Abstract
Sex determination (SD) involves mechanisms that determine whether an individual will develop into a male, female, or in rare cases, hermaphrodite. Crustaceans harbor extremely diverse SD systems, including hermaphroditism, environmental sex determination (ESD), genetic sex determination (GSD), and cytoplasmic sex determination (e.g., Wolbachia controlled SD systems). Such diversity lays the groundwork for researching the evolution of SD in crustaceans, i.e., transitions among different SD systems. However, most previous research has focused on understanding the mechanism of SD within a single lineage or species, overlooking the transition across different SD systems. To help bridge this gap, we summarize the understanding of SD in various clades of crustaceans, and discuss how different SD systems might evolve from one another. Furthermore, we review the genetic basis for transitions between different SD systems (i.e., Dmrt genes) and propose the microcrustacean Daphnia (clade Branchiopoda) as a model to study the transition from ESD to GSD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sex determination (SD) systems designate whether an individual will develop into a male, female, or in rare cases hermaphrodite. One potential advantage of having separate sexes (male and female) is being able to bring beneficial alleles from both individuals together when sexual reproduction occurs (McDonald et al. 2016; Muller 1932; Otto 2009). Of the two main types of SD systems, i.e., genetic sex determination (GSD) and environmental sex determination (ESD), the former is controlled by genetic factors as expansive as sex chromosomes, whereas the latter is driven by environmental cues (e.g., temperature and photoperiod). GSD can be further divided into XY and ZW systems depending on whether the males or females are heterogametic. XY systems are found in many species, such as humans, Drosophila, and C. elegans, in which the Y chromosome is extremely degenerate or absent (Bachtrog 2013; Blackmon et al. 2017). Contrary to the XY system, under the ZW system, females are heterogametic (ZW) and males are homogametic (ZZ). The ZW system is utilized by birds (Stevens 1997), reptiles (Ezaz et al. 2009), insects (Blackmon et al. 2017), and many crustaceans (Cui et al. 2015; Jiang and Qiu 2013; Parnes et al. 2003).
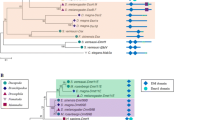
Crustaceans originated ~ 500 million years ago during the Precambrian period (Zhang et al. 2007), and now include ~ 67,000 species, such as crabs, lobsters, crayfish, shrimps, and water fleas. Although most crustaceans are aquatic and free-living, some are terrestrial (e.g., woodlice) or parasitic (e.g., fish lice). Crustaceans are phylogenetically close to insects, and together form the Pancrustacean clade (Budd and Telford 2009) (Fig. 1). Extant crustacean lineages can be classified into four primary clades: Branchiopoda (e.g., water fleas; and clams, fairy, and tadpole shrimps), Maxillopoda (e.g., barnacles and copepods), Malacostraca (e.g., shrimps, crabs, lobsters, and crayfish), and Ostracoda (e.g., sea firefly) (Fig. 1), with the number of species ranging from 800 in Branchiopoda to 25,000 in Malacostraca (Schwentner et al. 2017). Also, crustaceans contain two minor clades, Remipedia and Cephalocarida (e.g., horseshoe shrimp), each of which contains 12–17 species (Schwentner et al. 2017).
Phylogeny of extant crustaceans and modes of sex determination for each clade. The original phylogenetic tree was generated by Schwentner et al. (2017) based on the PhyloBayes analysis of 1077 decisive orthogroups and 301,748 amino acid positions with the Site-Heterogeneous CAT-GTR Model. We modified the tree to show only the major Pancrustacean clades. ESD environmental sex determination, GSD genetic sex determination; cytoplasmic (e.g., Wolbachia controlled SD system)
Crustaceans have diversified SD systems in each clade (Bauer 2000; Becking et al. 2017; Chandler et al. 2018; Hessler et al. 1995; Toyota et al. 2021; Yager 1991). In Malacostraca and Ostracoda, sex is almost exclusively controlled by genetic factors (Becking et al. 2017; Fang et al. 2020; Parnes et al. 2003), whereas sex determination in Maxillopoda and Branchiopoda is largely by environmental factors (Blaxter et al. 1998; Michaud et al. 2004; Toyota et al. 2015a, b). The two most primitive groups, Cephalocarida and Remipedia, reproduce exclusively by simultaneous hermaphroditism (Hessler et al. 1995; Yager 1991).
These highly variable SD systems provide a basis for studying how SD evolves in the diversified crustacean taxa. Moreover, some crustaceans (e.g., Lysmat) have unique sexual systems, such as protandric simultaneous hermaphroditism (Bauer 2000), which is important for the study of sex-allocation theory. In addition, transitions from ESD to GSD have occurred in lineages such as Daphnia (clade Branchiopoda), providing a valuable model for studying the rapid transition between system types. In this review, we survey our understanding of the SD systems in crustaceans and further discuss their evolutionary paths.
Sex determination systems in crustaceans
In the largest clade of crustaceans, Malacostraca (> 40,000 species), both ZW and XY SD systems are present. The ZW system is found in most shrimps and crayfishes (Jiang and Qiu 2013; Parnes et al. 2003), whereas the XY system is employed by groups, such as crabs and lobsters (Becking et al. 2017; Chandler et al. 2017; Fang et al. 2020; Mlinarec et al. 2016; Triño et al. 1999) (Table 1). The presence of both XY and ZW systems in species belonging to the same genera of Malacostraca implies the potential for rapid transitions between such systems (Becking et al. 2017). In addition, some species in the Malacostraca (e.g., Palaemon elegans) have been found to have multiple sex chromosomes: X1, X2, and Y, resulting in a SD system of X1X1X2X2♀/X1X2Y♂ (Torrecilla et al. 2017). Additionally, in Armadillidium vulgare (clade Malacostraca), SD is mediated by bacterial endosymbionts (i.e., Wolbachia), which can convert ZZ genetic males to phenotypic females (Cordaux et al. 2011; Rigaud et al. 1997).
Ostracoda is the second-largest clade of crustacean with more than 13,000 living species. In Ostracoda, most studied species have X0 SD system, with sex dimorphism being controlled by the dosage effect of genes on chromosome X (Jeffery et al. 2017; Sajuthi et al. 2015; Turgeon and Hebert 1995). Although a few Ostracoda species have a Y chromosome, it appears that the sex-determining locus is not located on the Y, and sex is determined by the number of X chromosomes (Dietz 1957; Havel et al. 1990).
In contrast to Malacostraca and Ostracoda, in which sex is nearly always determined by genetic factors, sex in Maxillopoda and Branchiopoda species is often at least in part influenced by environmental factors (Blaxter et al. 1998; Michaud et al. 2004; Toyota et al. 2015a, b). For example, the copepod Tigriopus californicus (clade Maxillopoda) produces more males at higher temperatures (Voordouw and Anholt 2002). Similarly, food concentration and quality determines the percentage of males in other copepods (Irigoien et al. 2000; Michaud et al. 2004). Furthermore, some barnacles and shrimps (clade Maxillopoda) produce male and female gametes at distinct life stages (Chiba 2007; Fukuhara 1999; Subramoniam 2013), which is a system known as sequential hermaphroditism. In Daphnia (clade Branchiopoda), females are generally produced under favorable conditions, whereas males are more likely to be produced in unfavorable conditions, such as crowding (Hebert 1978) or short photoperiod (Toyota et al. 2015a, b).
Although most crustaceans have separate sexes, Cephalocarida and Remipedia species are exceptions. They reproduce exclusively by simultaneous hermaphroditism in which individuals have both male and female sex organs, and generate both types of gametes (Hessler et al. 1995; Yager 1991). Simultaneous hermaphroditism has been observed also in a small number of decapod species (clade Malacostraca) (Bauer and Holt 1998; Fiedler 1998). Simultaneous hermaphroditism is not without disadvantages as each parent must grow and maintain two sets of reproductive machinery (Heath 1977). It has been suggested that simultaneous hermaphroditism evolved as a result of the scarcity of mating partners (Cabej 2013). Consistent with this, Remipedia have been reported to reside solely in submerged caves (Yager 1991) and are slow movers (Regier et al. 2010), which may make finding mating partners more challenging.
Evolution of sex determination in crustaceans
Because Cephalocarida and Remipedia are the most basal crustacean clades (Fig. 1), it has been proposed that they represent ancestral SD system in crustaceans, i.e., simultaneous hermaphroditism (Legrand et al. 1987; Rigaud 1991). A different type of hermaphroditism known as sequential hermaphroditism is more prevalent in crustaceans particularly in barnacles and shrimps (Chiba 2007; Fukuhara 1999; Subramoniam 2013). Species with sequential hermaphroditism create male and female gametes at distinct periods of development, and the two sexes are functionally and morphologically different. In crustaceans, sequential hermaphroditism is an example of phenotypic flexibility in response to environmental changes, supporting the size advantage theory (Warner 1988). Individual crustacea develop in size as they age because they tend to grow continually throughout their lives. According to the size advantage theory, sex change is preferable when the reproductive cost of one sex increases faster than that of the other as body size increases. Size change is employed to optimize the combined fitness of an individual’s male and female stages. Depending on which sex matures first, sequential hermaphroditism is classified as protandry or protogyny. The former is utilized by a vast majority of Malacostraca species (Bauer 1986) in which individuals mature first as males and later change sex to females (Bauer 2000). Females benefit from large size because egg formation demands a significant amount of energy, and the egg survival rate is closely associated with body size in crustaceans (Bauer 2020). Conversely, some isopods (clade Malacostraca) rely on protogyny in which larger males have a reproductive advantage over smaller ones, associated with protection of mates (Abe and Fukuhara 1996). Consistent with the size advantage theory, Nakashima (1987) discovered that A. dorsalis competes for females during the breeding season, with larger males being more successful at copulating than smaller males.
Simultaneous hermaphroditism may develop into sequential hermaphroditism if oogenesis is not synchronized with spermatogenesis (Bauer 2000; Hoffman 1972). In some cases, reproductive resources are limited in brooding animals due to a lack of brooding space, and resources may be totally dedicated to sperm production, resulting in sequential hermaphrodites (Heath 1979). Conversely, sequential hermaphrodites could evolve into simultaneous hermaphrodites if both male and female organs are generated concurrently during the female phase (Bauer 2000; Hoffman 1972). This typically happens when population density is low and difficult to find mating partners.
ESD, which is utilized by a large number of crustaceans, shares similar characteristics with sequential hermaphroditism (Breton et al. 2018). First, neither system requires genetic differentiation between the two sexes. Second, both ESD and sequential hermaphroditism typically result in a skewed sex ratio. Third, sex change is preferable in both systems when one sex has a greater reproductive fitness than the other. Fourth, both ESD and sequential hermaphroditism share a common sex determination mechanism—stress-related pathways. Social structure could lead to situations in which individuals within a group experience varying degree of stress. It has been demonstrated that social factors within mating groups influence sex change in crustaceans, such as hippolytid shrimp (Bauer and Baeza 2004; Lin and Zhang 2001) and pandalid shrimp (Carpenter 1978; Charnov and Hannah 2002), both of which are hermaphrodites. Due to these similarities, it has been postulated that ESD may evolve from sequential hermaphroditism by a heterochronic shift (Straková et al. 2020).
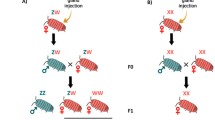
GSD, which is the most prevalent form of SD in crustaceans, may evolve from either hermaphroditism or ESD. In the case of hermaphroditism, distinct sexes could arise by male- or female-sterility mutations resulting in a system in which genetic females or males coexist with hermaphrodites (Charlesworth and Charlesworth 1978). Additional mutations beneficial to each sex will be retained in such a system if they are linked to the respective sex-determining gene (Edwards 1998). The sex-determining locus and its associated sex-specific genes could then lead to the establishment of a “proto-sex chromosome”, which may be maintained by chromosomal inversion or located in the centromere region, as both structural features minimize or eliminate recombination (Natri et al. 2019). GSD may also emerge from ESD as a result of a male- or female-sterility mutation, in which some females or males are genetically determined whereas others are subject to ESD. A system of this type could be maintained if it restores the sex ratio or lowers the cost of inbreeding (Edwards 1998; Reisser et al. 2017; Ye et al. 2019). It is believed that the shift from ESD to GSD is unidirectional as the majority of the well-supported transitions have occurred in the direction of ESD to GSD (Straková et al. 2020), with rare instances of GSD to ESD when male heterogamety disappeared (Bull 1981). When the environment is unpredictable or very variable, evolution from ESD to GSD is favored as GSD can maintain balanced sex ratios (Bull 1983). For example, snow skinks used ESD at low elevations, but shift to GSD at higher elevations where fluctuations in temperature are more pronounced (Pen et al. 2010).
In summary, while direct evidence for transitions among different SD systems in crustaceans is currently scarce, we infer putative evolutionary routes based on the available research. It has been proposed that crustaceans exhibited ancestral hermaphroditism in the form of simultaneous hermaphroditism and/or sequential hermaphroditism (Hessler et al. 1995; Yager 1991). ESD may have evolved from sequential hermaphroditism by a heterochronic shift of sex change during the early embryonic stage (Straková et al. 2020) (Fig. 2). Both hermaphroditism and ESD have the potential to progress to GSD with rare instances of reversion from GSD to ESD. We summarize the possible evolutionary routes for the SD systems in crustaceans in Fig. 2.
The role of Dmrt genes in sex determination
As the SD systems in crustaceans are capable of transitioning among each other, it is critical to understand the genetic basis for those transitions. Although the mechanism of sex determination varies across species, the underlying sex regulator genes appear to have converged on the doublesex and male abnormal-3 (Mab-3)-related transcription factor (Dmrt) gene family (Kopp 2012; Zarkower 2001) (Fig. 3). In mammals, development of male embryos is regulated by the Y chromosome-linked Sry gene, with Sry depletion resulting in ovary development (Sekido and Lovell-Badge 2009; Wilhelm et al. 2007). Sox9 (Sry-related box 9) is a downstream gene of Sry that is essential for testis development, and its absence results in male–female sex reversal (Foster et al. 1994). Dmrt1 is thought to be a pioneer factor for opening chromatin and allowing binding of Sox9 (Lindeman et al. 2021), and is expressed primarily in the testis following sex differentiation (Kim et al. 2007a, b). Loss of Dmrt1 function results in decreased Sox9 expression, which eventually results in sex reversal (Matson et al. 2011). The expression of Sox9 is maintained by a positive feedback loop with fibroblast growth factor 9 (Fgf9) (Piprek 2009), which together activates the male pathway (Kim et al. 2006a, b).
Sex switch and Dmrt genes in the sex determination pathways in diverse model species. DMRT doublesex and male abnormal-3-related transcription factors gene family, Sry sex-determining Region Y, Sox9 sry-related box 9, Xol-1 XO lethal protein 1, Tra transformer; Sxl sex lethal, MF methyl farnesoate, Dapalr doublesex1 alpha promoter-associated long non-coding RNA, Mab-3 male abnormal-3. Wtn4 Wnt family member 4. Arrows indicate positive regulation, and crossbars indicate repressive regulation
In C. elegans, sex is determined in a dose-dependent manner (males have a X chromosome and autosomes ratio of 0.5 (XO), whereas hermaphrodites have a ratio of 1.0 (XX)) by a master switch gene called Xol-1 (Luz et al. 2003). Xol-1 promotes male development, and loss-of-function mutations cause lethality in male animals (Miller et al. 1988). High levels of xol-1 expression in XO animals could repress the activity of sdc-1, sdc-2, and sdc-3 genes, whereas low xol-1 levels in XX animals permits the activation of the three sdc genes (Luz et al. 2003) (Fig. 3). Proteins from all three sdc genes function in suppression of her-1 (Yonker and Meyer 2003), which is a protein that promotes male development by inhibiting the function of Tra-2 (Perry et al. 1993; Pilgrim et al. 1995). Tra-2 inactivates the expression of the three fem (feminization) proteins (Gaudet et al. 1996; Pilgrim et al. 1995), which further inhibits the tra-1 activity in XO animals (Hodgkin 1986; Kimble et al. 1984). Mab-3, which is a member of the Dmrt gene family, is a Tra1 target gene that controls male sexual development and behavior in C. elegans (Yi et al. 2000). Tra1 represses the transcription of Mab-3 in XX animals resulting in hermaphrodites, where in XO animals Tra-1 is inactivated and the male fate was determined (Yi et al. 2000).
In Drosophila, the ratio of X chromosomes to autosomes determines whether the Sex lethal (Sxl) gene is on (in XX females) or off (in XY males) (Fig. 3). Sxl regulates the production of Tra protein in females (Salz and Erickson 2010). In males, due to the lack of Sxl, an mRNA with no long open reading frame is produced and functional protein is not generated (Fig. 3). Tra then controls sex-specific splicing of doublesex (Dsx) pre-mRNA (Bachtrog et al. 2014). Dsx was the first Dmrt gene discovered in insects, and its involvement in sex determination has been established (Burtis et al. 1991). The male and female-specific Dsx proteins then determine the unique somatic structures and external morphology in each sex (Baker et al. 2001; MacDougall et al. 1995).
Dmrt is also a critical sex determinant in crustaceans, such as D. magna (Kato et al. 2011a, b) and D. pulex (Xu et al. 2014). Male production is stimulated in D. magna by the hormone methyl farnesoate (MF) (Toyota et al. 2015a, b), which is then directly coupled to the methoprene-tolerant (Met) and steroid receptor coactivator (SRC) complex (Miyakawa et al. 2013). The binding sites of Met have been found in a bZIP transcription factor, Vrille (Mohamad Ishak et al. 2017), suggesting that the MF–met complex may directly activate Vrille (Fig. 3). Vrille then regulates the expression of Dsx1 and doublesex1 alpha promoter-associated long non-coding RNA (DAPALR) (Kato and Watanabe 2022) (Fig. 3). In addition, DAPALR could regulate Dsx1 expression in trans (Kato et al. 2018). Dsx1 is exclusively expressed in male Daphnia where its transcripts are found largely in male-specific structures (Kato et al. 2011a, b). Reduced Dsx1 expression in D. magna causes testes to develop an ovary-like morphology, and ectopic Dsx1 expression in female embryos leads to the development of male-like phenotypes (Kato et al. 2011a, b). Additionally, Dmrts have been identified in other crustaceans, such as Penaeidae (prawns), Palinuridae (lobsters), Palaemonidae (shrimp) and Portunidae (crabs) (Chandler et al. 2016). Moreover, their functional conservation has been demonstrated in crustaceans, such as the eastern spiny lobster (S. verreauxi) (Chandler et al. 2017).
The conservation of Dmrt genes in regulating SD allows for the study of SD evolution in crustaceans, particularly in species with little genetic background. First, Dmrt genes share a common function in sex determination and sex differentiation across taxa (Kopp 2012), which is to promote male-specific development and differentiation (Balciuniene et al. 2006; Kim et al. 2007a, b). This function is conserved in species with ESD (e.g., turtles) and GSD (e.g., Drosophila, mammals) systems (Kopp 2012). Second, Dmrt genes are expressed exclusively in developing gonads of all animals including mammals (Kim et al. 2003; Raymond et al. 2000), flies (Hempel and Oliver 2007), nematodes (Yi et al. 2000) and crustaceans (Farazmand et al. 2010; Kato et al. 2011a, b; Zhang and Qiu 2010). Third, Dmrt genes share a common DNA-binding domain (DM domain) that is highly conserved across phyla (Chandler et al. 2018; Raymond et al. 1998), whereas there is substantial sequence variation outside of the DM domain, sequences within the DM domain are extremely conserved (Chandler et al. 2018). The conserved nature of the DM domain enables the identification of genes that bind to it. Indeed, numerous genes associated with sex determination have been found in humans (Murphy et al. 2010) and Drosophila (Luo et al. 2011) via their physical or structural association with the DM domain. However, such studies are currently scarce in crustaceans.
Regardless of the conservation, the mechanisms by which Dmrt influences sex determination in each species may vary. For example, in insects, different isoforms of Dsx derived by alternative splicing are expressed in males and females, whereas in Daphnia, Dsx is expressed exclusively during male development (Kato et al. 2011a, b). It is worth mentioning that whereas Dmrt genes are not necessarily the master sex-determining genes, they often directly or indirectly interact with the master switch gene. Thus, identifying Dmrt genes will always be helpful in locating the SD locus and/or mechanisms in crustaceans.
Using Daphnia as a model to study the evolution of sex determination
Transitional species are of particular interest for shedding light on the evolution of sexual determination. Members of the genus Daphnia are now experiencing such a transition. Most Daphnia reproduce by cyclical parthenogenesis, with extended periods of parthenogenesis interspersed with sexual resting-egg production, generally on a yearly cycle (Hebert 1978). Parthenogenetic eggs may develop into females or males as sex determination is typically induced by environmental factors (e.g., short photoperiod). However, in some Daphnia populations, sex is also controlled by genetic factors (Reisser et al. 2017; Ye et al. 2019). Within such Daphnia populations, some females have lost the ability to produce males, resulting in the formation of non-male-producing (NMP) clones (Galimov et al. 2011; Tessier and Cáceres 2004). The coexistence of NMP clones and hermaphrodites (MP clones) in Daphnia creates a system called gynodioecy in which only females are genetically determined.
Because crosses between NMP and MP clones consistently produce a close to 1:1 ratio of NMP and MP offspring, whereas almost all offspring of MP × MP crosses exhibit MP phenotypes (Galimov et al. 2011; Innes and Dunbrack 1993), the presence of a dominant allele at a single locus underlying the NMP phenotype appears likely. To be more precise, all NMP clones are hypothesized to be WZ heterozygous at the locus conferring the NMP phenotype, whereas MP clones are thought to be ZZ homozygotes as is the case with classical W/Z sex determination systems. Additionally, it has been reported that the NMP phenotype is regulated by a single dominant allele contained within a 1.2 Mb non-recombining region in D. pulex (Ye et al. 2019) although it is largely unknown which genes within this region are involved in SD. The functional conservation of the Dmrt genes provides a good opportunity to connect genes in the 1.2 Mb region to the master sex switch gene (i.e., Dsx) in Daphnia.
To identify potential genes implicated in the shift from ESD to GSD, it is necessary to first understand the SD pathway in Daphnia. Typically, SD in Daphnia is regulated via environmental cues, such as short photoperiod or by adding exogenous juvenile hormone (JH). JH is an essential endocrine factor that regulates molting and metamorphosis in insects (Nijhout 1998). JH was found in Malacostraca (e.g., crab and crayfish) and Branchiopoda (e.g., Daphnia) species, but JH in those species lacks the epoxide group compared to that in insects (Laufer et al. 1987). It has been suggested that the JH in crustaceans, methyl farnesoate (MF), is important in molting and reproduction (Homola and Chang 1997). Additionally, JH has been shown to induce male production in Daphnia (Abe et al. 2015) and other cladoceran (clade Branchiopoda) species, such as Moina, Ceriodaphnia, and Bosmina (Kim et al. 2006a, b; Oda et al. 2005). Thus, it has been postulated that JH plays a general role in SD throughout the clade Branchiopoda (Kim et al. 2006a, b; Olmstead and Leblanc 2002).
In Daphnia, the SD pathway could be separated into two parts, namely the upstream and the downstream JH pathway (Fig. 4). The signal (e.g., short photoperiod) from the environment activates protein kinase C (PKC) in the upstream JH pathway. Then, PKC promotes the opening of the N-methyl-D-aspartic acid receptor (NMDAR) channel (Toyota et al. 2017) (Fig. 4). NMDAR is a type of ionotropic glutamate receptor that is required for male reproduction, and is believed to act as an upstream regulator of juvenile hormone acid O-methyltransferase (JHAMT) (Toyota et al. 2015a, b). JHAMT is utilized to synthesize JH from farnesoic acid in Daphnia (Toyota et al. 2015a, b), and it expresses at a higher level in male-producing conditions than in female-producing ones (Toyota et al. 2015a, b). JH was directly coupled to the methoprene-tolerant (Met) and steroid receptor coactivator (SRC) complex following its synthesis, and mutations within Daphnia Met were found to significantly alter the receptor's responsiveness (Miyakawa et al. 2013). Met's immediate target in insects is Krüppel homolog 1 (kr-h1) (Cui et al. 2014), but its function in Daphnia is still unknown (Toyota et al. 2018). In both D. magna and D. pulex, the sexual differentiation is eventually mediated by the Dsx1 gene (Kato et al. 2011a, b; Xu et al. 2014). Kato et al. discovered that the expression pattern for Dsx1 is male-specific in Daphnia, and that knocking down Dsx1 in male embryos or ectopic expression of Dsx1 in female embryos led to sex reversed traits (Kato et al. 2011a, b; Toyota et al. 2013). Dsx1 has been shown to be directly regulated by a bZIP transcription factor, Vrille (Mohamad Ishak et al. 2017), as well as by the doublesex1 alpha promoter-associated long non-coding RNA (DAPALR) (Kato et al. 2018) (Fig. 4).
Signaling cascades of the sex determination pathway in Daphnia (Branchiopoda). JH juvenile hormone, PKC protein kinase C, NMDAR N-methyl-D-aspartic acid receptor, JHAMT juvenile hormone acid O-methyltransferase, Met methoprene-tolerant, SRC steroid receptor coactivator, DAPALR doublesex1 alpha promoter-associated long non-coding RNA, Dsx doublesex. Regulation of DAPALR, Vrille, Dsx1 happened in embryos
The transition from ESD to GSD in D. pulex is thought to be induced by a factor downstream of the JH signaling pathway as the phenotype cannot be recovered by exogenous JH (Ye et al. 2019). Notably, a 1.2 Mb region containing 87 genes in NMP clones possesses NMP-specific substitutions, establishing the foundation for further investigation of the genetic components underlying the NMP trait. As a start, it is necessary to determine whether any of the 87 genes are direct targets of Met or Src. Additionally, the 1.2 Mb region in NMP clones may constitute a “proto-sex chromosome”, a hypothesis that may be validated by investigating whether the 87 genes in the NMP region express in a sex-specific manner. Additionally, if the SD transition involves the full 1.2 Mb non-recombining region, it will be worthwhile to determine whether this region is preserved by genome inversion. Finally, elucidating the SD pathway in Daphnia will provide an opportunity to investigate the transition from ESD to GSD in crustaceans.
Future perspectives
In this review, we summarize knowledge about the SD systems in crustaceans and discussed how these distinct systems might evolve from one another. However, due to the lack of research, genes in the SD pathway remain largely unknown in most crustaceans. Up till now, all ideas regarding SD transitions (e.g., from ESD to GSD) have been purely theoretical and require further validation. Daphnia could be an excellent model for studying the evolution of SD systems in crustaceans (and more broadly, arthropods) given its high-quality reference genomes, extensive research on the SD pathway, and powerful genetic editing tools such as CRISPR/Cas9. To begin, we could use Chip-seq data to identify genes that interact with Dsx1; second, transcriptomic data could be used to validate sex-specific genes in the NMP region; and third, we could compare high-quality genome assemblies from MP and NMP Daphnia clones to determine if structural variation (e.g., genome inversion) underlies the transition from ESD to GSD. Along with transitions between SD systems, transitions within the same SD system (e.g., XY to ZW) are widespread among crustacean genera (Becking et al. 2017). These transitions are particularly interesting because they provide insight into the rapid turnover within the same SD system. As such, new investigations aimed at identifying Dmrt genes, their targets, and genes that interact with the DM domain in genera with such transitions would be particularly exciting.
Data availability
N/A.
References
Abe M, Fukuhara H (1996) Protogynous hermaphroditism in the brackish and freshwater isopod, Gnorimosphaeroma naktongense (Crustacea: Isopoda, Sphaeromatidae). Zoolog Sci 13:325–329
Abe R, Toyota K, Miyakawa H, Watanabe H, Oka T, Miyagawa S, Nishide H, Uchiyama I, Tollefsen KE, Iguchi T (2015) Diofenolan induces male offspring production through binding to the juvenile hormone receptor in Daphnia magna. Aquat Toxicol 159:44–51
Alexander H, Richardson J, Edmands S, Anholt B (2015) Sex without sex chromosomes: genetic architecture of multiple loci independently segregating to determine sex ratios in the copepod Tigriopus californicus. J Evol Biol 28:2196–2207
Bachtrog D (2013) Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet 14:113–124
Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman T-L, Hahn MW, Kitano J, Mayrose I, Ming R (2014) Sex determination: why so many ways of doing it? PLoS Biol 12:e1001899
Baeza JA, Bauer RT (2004) Experimental test of socially mediated sex change in a protandric simultaneous hermaphrodite, the marine shrimp Lysmata wurdemanni (Caridea: Hippolytidae). Behav Ecol Sociobiol 55:544–550
Baker BS, Taylor BJ, Hall JC (2001) Are complex behaviors specified by dedicated regulatory genes? Reasoning from Drosophila. Cell 105:13–24
Balciuniene J, Bardwell VJ, Zarkower D (2006) Mice mutant in the DM domain gene Dmrt4 are viable and fertile but have polyovular follicles. Mol Cell Biol 26:8984–8991
Bauer RT (1986) Sex change and life history pattern in the shrimp Thor manningi (Decapoda: Caridea): a novel case of partial protandric hermaphroditism. Biol Bull 170:11–31
Bauer RT (2000) Simultaneous hermaphroditism in caridean shrimps: a unique and puzzling sexual system in the Decapoda. J Crustac Biol 20:116–128
Bauer RT (2020) Analysis of embryo production in a caridean shrimp guild from a tropical seagrass meadow. In: Crustacean egg production. CRC Press, pp 181–191
Bauer RT, Holt GJ (1998) Simultaneous hermaphroditism in the marine shrimp Lysmata wurdemanni (Caridea: Hippolytidae): an undescribed sexual system in the decapod Crustacea. Mar Biol 132:223–235
Becking T, Giraud I, Raimond M, Moumen B, Chandler C, Cordaux R, Gilbert C (2017) Diversity and evolution of sex determination systems in terrestrial isopods. Sci Rep 7:1–14
Blackmon H, Ross L, Bachtrog D (2017) Sex determination, sex chromosomes, and karyotype evolution in insects. J Hered 108:78–93
Blaxter JH, Douglas B, Tyler PA, Mauchline J (1998) The biology of calanoid copepods. Academic Press
Breton S, Capt C, Guerra D, Stewart D (2018) Sex-determining mechanisms in bivalves. In: Leonard J (ed) Transitions between sexual systems. Springer, Cham. https://doi.org/10.1007/978-3-319-94139-4-6
Budd GE, Telford MJ (2009) The origin and evolution of arthropods. Nature 457:812–817
Bull J (1981) Evolution of environmental sex determination from genotypic sex determination. Heredity 47:173–184
Bull JJ (1983) Evolution of sex determining mechanisms. The Benjamin/Cummings Publishing Company, Inc.
Burtis KC, Coschigano KT, Baker BS, Wensink PC (1991) The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J 10:2577–2582
Cabej NR (2013) Epigenetic control of animal development. Building the most complex structure on earth. Elsevier, Oxford, pp 121–192
Carpenter A (1978) Protandry in the freshwater shrimp, Paratya curvirostris (Heller, 1862) (Decapoda: Atyidae), with a review of the phenomenon and its significance in the Decapoda. J R Soc N Z 8:343–358
Chandler JC, Aizen J, Fitzgibbon QP, Elizur A, Ventura T (2016) Applying the power of transcriptomics: understanding male sexual development in decapod crustacea. Integr Comp Biol 56:1144–1156
Chandler JC, Fitzgibbon QP, Smith G, Elizur A, Ventura T (2017) Y-linked iDmrt1 paralogue (iDMY) in the Eastern spiny lobster, Sagmariasus verreauxi: The first invertebrate sex-linked Dmrt. Dev Biol 430:337–345
Chandler JC, Elizur A, Ventura T (2018) The decapod researcher’s guide to the galaxy of sex determination. Hydrobiologia 825:61–80
Charlesworth B, Charlesworth D (1978) A model for the evolution of dioecy and gynodioecy. Am Nat 112:975–997
Charnov E, Hannah R (2002) Shrimp adjust their sex ratio to fluctuating age distributions. Evol Ecol Res 4:239–246
Chiba S (2007) A review of ecological and evolutionary studies on hermaphroditic decapod crustaceans. Plankton Benthos Res 2:107–119
Cordaux R, Bouchon D, Grève P (2011) The impact of endosymbionts on the evolution of host sex-determination mechanisms. Trends Genet 27:332–341
Cui Y, Sui Y, Xu J, Zhu F, Palli SR (2014) Juvenile hormone regulates Aedes aegypti Krüppel homolog 1 through a conserved E box motif. Insect Biochem Mol Biol 52:23–32
Cui Z, Hui M, Liu Y, Song C, Li X, Li Y, Liu L, Shi G, Wang S, Li F (2015) High-density linkage mapping aided by transcriptomics documents ZW sex determination system in the Chinese mitten crab Eriocheir sinensis. Heredity 115:206–215
Dietz R (1957) Multiple Geschlechtschromosomen bei den cypriden Ostracoden, ihre evolution und ihr Teilungsverhalten. Chromosoma 9:359–440
Edwards AW (1998) Natural selection and the sex ratio: Fisher’s sources. Am Nat 151:564–569
Ezaz T, Moritz B, Waters P, Graves JAM, Georges A, Sarre SD (2009) The ZW sex microchromosomes of an Australian dragon lizard share no homology with those of other reptiles or birds. Chromosome Res 17:965–973
Fang S, Zhang Y, Shi X, Zheng H, Li S, Zhang Y, Fazhan H, Waiho K, Tan H, Ikhwanuddin M (2020) Identification of male-specific SNP markers and development of PCR-based genetic sex identification technique in crucifix crab (Charybdis feriatus) with implication of an XX/XY sex determination system. Genomics 112:404–411
Farazmand A, Inanloo K, Agh N (2010) Expression of DMRT family genes during gonadal differentiation in two species of Artemia (Branchiopoda, Anostraca) from Urmia Lake (Iran). Crustaceana 83:1153–1165
Fiedler GC (1998) Functional, simultaneous hermaphroditism in female-phase Lysmata amboinensis (Decapoda: Caridea: Hippolytidae). Pac Sci 52:161–169
Foster JW, Dominguez-Steglich MA, Guioli S, Kwok C, Weller PA, Stevanović M, Weissenbach J, Mansour S, Young ID, Goodfellow PN (1994) Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature 372:525–530
Fukuhara H (1999) Sex change in Crustacea. Aquabiology 125:487–494
Galimov Y, Walser B, Haag CR (2011) Frequency and inheritance of non-male producing clones in Daphnia magna: evolution towards sex specialization in a cyclical parthenogen? J Evol Biol 24:1572–1583
Gaudet J, VanderElst I, Spence AM (1996) Post-transcriptional regulation of sex determination in Caenorhabditis elegans: widespread expression of the sex-determining gene fem-1 in both sexes. Mol Biol Cell 7:1107–1121
Gomez ED (1975) Sex determination in Balanus (conopea) galeatus (L.) (Cirripedia Thoracica). Crustaceana 28:105–107
Goodwin EB, Ellis RE (2002) Turning clustering loops: sex determination in Caenorhabditis elegans. Curr Biol 12:R111–R120
Havel JE, Hebert PD, Delorme LD (1990) Genetics of sexual Ostracoda from a low Arctic site. J Evol Biol 3:65–84
Heath D (1977) Simultaneous hermaphroditism; cost and benefit. J Theor Biol 64:363–373
Heath D (1979) Brooding and the evolution of hermaphroditism. J Theor Biol 81:151–155
Hebert PD (1978) The population bilogy of Daphnia (Crustacea, Daphnidae). Biol Rev 53:387–426
Hempel LU, Oliver B (2007) Sex-specific Doublesex M expression in subsets of Drosophila somatic gonad cells. BMC Dev Biol 7:1–15
Hessler RR, Elofsson R, Hessler AY (1995) Reproductive system of Hutchinsoniella macracantha (Cephalocarida). J Crustac Biol 15:493–522
Hobaek A, Larsson P (1990) Sex determination in Daphnia magna. Ecology 71:2255–2268
Hodgkin J (1986) Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics 114:15–52
Hoffman DL (1972) The development of the ovotestis and copulatory organs in a population of protandric shrimp Pandalus platyceros Brandt from Lopez Sound, Washington. Biol Bull 142:251–270
Homola E, Chang ES (1997) Methyl farnesoate: crustacean juvenile hormone in search of functions. Comp Biochem Physiol B Biochem Mol Biol 117:347–356
Innes DJ, Dunbrack RL (1993) Sex allocation variation in Daphnia pulex. J Evol Biol 6:559–575
Irigoien X, Obermüller B, Head R, Harris R, Rey C, Hansen BW, Hygum B, Heath M, Durbin E (2000) The effect of food on the determination of sex ratio in Calanus spp.: evidence from experimental studies and field data. ICES J Mar Sci 57:1752–1763
Jeffery NW, Ellis EA, Oakley TH, Gregory TR (2017) The genome sizes of ostracod crustaceans correlate with body size and evolutionary history, but not environment. J Hered 108:701–706
Jiang XH, Qiu GF (2013) Female-only sex-linked amplified fragment length polymorphism markers support ZW/ZZ sex determination in the giant freshwater prawn Macrobrachium rosenbergii. Anim Genet 44:782–785
Kato Y, Watanabe H (2022) Regulation of doublesex1 expression for environmental sex determination in the cladoceran crustacean Daphnia. Front Cell Dev Biol 10:881255
Kato M, Takehana Y, Fukuda Y, Naruse K, Sakaizumi M, Hamaguchi S (2011a) An autosomal locus controls sex reversal in interspecific XY hybrids of the medaka fishes. Heredity 107:523–529
Kato Y, Kobayashi K, Watanabe H, Iguchi T (2011b) Environmental sex determination in the branchiopod crustacean Daphnia magna: deep conservation of a doublesex gene in the sex-determining pathway. PLoS Genet 7:e1001345
Kato Y, Perez CAG, Ishak NSM, Nong QD, Sudo Y, Matsuura T, Wada T, Watanabe H (2018) A 5′UTR-overlapping lncRNA activates the male-determining gene doublesex1 in the crustacean Daphnia magna. Curr Biol 28:1811–1817
Kim S, Kettlewell JR, Anderson RC, Bardwell VJ, Zarkower D (2003) Sexually dimorphic expression of multiple doublesex-related genes in the embryonic mouse gonad. Gene Expr Patterns 3:77–82
Kim K, Kotov AA, Taylor DJ (2006a) Hormonal induction of undescribed males resolves cryptic species of cladocerans. Proc R Soc Lond B Biol Sci 273:141–147
Kim Y, Kobayashi A, Sekido R, DiNapoli L, Brennan J, Chaboissier M-C, Poulat F, Behringer RR, Lovell-Badge R, Capel B (2006b) Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol 4:e187
Kim S, Bardwell VJ, Zarkower D (2007a) Cell type-autonomous and non-autonomous requirements for Dmrt1 in postnatal testis differentiation. Dev Biol 307:314–327
Kim S, Namekawa SH, Niswander LM, Ward JO, Lee JT, Bardwell VJ, Zarkower D (2007b) A mammal-specific Doublesex homolog associates with male sex chromatin and is required for male meiosis. PLoS Genet 3:e62
Kimble J, Edgar L, Hirsh D (1984) Specification of male development in Caenorhabditis elegans: the fem genes. Dev Biol 105:234–239
Kopp A (2012) Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet 28:175–184
Laufer H, Borst D, Baker F, Reuter C, Tsai L, Schooley D, Carrasco C, Sinkus M (1987) Identification of a juvenile hormone-like compound in a crustacean. Science 235:202–205
Legrand J, Legrand-Hamelin E, Juchault P (1987) Sex determination in Crustacea. Biol Rev 62:439–470
Lin J, Zhang D (2001) Reproduction in a simultaneous hermaphroditic shrimp, Lysmata wurdemanni: any two will do? Mar Biol 139:919–922
Lindeman RE, Murphy MW, Agrimson KS, Gewiss RL, Bardwell VJ, Gearhart MD, Zarkower D (2021) The conserved sex regulator DMRT1 recruits SOX9 in sexual cell fate reprogramming. Nucleic Acids Res 49:6144–6164
Luo SD, Shi GW, Baker BS (2011) Direct targets of the D. melanogaster DSXF protein and the evolution of sexual development. Development 138:2761–2771
Luz JG, Hassig CA, Pickle C, Godzik A, Meyer BJ, Wilson IA (2003) XOL-1, primary determinant of sexual fate in C. elegans, is a GHMP kinase family member and a structural prototype for a class of developmental regulators. Genes Dev 17:977–990
MacDougall C, Harbison D, Bownes M (1995) The developmental consequences of alternate splicing in sex determination and differentiation in Drosophila. Dev Biol 172:353–376
Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D (2011) DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 476:101–104
McDonald MJ, Rice DP, Desai MM (2016) Sex speeds adaptation by altering the dynamics of molecular evolution. Nature 531:233–236
Michaud M, de Meeûs T, Renaud F (2004) Environmental sex determination in a parasitic copepod: checking heterogeneity and unpredictability of the environment. Mar Ecol Prog Ser 269:163–171
Miller LM, Plenefisch JD, Casson LP, Meyer BJ (1988) xol-1: a gene that controls the male modes of both sex determination and X chromosome dosage compensation in C. elegans. Cell 55:167–183
Miyakawa H, Toyota K, Hirakawa I, Ogino Y, Miyagawa S, Oda S, Tatarazako N, Miura T, Colbourne JK, Iguchi T (2013) A mutation in the receptor methoprene-tolerant alters juvenile hormone response in insects and crustaceans. Nat Commun 4:1856
Mlinarec J, Porupski I, Maguire I, Klobučar G (2016) Comparative karyotype investigations in the white-clawed crayfish Austropotamobius pallipes species complex and stone crayfish A. Torrentium (Decapoda: Astacidae). J Crustac Biol 36:87–93
Mohamad Ishak NS, Nong QD, Matsuura T, Kato Y, Watanabe H (2017) Co-option of the bZIP transcription factor Vrille as the activator of Doublesex1 in environmental sex determination of the crustacean Daphnia magna. PLoS Genet 13:e1006953
Muller HJ (1932) Some genetic aspects of sex. Am Nat 66:118–138
Muller HJ (1964) The relation of recombination to mutational advance. Mutat Res 1:2–9
Murphy MW, Sarver AL, Rice D, Hatzi K, Ye K, Melnick A, Heckert LL, Zarkower D, Bardwell VJ (2010) Genome-wide analysis of DNA binding and transcriptional regulation by the mammalian Doublesex homolog DMRT1 in the juvenile testis. Proc Natl Acad Sci USA 107:13360–13365
Nakashima Y (1987) Sex change in crustaceans. Sex change in fishes. Tokai University Press, Tokyo, p 284
Natri HM, Merilä J, Shikano T (2019) The evolution of sex determination associated with a chromosomal inversion. Nat Commun 10:1–13
Nijhout HF (1998) Insect hormones. Princeton University Press
Oda S, Tatarazako N, Watanabe H, Morita M, Iguchi T (2005) Production of male neonates in four cladoceran species exposed to a juvenile hormone analog, fenoxycarb. Chemosphere 60:74–78
Olmstead AW, LeBlanc GA (2002) Juvenoid hormone methyl farnesoate is a sex determinant in the crustacean Daphnia magna. J Exp Zool A Ecol Genet Physiol 293:736–739
Otto SP (2009) The evolutionary enigma of sex. Am Nat 174:S1–S14
Parnes S, Khalaila I, Hulata G, Sagi A (2003) Sex determination in crayfish: are intersex Cherax quadricarinatus (Decapoda, Parastacidae) genetically females? Genet Res 82:107–116
Pen I, Uller T, Feldmeyer B, Harts A, While GM, Wapstra E (2010) Climate-driven population divergence in sex-determining systems. Nature 468:436–438
Perry M, Li W, Trent C, Robertson B, Fire A, Hageman J, Wood W (1993) Molecular characterization of the her-1 gene suggests a direct role in cell signaling during Caenorhabditis elegans sex determination. Genes Dev 7:216–228
Pilgrim D, McGregor A, Jäckle P, Johnson T, Hansen D (1995) The C. elegans sex-determining gene fem-2 encodes a putative protein phosphatase. Mol Biol Cell 6:1159–1171
Piprek R (2009) Genetic mechanisms underlying male sex determination in mammals. J Appl Genet 50:347–360
Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D (1998) Evidence for evolutionary conservation of sex-determining genes. Nature 391:691–695
Raymond CS, Murphy MW, O’Sullivan MG, Bardwell VJ, Zarkower D (2000) Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev 14:2587–2595
Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, Wetzer R, Martin JW, Cunningham CW (2010) Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 463:1079–1083
Reisser CM, Fasel D, Hurlimann E, Dukic M, Haag-Liautard C, Thuillier V, Galimov Y, Haag CR (2017) Transition from environmental to partial genetic sex determination in Daphnia through the evolution of a female-determining incipient W chromosome. Mol Biol Evol 34:575–588
Rigaud T (1991) Déterminisme extrachromosomique du sexe chez Armadillidium vulgare Latr. (Crustacé, Isopode: modification du sex ratio par une bactérie endocytobiotique et conséquences sur l'évolution des génotypes sexuels dans les populations. PhD diss., Tours
Rigaud T, Juchault P, Mocquard JP (1997) The evolution of sex determination in isopod crustaceans. BioEssays 19:409–416
Sajuthi A, Carrillo-Zazueta B, Hu B, Wang A, Brodnansky L, Mayberry J, Rivera AS (2015) Sexually dimorphic gene expression in the lateral eyes of Euphilomedes carcharodonta (Ostracoda, Pancrustacea). EvoDevo 6:1–14
Salz H, Erickson JW (2010) Sex determination in Drosophila: The view from the top. Fly 4:60–70
Sassaman C, Weeks SC (1993) The genetic mechanism of sex determination in the conchostracan shrimp Eulimnadia texana. Am Nat 141:314–328
Schwentner M, Combosch DJ, Nelson JP, Giribet G (2017) A phylogenomic solution to the origin of insects by resolving crustacean-hexapod relationships. Curr Biol 27:1818–1824
Sekido R, Lovell-Badge R (2009) Sex determination and SRY: down to a wink and a nudge? Trends Genet 25:19–29
Stevens L (1997) Sex chromosomes and sex determining mechanisms in birds. Sci Prog 80:197–216
Straková B, Rovatsos M, Kubička L, Kratochvíl L (2020) Evolution of sex determination in amniotes: Did stress and sequential hermaphroditism produce environmental determination? BioEssays 42:2000050
Subramoniam T (2013) Origin and occurrence of sexual and mating systems in Crustacea: a progression towards communal living and eusociality. J Biosciences 38:951–969
Tessier AJ, Cáceres CE (2004) Differentiation in sex investment by clones and populations of Daphnia. Ecol Lett 7:695–703
Torrecilla Z, Martínez-Lage A, Perina A, González-Ortegón E, González-Tizón AM (2017) Comparative cytogenetic analysis of marine Palaemon species reveals a X 1 X 1 X 2 X 2/X 1 X 2 Y sex chromosome system in Palaemon elegans. Front Zool 14:1–9
Toyota K, Kato Y, Sato M, Sugiura N, Miyagawa S, Miyakawa H, Watanabe H, Oda S, Ogino Y, Hiruta C (2013) Molecular cloning of doublesex genes of four cladocera (water flea) species. BMC Genomics 14:1–13
Toyota K, Miyakawa H, Hiruta C, Furuta K, Ogino Y, Shinoda T, Tatarazako N, Miyagawa S, Shaw JR, Iguchi T (2015a) Methyl farnesoate synthesis is necessary for the environmental sex determination in the water flea, Daphnia pulex. J Insect Physiol 80:22–30
Toyota K, Miyakawa H, Yamaguchi K, Shigenobu S, Ogino Y, Tatarazako N, Miyagawa S, Iguchi T (2015b) NMDA receptor activation upstream of methyl farnesoate signaling for short day-induced male offspring production in the water flea, Daphnia pulex. BMC Genomics 16:186
Toyota K, Sato T, Tatarazako N, Iguchi T (2017) Protein kinase C is involved with upstream signaling of methyl farnesoate for photoperiod-dependent sex determination in the water flea Daphnia pulex. Biol Open 6:161–164
Toyota K, Tatarazako N, Iguchi T (2018) Environmental control of sex differentiation in Daphnia. In: Kobayashi K, Kitano T, Iwao Y, Kondo M (eds) Reproductive and developmental strategies: the continuity of life. Springer, Japan, Tokyo, pp 247–265
Toyota K, Miyakawa H, Hiruta C, Sato T, Katayama H, Ohira T, Iguchi T (2021) Sex determination and differentiation in decapod and cladoceran crustaceans: an overview of endocrine regulation. Genes 12:305
Triño AT, Millamena OM, Keenan C (1999) Commercial evaluation of monosex pond culture of the mud crab Scylla species at three stocking densities in the Philippines. Aquaculture 174:109–118
Turgeon J, Hebert P (1995) Genetic characterization of breeding systems, ploidy levels and species boundaries in Cypricercus (Ostracoda). Heredity 75:561–570
Voordouw MJ, Anholt BR (2002) Environmental sex determination in a splash pool copepod. Biol J Linn Soc 76:511–520
Warner RR (1988) Sex change and the size-advantage model. Trends Ecol Evol 3:133–136
Wilhelm D, Palmer S, Koopman P (2007) Sex determination and gonadal development in mammals. Physiol Rev 87:1–28
Xu S-L, Zhou W, Chen P, Zhou J-K, Zou X, Wang C-L, Wang D-L, Zhao Y-L (2014) Identification and expression analysis of a doublesex1 gene in Daphnia pulex during different reproductive stages. Dev Genes Evol 224:147–157
Yager J (1991) The reproductive biology of two species of Remipedes. In: Bauer RT, Martin JW (eds) Crustacean sexual biology. Columbia University Press, New York, pp 271–289
Ye Z, Molinier C, Zhao C, Haag CR, Lynch M (2019) Genetic control of male production in Daphnia pulex. Proc Natl Acad Sci 116:15602–15609
Yi W, Ross JM, Zarkower D (2000) Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development 127:4469–4480
Yonker SA, Meyer BJ (2003) Recruitment of C. elegans dosage compensation proteins for gene-specific versus chromosome-wide repression. Development 130:6519–6532
Zarkower D (2001) Establishing sexual dimorphism: conservation amidst diversity? Nat Rev Genet 2:175–185
Zhang E-F, Qiu G-F (2010) A novel Dmrt gene is specifically expressed in the testis of Chinese mitten crab, Eriocheir sinensis. Dev Genes Evol 220:151–159
Zhang X, Siveter DJ, Waloszek D, Maas A (2007) An epipodite-bearing crown-group crustacean from the lower Cambrian. Nature 449:595–598
Acknowledgements
We thank Wen Wei for helpful discussions. This work was financially supported by NIH grant R35-GM122566-01 to M.L. and NIH Enabling Discovery through GEnomics (EDGE) grant IOS-1922914 to M.L. and Andrew Zelhof (Indiana University).
Author information
Authors and Affiliations
Contributions
ZY conceived and designed the research; ZY, TB, YW, and RS analyzed data and performed research; ZY and ML wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors confirm that there are no known conflicts of interest associated with this publication.
Animal and human rights statement
N/A.
Additional information
Edited by Jiamei Li.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ye, Z., Bishop, T., Wang, Y. et al. Evolution of sex determination in crustaceans. Mar Life Sci Technol 5, 1–11 (2023). https://doi.org/10.1007/s42995-023-00163-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42995-023-00163-4