for non-inhibited samples. Protective clothing and face mask were worn for the pre-PCR laboratory steps, utilizing sterile equipment and strict decontamination measures in spaces dedicated to eDNA extraction and PCR preparation. PCRs were conducted in 96-well plates with two replicates per DNA extract. The remaining plate positions were evenly distributed and assigned to negative controls from PCR and from the extraction (30 mL of 96% ethanol processed along with other samples).
The PCR2 products were pooled equimolarly based on fluorometric quantification (QuantusTM Fluorometer, QuantiFluor® ONE dsDNA System, Promega) and 1% agarose gel electrophoresis (Gel Logic 212 Pro, MI software, Carestream) and purified using 0.8× SPRIselect beads (Beckman Coulter). The final 15 pM sequencing library included 10% PhiX and was analyzed on the Illumina MiSeq platform with Reagent Kit v3 (2 × 300 bp) at the Institute of Chemistry, Slovak Academy of Sciences.
Data processing
The raw sequencing reads were demultiplexed, and the primers were trimmed using Cutadapt v4.146 with the default filtering parameters. Pairs of reads were then merged by PEAR v0.9.1147 with a quality threshold of 26 and 5 bp minimum overlap to account for decreasing quality at the end of the 300 bp MiSeq reads. Vsearch v2.22.148 was utilized to keep only reads with a maximum of 2 expected errors, and to dereplicate and denoise the reads, setting minsize at 6 and alpha at 2, considering the data size. After length filtering (400–440 bp to capture invertebrate diversity), only sensible protein-coding sequences according to the invertebrate mitochondrial codon table were retained using metaMATE49. Subsequently, chimera removal, greedy clustering of OTUs (operational taxonomic units), and read mapping at a 97% identity threshold were performed48, followed by default filtering with LULU v0.1.050.
Taxonomy for the resulting OTUs was assigned by alignment against all records in the Barcode of Life Data System51 via BOLDigger v2.1.052 with the API correction option. Nontarget sequences were discarded by retaining only animal phyla and only OTUs with > 85% similarity to their best hit. Besides typical benthic macroinvertebrates, whose capture was favored by our experimental setup (kick-net mesh size, PCR primers), the resulting taxon list contained other groups as well. Thus, the remaining OTUs were further reviewed and labeled by basic ecological attributes to test whether these affect the availability of the taxon’s DNA in fixative. The labeling considered three groupings: the recorded animal taxa were first split into aquatic vs. terrestrial, and the aquatic fauna was further labeled as macrofauna vs. meiofauna, and planktonic vs. permanent benthic vs. temporal benthic (i.e., imagoes terrestrial but immature stages benthic, see Supplementary Table S2).
For each OTU occurrence, double the maximum number of that OTU’s reads found in negative controls was subtracted from each sample. Occurrences with read abundance less than 0.01% of their respective sample were removed along with those unconfirmed from any other replicate per lake. To prevent zero inflation, the long-format dataset was stripped of lines representing OTU occurrence in a lake where that OTU was absent from all replicates. Read proportions and OTU counts among the taxa were visualized for an overall portrayal of the dataset employing the R statistical software v4.3.153 with the Tidyverse package v2.0.054 and readxl v1.4.255 (see Supplementary Data). The sufficiency of read coverage was inspected from rarefaction curves utilizing the package vegan v2.6-456. The rarefaction was calculated before filtering by replicate consistency and abundance, because it would remove rare OTUs (leading to minor differences in outcomes: slightly flattened curves or earlier endpoints).
Statistical analysis
The effects of storage duration on the metabarcoding results (OTU richness, detection probability of individual taxa, proportions of nontarget reads) and on the concentrations of ethanol and DNA were tested using generalized linear mixed models (GLMMs) from the R package glmmTMB v1.1.757. The P values for the model coefficients were determined from Wald Z statistics (two-tailed tests), and the common significance (alpha) level of 0.05 was accepted for all tests in this work. To adjust the P values of factor variables with three levels, pairwise comparisons of the model effects were performed with Tukey post hoc tests in the package lsmeans v2.3.0-058. Diagnostic plots of data normality were checked, alongside zero inflation and dispersion, using the DHARMa package v0.4.659. Model outcomes were exported with the package modelsummary v1.4.160, effects were visualized using the packages lsmeans58 or effects v4.2-261,62,63, and the significance of each term in the initial model formula was tested (package car v3.1-261) to retain only relevant variables in the final models.
More specifically, OTU richness was regressed against subsampling day, ecological group, DNA concentration and read coverage. The latter two parameters were included to control for their potential effect via availability of sample’s DNA for PCR and sequencing, and their values were scaled by natural logarithm. Lake itself was set as the parameter for random effects in the GLMMs, considering that we collected one benthic sample per lake, which may thus embody substantial portion of variability in community composition, substrate, concentrations, and states of species’ DNA. Subsampling day and ecological group were the categorical variables of interest, possibly determining the composition of DNA in fixative. The groups neuston and nekton were data deficient and thus excluded. As for the model family, negative binomial distribution with quadratic parametrization was selected to remove the assumptions of fixed dispersion. Further, individual detection probabilities were regressed similarly but with binomial distribution because presence/absence served as the response variable.
In addition, effects on the concentrations of DNA and ethanol, and on the proportion of nontarget reads were modeled using Gaussian distribution. The models also considered the actual amount of substrate and the substrate: fixative volume ratio, to correct for volume approximations from the fieldwork. These two metrics were estimated post hoc from the height of the two phases settled in sample bottles after substrate homogenization (performed for a separate bulk DNA metabarcoding experiment, beyond the scope of the present article). Interactions between subsampling day and ecological group, which could reveal among-group differences in DNA availability across time, did not reach sufficient significance to be retained in the final models.
The contribution of subsampling day to the variation in community composition was determined by the PERMANOVA (permutational multivariate analysis of variance) in the adonis2 function from the package vegan56, using the Jaccard similarity index on presence/absence data, which is a common approach for metabarcoding data14,20. The significance of the sample parameters was assessed through the marginal effect in the ANOVA for multidimensional data, where the computed pseudo-F statistics with their P values were determined in the permutation test with a restricted permutation design (999 permutations in mirrored time series among the 12 “plots” of extraction replicates per lake “block” but none “within”)56.
To confirm whether the results of fixative DNA metabarcoding reflect lake environmental conditions, a similar assessment was performed for environmental parameters, retaining only the less correlated and more explanatory parameters (|R| ≤ 0.65, packages corrplot v0.9264 and Hmisc v5-1.065), with the permutation design adjusted for the lake level (999 free permutations among the 15 “plots” of lakes but none “within”). The PERMANOVA was supplemented by Principal Coordinates Analysis (PCoA) with a passive projection of the environmental and sample variables56. The significance of the relationships between individual variables and the first two ordination axes was evaluated through the permuted P value based on the permutation test for the coefficient of determination (R2, permutation design for the lake level or for the respective level of sample replication). Another PCoA for the dataset excluding terrestrial OTUs was produced to inspect the role of aquatic-only taxa in the assessment. Finally, the Mantel test56 and coinertia analysis were performed with the package ade4 v1.7-2266 to evaluate the similarity between the PCoA for lake water chemical composition and the PCoA for fixative DNA metabarcoding (occurrences merged by lake). In addition, similarity percentage analysis was performed to identify the characteristic OTUs differentiating the observed lake clusters56.
Results
Distribution of reads and taxa
Sequencing yielded 15,037,762 demultiplexed reads for the 180 samples, with an average of 83,543 per sample (± 43,817 SD), and the final filtered dataset contained 6,507,580 reads (mean 36,153 ± 20,016 SD per sample) distributed in 827 OTUs (mean 73 ± 37 SD, see Supplementary Data). As for phyla, the dataset consisted mostly of Arthropoda (50% reads, 78% OTUs) and Annelida (44% reads, 7% OTUs), while the order Diptera was exceptionally abundant both in terms of reads and OTUs (Fig. 2). Regarding the taxonomic resolution, 438 OTUs were identified to 406 different species. Further 137 OTUs were assigned only to the genus level, 104 to family, 144 to order, and the remaining 4 OTUs matched records without information on order or class. Nontarget sequences would comprise 32% reads (mean 31% ± 16 SD) and 88% OTUs (mean 74% ± 13 SD) if they were retained in the dataset, with a slightly lower read loss on day 1 (by 2.3% ± 0.8 SE, P = 0.007). Target OTUs were observed at the expected fragment length (418 bp), except for Rotifera at 421 bp, Tricladida at 415 bp, Thripidae and Hymenopteran Encyrtidae, Eulophidae, Mymaridae, Pteromalidae at 412 bp, Platygastridae at 409 bp, and only six OTUs deviated (three Diptera at 417 bp, three unidentified Coleoptera at 415 or 412 bp).
All 180 samples received enough read coverage to reach a long rarefaction plateau or at least a similar degree of richness as their plateaued replicates (Supplementary Fig. S1), despite several samples obtaining less than 20,000 reads. Only one PCR replicate of lake VS02 is questionable: an outlier with < 2000 reads remained at 25 OTUs as replicates from days 1 and 28 rather than climbing to ∼ 65 OTUs with other replicates from day 14. Nevertheless, comparable discrepancies were observed even among replicates with high read coverage.
Models of taxonomic richness and detection probability
The main regressions provided significant evidence for an increase in overall OTU richness on day 14 against day 1 (by 3.9 OTUs, P = 0.018) with a tendency against day 28 (+ 3.0 OTUs, P = 0.094, Fig. 3a, Supplementary Table S3), as well as for a “peak” in detection probability on day 14 against days 1 (by 3.2–3.9%, P < 0.001) and 28 (by 2.5–3.5%, P: 0.002–0.007, Fig. 3b, Supplementary Table S4) across all groupings. In general, larger portions of OTUs were recorded for the groups aquatic, macrofauna, and benthic fauna than for their counterparts (Fig. 3a). Moreover, these OTUs exhibited significantly elevated detection probabilities: by 5.8% for aquatic fauna (P < 0.001), by 17.3% for macroscopic fauna (P < 0.001), by 3.0% for temporal over permanent benthic fauna (P = 0.006) and by 12.1% for temporal benthic over planktonic fauna (P < 0.001, see Fig. 3b and Supplementary Table S4). The interactions between day and group were omitted in the final models because they did not substantially improve these models (Supplementary Tables S3 and S4), and the lowest P value obtained for an interaction was 0.099. Likewise negligible was the contribution of subsampling day to the explained variability in the case of OTU richness models that included the ecological classification of taxa, but the variable was retained for reference to the presence/absence models (Fig. 3). Final model diagnostics were satisfactory, although the OTU richness models exhibited some deviations in the residual plots (Supplementary Figs. S3 and S4).
Effects of subsampling day on the richness (a) and detection probability (b) of metazoan OTUs determined from fixative DNA metabarcoding. The taxa were grouped by their ecological characteristics as indicated at the top, the number of observations (n) is given within each plot, the red arrows are an estimate for the significance of differences (arrows overlap where P > 0.05), and the blue error bars represent 95% confidence intervals. See Supplementary Tables S3 and S4 for the exact values. B.Perm = permanent benthic fauna, B.Temp = temporal benthic fauna.
Read coverage and DNA concentration had a positive effect across all the main models (Supplementary Tables S3, S4, Supplementary Fig. S4). Conversely, the ethanol concentration and amount of substrate had no evident contribution to the OTU richness or presence/absence models and were thus omitted from the final models (Supplementary Tables S3, S4). In the additional models, the concentration of ethanol (86.8% ± 2.8 SD, Supplementary Table S5) exhibited minor differences among the subsampling days, where the decrease was evident only between days 1 and 28, by 0.6% (± 0.2 SE, P = 0.028, n = 54, Supplementary Fig. S4). Although the DNA concentration exhibited no substantial effect of subsampling day, it tended to increase with increasing ratios of substrate to fixative (2.7 ± 1.6 SE, log scale, P = 0.09, n = 90).
Community composition and environmental factors
The PERMANOVA further confirmed the contributions of subsampling day, read coverage, and DNA concentration to the variation in the detected fauna (P = 0.066, 0.001, and 0.023, respectively, Table 2a), but their R2 values were lower by two orders of magnitude than those for the lake term, which explained more than 68% of the variation. The dispersion test67 revealed no significant evidence of unequal dispersion in subsampling day (P = 0.635). Further attribution of the variation among lakes to individual environmental parameters exposed their extensive correlations (Supplementary Fig. S5). For the sake of validity, PERMANOVA was performed “by margin,” which fails when terms are correlated. Hence, only four representative environmental parameters were selected, and they explained 6–10% of the variation each, while the residual increased above 65% (Table 2b).
The PCoA diagrams displayed two main clusters corresponding to the 13 (sub)alpine lakes vs. two dystrophic forest lakes (Figs. 4 and 5). The clusters were separated along the main axis, which was correlated especially with the concentrations of orgN, DOC, and TP (Supplementary Table S6). The 13-lake cluster was depicted with NE01 as an outlier in all the plots and more homogeneously after excluding terrestrial fauna (Figs. 4 and 5, Supplementary Fig. S6). Moreover, the similarity of the chemical and metabarcoding datasets was confirmed by the Mantel statistic being higher than any of the simulated values (Fig. 5), both with and without the terrestrial fauna. The contour lines underlying the PCoA diagrams are only a smooth representation of the modeled altitude and were added for easier orientation, not to be interpreted strictly.
PCoA diagram showing the differences in lake community composition based on the OTU data of whole fauna determined from fixative DNA metabarcoding. The symbols refer to the fixation intervals, the ellipses mark the standard deviation from the lake centroids, and lake altitude was fitted by a generalized additive model.
Fit between the PCoA of the whole-fauna OTU dataset from fixative DNA metabarcoding (a, occurrences merged by lake) and lake water chemical composition (b), validated with the Mantel test (c) and visualized through coinertia in a common ordination plane (d, eigenvalues show the prominence of the two displayed axes). The orientation of the PCoA2 axis is flipped (a, b) to correspond with Fig. 4 and the coinertia plot (d).
The similarity percentage analysis highlighted that the most distinctive OTUs of the forest lakes (FU07 and FU08) were assigned to the daphniid Simocephalus vetulus, followed by the chironomid Psectrocladius sp., the mayfly Leptophlebia vespertina, Ceriodaphnia quadrangula, the newt Ichthyosaura alpestris, certain chaetonotid OTUs, etc. Another notable outcome is the absence of the clitellate OTUs assigned to Stylodrilus heringianus and Nais sp. In contrast, the Nais OTUs assigned to the species N. communis and N. pseudobtusa did occur in the forest lakes, while three species of the abovementioned Psectrocladius did not (P. oligosetus, P. platypus, P. oxyura). The outlier NE01 differed inter alia by a unique OTU within the annelid genus Tubifex.
Discussion
DNA metabarcoding of fixative ethanol from benthic samples has shown potential to facilitate voucher-preserving biodiversity assessment and localization of novel genetic lineages. The outcomes of our chronological experiment provided firm support for the hypothesis that storage duration affects metabarcoding results. However, the effect size was relatively low, so the results can be considered highly consistent across the first month of storage. For instance, if an OTU was detected in a lake, it was around 3% more likely to be found in the subsamples from day 14, which were also richer overall by approximately 3 OTUs. This is in line with the findings of Martins et al.20 that richness rises in the first two weeks of storage by up to 10 species, with signs of an optimum around day 7. Further storage until day 28 in our experiment was no longer favorable, as the estimates returned to the levels of day 1.
Secondarily, deducing any steeper slopes of this effect in the ecological groups with the largest estimates (meiofauna and plankton in the initial models) would lack statistical support from the interactions, even in the superior models of detection probability. With over 10,000 observations of presence/absence from individual OTUs, the detection probability models obtained excellent diagnostic plots and proved preferable for inspecting weak trends. In contrast, the OTU richness models with only 180 data points per group captured the effect of storage duration only in the overall model, where statistical power was not depleted by the grouping variable. Thus, for effects on richness to be evident in the present study design, species turnover has to be much more dramatic than what was anticipated for our DNA extracts that differed only by the storage duration of the material, sampled mostly from oligotrophic ecosystems. The subtle effects of storage duration are reflected even in the statistically significant yet minute contribution of subsampling day to explaining the variability in community composition (R2 = 0.3%, P = 0.049), which resembles the outcomes from Martins et al.20 (R2 = 0.5%, P = 0.0001; despite differences in formula and permutational design).
Further, the ethanol concentration decreased only marginally throughout the observed period and should stabilize once the remaining water-binding structures within the sample reach an equilibrium with the fixative. The observed 10% decrease from the stock ethanol concentration is slightly greater than that reported by Elbrecht et al.26 but poses no concern for DNA stability when starting with 96% stock ethanol. Hence, our data do not provide a rationale for the modeled decline in OTU richness and detectability after day 14. Interestingly, the effects for DNA concentration imply an opportunity for fine-tuning the metabarcoding outcomes by decreasing the volume of fixative added to the sample. For that purpose, ethanol could be added according to more precise sample measurements, e.g., of its weight. Still, the amounts we added proved sufficient for maintaining a high ethanol concentration. In addition, the comforting findings about the long-term stability of DNA in bulk samples27,68 fit our experience from an earlier season, where fixative DNA extracts were suitable for COI metabarcoding despite processing the fixative after two years since subsampling. Nonetheless, it is worth noting that storing our samples at − 20 °C might have played a key role in the extracts’ performance, as Zenker et al.24 attributed the poor quality of their fixative DNA to suboptimal storage conditions.
Depicting environmental gradients via fixative DNA from benthic samples appears viable, which contrasts the work of Chimeno et al.33 with terrestrial fauna from Malaise traps. We obtained an impressive match between the results from metabarcoding and chemical parameters. We also confirmed that the fauna of the two dystrophic forest lakes clearly diverged from the remaining 13 sites, whose substrate contained mainly gravel, sand, and fine organic sediment, in contrast to vegetation and soil in the forest lakes. Moreover, the gap is relatable to orgN, DOC, and TP, which quantify the amount of organic matter accumulated in watersheds (soil, vegetation), especially in the forest zone37. Furthermore, even pH stood out in the PERMANOVA, presumably because it represented the large group of correlated variables. Likewise, the importance of altitude and lake size for community composition probably comes from their confounding link to other environmental parameters, such as vegetation zone, food availability, temperature, precipitation, ice cover, migration barriers, etc.69,70.
Although our goal for the environmental analyses was limited to simply presenting their feasibility via fixative DNA, deeper relationships among the lakes can already be outlined from the obtained outcomes. Both whole-fauna and aquatic-only ordination diagrams agree on the unique placement of the Vyšné Terianske pleso (NE01), which indeed has specific attributes. Lying at a high altitude with a relatively small hydrological catchment rich in soils, this lake has unusually high levels of DOC, orgN, TP, primary production (chlorophyll a, turbid water), abundance of organic sediment, and slow recovery from acidification4,37,71. On the other hand, signs of the strong acidification history are less pronounced in the large lake FU10 (Vyšné Wahlengergovo pleso), whose catchment contains eroding taluses (rather than acid-rich soils) that act as a source of Ca2+ and alkalinity, fostering the lake’s already neutral pH4.
Particularly noteworthy is the position of lakes TE01, NE02, and FU10, whose morphology is so similar that they were chosen as promising candidates for studies about altitudinal gradients72. Hence, it is remarkable that all the PCoA plots place these three sites on a straight line, ordered by altitude, even despite the major redistribution caused by the exclusion of terrestrial fauna. Once terrestrial fauna was excluded or the occurrences were merged per lake, the distance of the Western Tatras became better highlighted (Supplementary Fig. S6, Fig. 5a). In addition, the two subalpine Temnosmrečinské lakes were illustrated further from their alpine neighbors when considering the aquatic-only, chemical or merged datasets (Supplementary Fig. S6, Fig. 5) compared to the whole-fauna PCoA (Fig. 4). This difference might reflect the resistance of TE01 and TE02 against acidification, which arises from their dominant Ca2+ concentrations and high alkalinity37,73. These observations match the assumed isolation and slow recovery of alpine populations of aquatic invertebrates owing to orographic barriers and limited migration potential5,6,71, as well as the possibility of sharing airborne DNA from terrestrial taxa in space and time74,75.
The quick, nondestructive analysis applied here could be followed by targeted barcoding of candidate vouchers from designated bulk samples. This enhancement would promote more efficient filling of gaps in reference barcode databases, especially for cases like benthic macroinvertebrates, as discussed below. Exhaustive haplotype analyses are beyond the scope of this article, but a brief review of the taxonomic assignments was sufficient to identify promising barcoding targets. For instance, the clitellate Tubifex in the lake NE01 formed a unique OTU, which did not match any BOLD record above the 98% similarity threshold. The genus name was assigned based solely on private records, as the closest published record reached only 91.3% similarity. Another candidate is the trichopteran in TE01 that could be assigned only to the family level (Limnephilidae) because it matched private records with incomplete taxonomy. For similar reasons, species information was missing for another limnephilid OTU of genus Acrophylax and a plecopteran Protonemura.
Further, the chironomid Psectrocladius sp. in the forest lakes FU07 and FU08 closely matched three BOLD records from two clusters (BINs), but the next reference was only 91% similar. Although the three records provide insufficient evidence for expanding the list of glacial relicts in the High Tatras38,39,40, they suggest the Arctic distribution of this Psectrocladius OTU: one record is from the Canadian shore of Lake Superior76, another from Norwegian Russevatn near Svanvik77, and the third was private but it also seemed to originate in Scandinavia. In addition, the identification of the clitellate Peipsidrilus saamicus, unusual for the region yet detected here with high consistency at six sites, was based solely on private matches at 99.8 or 86.7% similarity, all sharing the same taxonomy. When bulk or water-eDNA metabarcoding yields such incomplete identifications, the solution is another sampling with no guarantee of capturing the target individuals. On the other hand, nondestructive metabarcoding keeps the vouchers ready for any expert willing to accept the challenge of species identification and resolving the taxonomic questions arising from genetic diversity.
In contrast to the proposed targeted barcoding, “reverse” barcoding of numerous unknown specimens with little preselection has recently become an affordable way to efficiently gather barcodes from “dark taxa”78. However, prescreening and targeted barcoding might be more desirable when searching for a needle in the haystack of, e.g., benthic macroinvertebrates from Central Europe, where unique lineages are expected79 but at a lower rate than in unexplored regions or taxa. Moreover, feeding BOLD with identical yet poorly identified specimens has negative side effects. For example, the unique OTU of Tubifex was reidentified later and the few informative records already became hidden by 17 perfect matches to Clitellata incertae sedis (Supplementary Table S7). Only after another swarm of anonymous hits came the first published record (as the 68th hit), thus unavailable to BOLDigger, which relied merely on the top 20 hits (most recently up to 100 hits) because of problematic access to the BOLD ID engine51,52. Therefore, we propose the targeted barcoding upon fixative screening to prevent such cases when large numbers of identical references waste resources, overwhelm the database and complicate taxonomic assignment for later metabarcoding studies.
It is still a matter of discussion whether fixative-based analyses suit freshwater fauna better than terrestrial or marine fauna17,18,19,20,21,28,29,30,31,32,33. Higher detectability of soft-bodied families over heavily sclerotized families has been reported by Martins et al.28, in line with Carew et al.68, Zizka et al.18. Our observation of disproportionately high read abundance for the relatively few OTUs of soft-bodied annelids also supports this trend (Fig. 2). Until now, suitability of fixative DNA for meiofauna has not been thoroughly explored, as nondestructive benefits of such analysis fade when minuscule specimens become lost in extraction. Here, we classified OTUs by size and habitat, revealing higher detection probability of OTUs from macrofauna over OTUs from meiofauna (by 17%), aquatic over terrestrial fauna (by 6%), and benthic over planktonic fauna (by ~ 10%). The slightly elevated probabilities for temporal over permanent benthic fauna (by 3%) might be a consequence of the large abundance or body mass of the specimens (chironomids, caddisflies, etc.).
Benthic macroinvertebrates should innately comprise the majority of animal DNA in our benthic samples and thus be detected consistently among respective sample replicates. Therefore, the OTUs of meiofauna that we captured were likely just a fraction of the group’s true diversity present across the ecosystems80,81. Any work aiming to extensively detect all the faunal components would require a more complex sampling strategy, e.g., combining kick nets, plankton nets, surface sediment and Malaise traps30,33,80,81. Previous validation of the BF3/BR2 primers even for terrestrial arthropods45 speaks in favor of our model reliability, but the detection of other taxa such as nematodes would benefit from specifically optimized primers81. Even then, gaps in reference databases may hinder their taxonomic assignment80. Indeed, we have identified only three OTUs of nematodes, only to order level, and it was not sufficient to classify them in our grouping system. Our models of detection probability are only partially resistant against these biases, so the estimated 17% higher consistency for aquatic macroinvertebrate OTUs over meiofauna should be treated with caution.
Terrestrial arthropods, on the contrary, were observed in remarkably high numbers in our kick nets (specimens trapped in water, fragments in substrate). Their relatively high detection probabilities were thus in line with expectations, considering the obtained low OTU richness (Fig. 3). Still, if we detected a terrestrial OTU in a lake, on average it appeared less consistently across that lake’s replicates than did aquatic OTUs (by the abovementioned 6%). These results add to the prior knowledge about the role of body size and sclerotization in nondestructive DNA metabarcoding of benthic macroinvertebrates18,28,68, and will be further elaborated by comparison with several other DNA sources sampled in parallel.
Conclusion
Fixative DNA metabarcoding was proven to be a feasible method for community analysis of benthic samples from alpine lakes without the need for laborious and destructive sample processing. The fit between the dissimilarity in the composition of fauna and the in-lake concentrations of DOC, orgN, and TP suggested that catchment characteristics predetermined the composition of aquatic biota. In addition, the analysis served as a preliminary voucher-preserving screening of genetic lineages captured in precious samples from remote sites, and it was able to locate specimens relevant for targeted filling of gaps in barcode reference libraries. We have also confirmed and extended the previous findings that metabarcoding results are consistent throughout the first weeks of storage and that subsampling of the original storage ethanol is slightly more beneficial after 1–2 weeks rather than after a day or a month of fixation, if compatible with the requirements of particular studies. Furthermore, considerate retaining of the original fixative can be preferred over its wasteful replacement, provided that excess water is removed from the benthic samples before fixation. Likewise, future applications may profit from limiting the initial amount of concentrated fixative to increase the DNA concentration. Subsequent proper storage conditions or even adding more fixative upon the subsampling step should ensure the preservation of DNA. Moreover, our results emphasize that subtle trends are easily overlooked when simply modeling gross whole-sample metrics. Where appropriate, ordination analyses with proper permutational design can be supplemented by models of species-level responses in order to utilize the robustness of ecological datasets more efficiently.
Data availability
The demultiplexed FASTQ files from the MiSeq analysis have been uploaded to the NCBI Sequence Read Archive (Project ID PRJNA1070890). The data generated or analyzed during this study are included in this published article and its Supplementary Information files, which also include the R script for the analysis.
References
Birk, S. et al. Three hundred ways to assess Europe’s surface waters: An almost complete overview of biological methods to implement the Water Framework Directive. Ecol. Indic. 18, 31–41 (2012).
Sánchez-Bayo, F. & Wyckhuys, K. A. G. Worldwide decline of the entomofauna: A review of its drivers. Biol. Conserv. 232, 8–27 (2019).
Fjellheim, A. et al. Diversity and distribution patterns of benthic invertebrates along alpine gradients. A study of remote European freshwater lakes. Adv. Limnol. 62, 167–190 (2009).
Kopáček, J. et al. Climate change accelerates recovery of the Tatra Mountain lakes from acidification and increases their nutrient and chlorophyll a concentrations. Aquat. Sci. 81, 70 (2019).
Čiamporová-Zaťovičová, Z. & Čiampor, F. Alpine lakes and ponds – a promising source of high genetic diversity in metapopulations of aquatic insects. Inland Waters 7, 109–117 (2017).
Bitušík, P., Svitok, M., Novikmec, M., Trnková, K. & Hamerlík, L. Biological recovery of acidified alpine lakes may be delayed by the dispersal limitation of aquatic insect adults. Hydrobiologia 790, 287–298 (2017).
Hebert, P. D. N., Cywinska, A., Ball, S. L. & DeWaard, J. R. Biological identifications through DNA barcodes. Proc. R. Soc. B: Biol. Sci. 270, 313–321 (2003).
Deiner, K. et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 26, 5872–5895 (2017).
Leese, F. et al. Chapter two—why we need sustainable networks bridging countries, disciplines, cultures and generations for aquatic biomonitoring 2.0: A perspective derived from the DNAqua-Net COST action. In Adv. Ecol. Res. Vol. 58 (eds. David A. B. et al.) 63–99 (Academic Press, 2018).
Pereira-da-Conceicoa, L. et al. Metabarcoding unsorted kick-samples facilitates macroinvertebrate-based biomonitoring with increased taxonomic resolution, while outperforming environmental DNA. Environ. DNA 3, 353–371 (2021).
Weigand, H. et al. DNA barcode reference libraries for the monitoring of aquatic biota in Europe: Gap-analysis and recommendations for future work. Sci. Total Environ. 678, 499–524 (2019).
Zizka, V. M. A., Geiger, M. F. & Leese, F. DNA metabarcoding of stream invertebrates reveals spatio-temporal variation but consistent status class assessments in a natural and urban river. Ecol. Indic. 115, 106383 (2020).
Beermann, A. J., Werner, M. T., Elbrecht, V., Zizka, V. M. A. & Leese, F. DNA metabarcoding improves the detection of multiple stressor responses of stream invertebrates to increased salinity, fine sediment deposition and reduced flow velocity. Sci. Total Environ. 750, 141969 (2021).
Buchner, D. et al. Analysis of 13,312 benthic invertebrate samples from German streams reveals minor deviations in ecological status class between abundance and presence/absence data. PLOS ONE 14, e0226547 (2019).
Cordier, T. et al. Supervised machine learning outperforms taxonomy-based environmental DNA metabarcoding applied to biomonitoring. Mol. Ecol. Resour. 18, 1381–1391 (2018).
Elbrecht, V., Peinert, B. & Leese, F. Sorting things out: Assessing effects of unequal specimen biomass on DNA metabarcoding. Ecol. Evol. 7, 6918–6926 (2017).
Hajibabaei, M., Spall, J. L., Shokralla, S. & van Konynenburg, S. Assessing biodiversity of a freshwater benthic macroinvertebrate community through non-destructive environmental barcoding of DNA from preservative ethanol. BMC Ecol. 12, 28 (2012).
Zizka, V. M. A., Leese, F., Peinert, B. & Geiger, M. F. DNA metabarcoding from sample fixative as a quick and voucher-preserving biodiversity assessment method. Genome 62, 122–136 (2018).
Erdozain, M. et al. Metabarcoding of storage ethanol vs. conventional morphometric identification in relation to the use of stream macroinvertebrates as ecological indicators in forest management. Ecol. Indic. 101, 173–184 (2019).
Martins, F. M. S. et al. Have the cake and eat it: Optimizing nondestructive DNA metabarcoding of macroinvertebrate samples for freshwater biomonitoring. Mol. Ecol. Resour. 19, 863–876 (2019).
Wang, Y. et al. Environmental DNA of preservative ethanol performed better than water samples in detecting macroinvertebrate diversity using metabarcoding. Divers. Distrib. 27, 1989–2002 (2021).
Meissner, K., Aroviita, J., Baattrup-Pedersen, A. & Buchner, D. Metabarcoding for Use in Nordic Routine Aquatic Biomonitoring: A Validation Study (Nordic Council of Ministers, 2021).
Majaneva, M., Diserud, O. H., Eagle, S. H., Hajibabaei, M. & Ekrem, T. Choice of DNA extraction method affects DNA metabarcoding of unsorted invertebrate bulk samples. Metabarcoding Metagenom. 2, e26664 (2018).
Zenker, M. M., Specht, A. & Fonseca, V. G. Assessing insect biodiversity with automatic light traps in Brazil: Pearls and pitfalls of metabarcoding samples in preservative ethanol. Ecol. Evol. 10, 2352–2366 (2020).
Couton, M. et al. High-throughput sequencing on preservative ethanol is effective at jointly examining infraspecific and taxonomic diversity, although bioinformatics pipelines do not perform equally. Ecol. Evol. 11, 5533–5546 (2021).
Elbrecht, V., Vamos, E. E., Meissner, K., Aroviita, J. & Leese, F. Assessing strengths and weaknesses of DNA metabarcoding-based macroinvertebrate identification for routine stream monitoring. Methods Ecol. Evol. 8, 1265–1275 (2017).
Nichols, S. J. et al. Towards routine DNA metabarcoding of macroinvertebrates using bulk samples for freshwater bioassessment: Effects of debris and storage conditions on the recovery of target taxa. Freshw. Biol. 65, 607–620 (2020).
Martins, F. M. S. et al. Modelling technical and biological biases in macroinvertebrate community assessment from bulk preservative using multiple metabarcoding markers. Mol. Ecol. 30, 3221–3238 (2021).
Derycke, S. et al. Detection of macrobenthos species with metabarcoding is consistent in bulk DNA but dependent on body size and sclerotization in eDNA from the ethanol preservative. Front. Mar. Sci. 8, 859 (2021).
Marquina, D., Esparza-Salas, R., Roslin, T. & Ronquist, F. Establishing arthropod community composition using metabarcoding: Surprising inconsistencies between soil samples and preservative ethanol and homogenate from Malaise trap catches. Mol. Ecol. Resour. 19, 1516–1530 (2019).
Kirse, A., Bourlat, S. J., Langen, K., Zapke, B. & Zizka, V. M. A. Comparison of destructive and nondestructive DNA extraction methods for the metabarcoding of arthropod bulk samples. Mol. Ecol. Resour. 23, 92–105 (2023).
Persaud, S. F., Cottenie, K. & Gleason, J. E. Ethanol eDNA reveals unique community composition of aquatic macroinvertebrates compared to bulk tissue metabarcoding in a biomonitoring sampling scheme. Diversity 13, 34 (2021).
Chimeno, C. et al. Depicting environmental gradients from Malaise trap samples: Is ethanol-based DNA metabarcoding enough?. Insect Conserv. Divers. 16, 47–64 (2023).
Buchner, D., Haase, P. & Leese, F. Wet grinding of invertebrate bulk samples—a scalable and cost-efficient protocol for metabarcoding and metagenomics. Metabarcoding Metagenom. 5, e67533 (2021).
Batovska, J., Piper, A. M., Valenzuela, I., Cunningham, J. P. & Blacket, M. J. Developing a non-destructive metabarcoding protocol for detection of pest insects in bulk trap catches. Sci. Rep. 11, 7946 (2021).
Leese, F. et al. Improved freshwater macroinvertebrate detection from environmental DNA through minimized nontarget amplification. Environ. DNA 3, 261–276 (2021).
Kopáček, J. et al. Catchment biogeochemistry modifies long-term effects of acidic deposition on chemistry of mountain lakes. Biogeochemistry 125, 315–335 (2015).
Bitušík, P., Dobríková, D., Pıpík, R. & Hamerlík, L. Relict chironomid communities surviving in the coldest High Tatra Mountain lakes confirmed by a palaeolimnological survey. Biologia 72, 965–969 (2017).
Hamerlík, L., Thomková, K. & Bitušík, P. First record of Rheocricotopus (s. str.) reduncus Sæther & Schnell, 1988 (Diptera: Chironomidae) from Slovakia: A new glacial relict found in the Tatra Mountains. Lauterbornia 79, 58–60 (2015).
Lindholm, M., d’Auriac, M. A., Thaulow, J. & Hobæk, A. Dancing around the pole: holarctic phylogeography of the Arctic fairy shrimp Branchinecta paludosa (Anostraca, Branchiopoda). Hydrobiologia 772, 189–205 (2016).
Gregor, V. & Pacl, J. Hydrology of the Tatra mountain lakes. Acta Hydrol. Slovaca 6, 161–187 (2005).
Kopáček, J., Stuchlík, E. & Hardekopf, D. Chemical composition of the Tatra Mountain lakes: Recovery from acidification. Biologia 61, 21–33 (2006).
Frost, S. A., Huni, A. & Kershaw, W. Evaluation of kicking technique for sampling stream bottom fauna. Can. J. Zool. 49, 167–173 (1971).
Elbrecht, V. & Steinke, D. Scaling up DNA metabarcoding for freshwater macrozoobenthos monitoring. Freshw. Biol. 64, 380–387 (2019).
Elbrecht, V. et al. Validation of COI metabarcoding primers for terrestrial arthropods. PeerJ 7, e7745 (2019).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Zhang, J., Kobert, K., Flouri, T. & Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620 (2013).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 4, e2584 (2016).
Andújar, C. et al. Validated removal of nuclear pseudogenes and sequencing artefacts from mitochondrial metabarcode data. Mol. Ecol. Resour. 21, 1772–1787 (2021).
Frøslev, T. G. et al. Algorithm for post-clustering curation of DNA amplicon data yields reliable biodiversity estimates. Nat. Commun. 8, 1188 (2017).
Ratnasingham, S. & Hebert, P. D. N. BOLD: The barcode of life data system. Mol. Ecol. Notes 7, 355–364 (2007).
Buchner, D. & Leese, F. BOLDigger—a Python package to identify and organise sequences with the Barcode of Life Data systems. Metabarcoding Metagenom. 4, e53535 (2020).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org/ (2014).
Wickham, H. et al. Welcome to the tidyverse. J. Open Sourc. Softw. 4, 1686 (2019).
Wickham, H. & Bryan, J. readxl: Read Excel Files. version 1.4.2. https://rdrr.io/cran/readxl/ (2023).
Oksanen, J. et al. vegan: Community Ecology Package. R package version 2.6-4. https://rdrr.io/cran/vegan/ (2022).
Brooks, M. E. et al. glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. R J. 9, 378–400 (2017).
Lenth, R. V. Least-squares means: The R package lsmeans. J. Stat. Softw. 69, 1–33 (2016).
Hartig, F. DHARMa: Residual Diagnostics for Hierarchical (Multi-Level/Mixed) Regression Models. R package version 0.4.6. https://rdrr.io/cran/DHARMa/ (2022).
Arel-Bundock, V. modelsummary: Data and model summaries in R. J. Stat. Softw. 103, 1–23 (2022).
Fox, J. & Weisberg, S. An R Companion to Applied Regression (Sage, 2019).
Fox, J. & Weisberg, S. Visualizing fit and lack of fit in complex regression models with predictor effect plots and partial residuals. J. Stat. Softw. 87, 1–27 (2018).
Fox, J. Effect displays in R for generalised linear models. J. Stat. Softw. 8, 1–27 (2003).
Wei, T. & Simko, V. R package 'corrplot': Visualization of a Correlation Matrix. R package version 0.92. https://rdrr.io/cran/corrplot/ (2021).
Harrell Jr, F. E. Hmisc: Harrell Miscellaneous. R package version 5.1-0. https://rdrr.io/cran/Hmisc/ (2023).
Dray, S. & Dufour, A. B. The ade4 package: Implementing the duality diagram for ecologists. J. Stat. Softw. 22, 1–20 (2007).
Anderson, M. J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46 (2001).
Carew, M. E., Coleman, R. A. & Hoffmann, A. A. Can non-destructive DNA extraction of bulk invertebrate samples be used for metabarcoding?. PeerJ 6, e4980 (2018).
Catalan, J. et al. Ecological thresholds in European alpine lakes. Freshw. Biol. 54, 2494–2517 (2009).
Karlsson, J., Jonsson, A. & Jansson, M. Productivity of high-latitude lakes: Climate effect inferred from altitude gradient. Glob. Chang. Biol. 11, 710–715 (2005).
Stuchlík, E. et al. Complexity in the Biological recovery of tatra mountain lakes from acidification. Water Air Soil Pollut. 228, 5 (2017).
Čiamporová-Zaťovičová, Z., Hamerlík, L., Šporka, F. & Bitušík, P. Littoral benthic macroinvertebrates of alpine lakes (Tatra Mts) along an altitudinal gradient: A basis for climate change assessment. Hydrobiologia 648, 19–34 (2010).
Krno, I. et al. Littoral benthic macroinvertebrates of mountain lakes in the Tatra Mountains (Slovakia, Poland). Biologia 61, 147–166 (2006).
Klepke, M. J., Sigsgaard, E. E., Jensen, M. R., Olsen, K. & Thomsen, P. F. Accumulation and diversity of airborne, eukaryotic environmental DNA. Environ. DNA 4, 1323–1339 (2022).
Roger, F. et al. Airborne environmental DNA metabarcoding for the monitoring of terrestrial insects—a proof of concept from the field. Environ. DNA 4, 790–807 (2022).
Hebert, P. D. N. et al. Counting animal species with DNA barcodes: Canadian insects. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150333 (2016).
Ekrem, T., Stur, E., Orton, M. G. & Adamowicz, S. J. DNA barcode data reveal biogeographic trends in Arctic non-biting midges. Genome 61, 787–796 (2018).
Srivathsan, A. et al. ONTbarcoder and MinION barcodes aid biodiversity discovery and identification by everyone, for everyone. BMC Biol. 19, 217 (2021).
Bozáňová, J., Čiamporová Zatovičová, Z., Čiampor-Jr, F., Mamos, T. & Grabowski, M. The tale of springs and streams: How different aquatic ecosystems impacted the mtDNA population structure of two riffle beetles in the Western Carpathians. PeerJ 8, e10039 (2020).
Macher, J.-N. et al. Enhancing metabarcoding efficiency and ecological insights through integrated taxonomy and DNA reference barcoding: A case study on beach meiofauna. BioRxiv 2024, 587521 (2024).
Pawlowski, J. et al. Assessing the potential of nematode metabarcoding for benthic monitoring of offshore oil platforms. Sci. Total Env. 933, 173092 (2024).
Acknowledgements
This work was financially supported by the Slovak National Grant Agency (VEGA), project no. 2/0084/21. J.K.’s chemical analyses were supported by the Czech Science Foundation, project no. P503-23-06379S. We would like to thank Dr. Róbert Kubinec and Dr. Mária Šedivá for facilitating the ethanol concentration measurements and DNA sequencing, respectively.
Author information
Authors and Affiliations
Contributions
F.Č., Z.Č.-Z., and O.V. conceived the experiment and conducted the fieldwork. O.V. performed the laboratory work, analyzed the results, and prepared the manuscript. P.M. and K.T. took part in the sampling and processing, P.B. provided advice on the statistics, and J.K. contributed with the background on lake environmental conditions. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Vargovčík, O., Čiamporová-Zaťovičová, Z., Beracko, P. et al. Environmental gradients and optimal fixation time revealed with DNA metabarcoding of benthic sample fixative. Sci Rep 14, 18396 (2024). https://doi.org/10.1038/s41598-024-68939-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-68939-x
- Springer Nature Limited