Abstract
Patients with cancer have been disproportionately affected by the novel coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Knowledge collected during the last three decades of cancer research has helped the medical research community worldwide to respond to many of the challenges raised by COVID-19, during the pandemic. The review, briefly summarizes the underlying biology and risk factors of COVID-19 and cancer, and aims to present recent evidence on cellular and molecular relationship between the two diseases, with a focus on those that are related to the hallmarks of cancer and uncovered in the first less than three years of the pandemic (2020–2022). This may not only help answer the question “Why cancer patients are considered to be at a particularly high risk of developing severe COVID-19 illness?”, but also helped treatments of patients during the COVID-19 pandemic. The last session highlights the pioneering mRNA studies and the breakthrough discovery on nucleoside-modifications of mRNA by Katalin Karikó, which led to the innovation and development of the mRNA-based SARSCoV-2 vaccines saving lives of millions and also opened the door for a new era of vaccines and a new class of therapeutics.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer is a leading cause of death worldwide, and this disease accounted for nearly 10 million deaths in 2020 (Global Cancer Observatory: Cancer Today. Lyon: International Agency for Research on Cancer 2021). Since the outbreak of the Coronavirus Disease 2019 (COVID-19) pandemic, it caused more than 6.45 million deaths and more than 596 million cases worldwide (World Health Organization, WHO 2020). It is well known that patients with cancer are especially vulnerable to this newly emerging disease (e.g., Dai et al. 2020; Yang et al. 2020). Consequently, the COVID-19 pandemic has affected virtually all aspects of cancer medical care and research (Vrdoljak et al. 2020; American Association for Cancer Research 2022).
On the contrary, major breakthroughs of cancer research against cancer that are generated over decades, among others, in biology/genetics, immunology, vaccinology, and drug development have uniquely positioned cancer researchers and physicians to respond to the many challenges posed by COVID-19. They could extensively assist the development of COVID-19 diagnostics, treatments, and vaccines, from early phase of the pandemic (Bakouny et al. 2020; Elkrief et al. 2022; American Association for Cancer Research 2022).
The main area that merits special attention, where researchers have gained knowledge from the biology of cancer, is the immune response to COVID-19 (e.g., Bakouny et al. 2020; American Association for Cancer Research 2022). For example, the vast amount of knowledge on human papilloma viruses, HPVs and vaccines (zur Hausen 2008) and adaptation of the tests routinely used to monitor T-cell responses to proteins of HPVs could be well utilized for determining the effectivity of COVID-19 vaccines in eliciting immune responses with special regard to cancer patients undergoing treatment.
In a broader sense, cancer immunology is a key area of cancer research and medicine that have contributed very important elements to the understanding of COVID-19. In 2013, Science nominated cancer immunotherapy as the Breakthrough of the Year (Couzin-Frankel 2013). In the past decade, advanced cancer immunotherapies—chimeric antigen receptor (CAR)-modified T cell therapy and inhibitors of immune regulatory checkpoints—have revolutionized the treatment of an increasingly broad array of cancer types (Schuster et al. 2017; Ribas and Wolchok 2018). Knowledge drawn from CAR T-cell therapy has largely contributed to our understanding of “cytokine storm” in severe COVID-19 (discussed in later sessions).
Beyond scientific discoveries, state-of-the-art technologies, for instance, next-generation sequencing (NGS) (Mardis 2008), which is routinely used worldwide to detect a broad range of cancer-related pathogenic genetic alterations, were effectively applied to identify the genomic sequence and novel variants of SARS-CoV-2, in early phase of the pandemic.
This review provides a brief summary of the underlying molecular biology of COVID-19 and cancer, presents recent evidence on cellular and molecular relationship between the two diseases, covering common risk factors, shared molecular markers and overlapping signaling pathways/cancer hallmarks and illustrates how have the insights from cancer by research community may be applied to mitigate this new disease. To this end, the review examines the mRNA research conducted over decades that have led to the tremendous success of development of the mRNA-based SARSCoV-2 vaccines and novel mRNA-based cancer therapies.
SARS-CoV-2 and COVID-19
The novel coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported in December 2019 in Wuhan, China. World Health Organization, WHO declared the COVID-19 outbreak to be a pandemic in March 2020. SARS-CoV-2 belongs to a large family of coronaviruses (CoVs) that exist widely in nature (Hartenian et al. 2020). Three out of the seven coronaviruses that are capable of infecting humans have caused the deadly global outbreaks of severe acute respiratory syndrome (SARS) in 2002–2004, Middle East respiratory syndrome (MERS) in 2012, and COVID-19 (Hartenian et al. 2020).
The genetic material of SARS-CoV-2 is RNA that encodes four major structural proteins: envelope proteins, nucleocapsid, membrane, and the spike. However, CoVs, SARS-CoV-2 possesses higher infectivity and pathogenicity than other CoVs, what is largely due to its special key sites within the receptor binding domain of spike protein which are different from other types of CoVs (Yi et al. 2020).
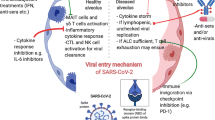
In the early phase of the pandemic, great attention was paid to understand the mechanisms by which SARS-CoV-2 enters the host cell—another area where researchers have relied on knowledge from cancer biology (Hoffmann et al. 2020). The first known entry factor is a receptor called angiotensin-converting enzyme 2 (ACE2) located on the surface of certain human cells in the nasal passages, lungs, and gastrointestinal tract, among others (Hoffmann et al. 2020). To infect a human host, the spike protein of SARS-CoV-2 attaches to the ACE2 receptor, which is abundantly expressed in the lung, heart, kidney, and intestine and induces an unwarranted immune response (Kuba et al. 2005). This could be an explanation for the vast array of symptoms that is manifested in patients with COVID-19. The other entry factor of SARS-CoV-2 internalization, on the surface of the host cell is a protein, called transmembrane serine protease 2 (TMPRSS2). The enzyme is needed to cleave SARS-CoV-2 spike protein and facilitate viral entry in human cells (Hoffmann et al 2020).
COVID-19 has been referred to as a disease of the lungs, the main target of severe COVID-19. However, it has turned out that not only the lung but an increasing number of other organs may suffer from COVID-19. These extrapulmonary manifestations frequently affect major organs as the heart, brain, kidneys, liver, but intestines, blood vessels, blood, and immune system may also be affected (Gupta et al. 2020). Many effects of COVID-19 could be potentially long-lasting, leading to development of Post COVID-19 or Long COVID-19 syndrome, which represents an emerging global crisis and consequently it is a hot area of contemporary research efforts (Gupta et al. 2020; Soriano et al. 2022).
Risk factors of COVID-19 susceptibility and severity
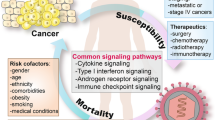
SARS-CoV-2 infections show a significant inter-individual variability, ranging from life-threatening disease to asymptomatic infections (Buitrago-Garcia et al. 2020). Environmental, clinical and social risk factors, host factors (e.g., increasing age, being a man and higher body-mass index) together with the genetic make-up of an individual may explain the variability in disease severity observed across individuals (Zhou et al. 2020).
Researchers are extensively investigating the role of genetic risk factors that are associated with a person’s susceptibility to COVID-19 (Callaway 2021). From early period of the COVID-19 pandemic, several large concerted efforts have been launched to identify the genetic determinants of COVID-19 susceptibility and severity. These studies have uncovered several genetic alterations associated with severe COVID-19 (Severe Covid-19 GWAS Group 2020; Tangye et al. 2021). There are many genes that are able to boost our defense against various viral infections, for example members of the interferon pathway. It is shown that inborn errors of, and autoantibodies directed against, type I interferons (IFNs) are responsible for about 20% of severe COVID-19 cases among SARS-CoV-2-infected individuals (Zhang et al 2020a).
Most recently, the OAS1/2/3 cluster (a subgroup of interferon-stimulated genes) has been identified as an important risk factor locus for severe COVID-19 among individuals of European ancestry (Pairo-Castineira et al. 2021. Moreover, Zeberg and Pääbo recently reported that both a major genetic risk factor for severe COVID-19 and a genomic region associated with protection against severe COVID-19 are inherited from Neandertals (Zeberg and Pääbo 2020, 2021).
Describing the role of a missing transposable element in SARS-CoV-2 infected cells, a most recent study provides further evidence on the significance of IFN-related genes in COVID-19, giving also a possible functional explanation for SARS-CoV-2 success in the combat against the host immune system (Sorek et al. 2022).
Increasing evidence support that SARS-CoV-2 entry factors (e.g., ACE2) influence COVID-19 risk and yields risk scores associated with severe disease (Hou et al. 2020). In addition, several novel genes of currently unknown significance were identified. Using whole-genome sequencing (WGS), the GenOMICC (Genetics of Mortality in Critical Care) project has discovered and replicated 23 independent variants that significantly predispose to severe COVID-19 (Kousathanas et al. 2022).
More recently, a large global effort has also launched to exploit genetic determinants of protective effects against SARS-CoV-2 infection. This was achieved by genetically analyzing individuals who show natural resistance to SARS-CoV-2 infection (Andreakos et al. 2022). Notably, early in the pandemic, several studies have provided convincing data that individuals with certain blood types were less susceptible to COVID-19 compared to those with other blood types (Barnkob et al. 2020; Severe Covid-19 GWAS Group 2020). However, according to a more recent research, blood type is not associated with disease susceptibility or severity (Anderson et al. 2021).
Genome-Wide Association Study (GWAS) have identified dozens of genomic regions associated with susceptibility and severity of COVID-19 (e.g., Severe COVID-19 GWAS Group 2020; Pairo-Castineira et al. 2021). However, the causal variants in these genomic regions have not been identified yet, which is hindering our ability to understand the pathophysiology of COVID-19.
Biology of cancer development
There is compelling evidence that cancer is, in essence, a genetic disease caused by the accumulation of a series of genetic and/or epigenetic changes that lead to changes in gene expression, and eventually to cancer. In a long-term microevolution process, a cell population develops that can ignore the normal controls of proliferation, differentiation and death and has the ability to transform to cancer (e.g., Nowell 1974; Fearon and Vogelstein 1990; Baylin et al. 2000).
Cancer is a multifactorial disease: beyond host factors (for example, age, comorbidities) several other factors can enhance development of the disease. The genetic events on the cancer pathway are arising during normal cell multiplication, and by endogenous mutagens or because of environmental exposures, or lifestyle factors (International Agency for Research on Cancer: http://monographs.iarc.fr).
In the past fifteen years the availability of the human genome sequence (International Human Genome Sequencing Consortium 2004), then the construction of HapMap containing single nucleotide polymorphisms (SNPs) (International HapMap Consortium 2005), together with the huge progress in next-generation sequencing (NGS) (Mardis 2008) and other novel genomic technologies now offer an unprecedented opportunity for the identification of all the genetic changes associated with a human cancer. Large-scale sequencing analyses have identified "cancer genes" (Futreal et al. 2004). These are mutated genes that are causally implicated in tumorigenesis. Cancer genes that are broadly grouped into oncogenes and (inactivated) tumorsuppressor genes arised from normal cells by intragenic mutations that can drive oncogenesis via genetically determined molecular pathways (e.g., Kuenzi and Ideker 2020).
It is well established that each tumor contains a collection of various cancer cell populations with a wide range of genetic variations. So, when we think about genetics and genomics of cancer, it is really complex, inborn/inherited (germline) and somatic mutations act together. In the cancer pathway, there are mostly (about 90%) somatic genetic mutations of oncogenes and tumor suppressor genes which acquired somatically during an individual’s lifetime, but in the hereditary cancer syndromes, one of the mutations is inherited. The cardinal feature of the syndromic hereditary cancers is a germline mutation associated with an increased risk for certain cancers and transmission to offspring through the maternal or paternal side. Most importantly, they often have an early age at onset and exhibit positive family history due to an autosomal dominant inheritance pattern (Ponder 1990). In 2022, we know over 125 hereditary cancer syndromes where pathogenic germline mutations of susceptibility genes put very high risk of cancer (Online Mendelian Inheritance in Man®, OMIM® 2022). With few exceptions, hereditary cancers of the inherited cancer syndromes are due to predisposing genetic variations (mutations) of the tumor suppressor genes. BRCA1 gene, the first’breast cancer susceptibility gene’ (BRCA1 gene with pathogenic germline mutation) was discovered by Mary-Claire King, American geneticist in 1990 (Hall et al. 1990). Identification of several dozens of genetic susceptibility genes to cancer followed in the next decades, including that of the EPCAM (also known as TACSTD1) gene with a new class of mutation predisposing to Lynch-syndrome (a hereditary non-polyposis colorectal cancer syndrome) that is discovered by our Lab (Kovács et al. 2009).
Elucidation of the genetic risk factors (inherited predisposition) to cancer has been one of the most successful areas of biomedical research (Oláh 2021). Studying germline susceptibility to cancer we not only gained new insights into cancer biology, but could also make risk assessment, and translate the new knowledge to clinical oncology practice. Genetic testing for inherited mutations in strongly predisposing genes of breast, ovarian, colorectal and other types of cancer is now part of the standard of clinical care for families suspected to have a strongly predisposing germline mutation. (Singer et al. 2019).
Over the past two decades, an expanding new knowledge has been generated on the complex nature of cancer genome (including also germline genome). Large international collaborative studies on genomics and epidemiology of cancer, for example by The Breast Cancer Association Consortium (2021) and The Consortium of Investigators of Modifiers of BRCA1/2, CIMBA (Chenevix-Trench et al. 2007) have made a great progress in uncovering the complex architecture of cancer susceptibility and cancer risk that involves germline pathogenic variants in high and moderate risk genes and polygenetic factors (Zhang et al. 2020b). It is now widely accepted view that genetic susceptibility to cancer—particularly in genetically unexplained families—is linked to variant patterns rather than single alternative variants (Mavaddat et al. 2015; Lakeman et al. 2019). For example, the rare, but strongly predisposing germline mutations of BRCA1/2 genes confer very high (around 70%) lifetime risk for breast or ovarian cancer (Kuchenbaecker et al. 2017), and the combined contribution to overall breast cancer incidence of these mutations is less than 10%. By contrast, a set of weakly predisposing, but common alleles could account for up to 40% of breast cancer incidence (Michailidou et al 2017).
It is widely demonstrated that the tumor(s) of every cancer patient have a unique combination of genetic variations (mutations). The identification of the critical somatic genetic differences is particularly important in determining the appropriate treatment strategies for each patient. The expanding new knowledge on both germline and somatic genome is rapidly improving our understanding of cancer biology and has the ability to transform the practice of oncology at a rapid pace not seen before (Meric-Bernstam et al. 2013).
Hallmark features of cancer
Hanahan and Weinberg in their prominent "Hallmarks of Cancer" position paper have laid down the framework of knowledge on cancer as a disease (Hanahan and Weinberg 2000). In this review, they identified six hallmark features (capabilities) of the cancer cells that are essential to be acquired during development and progression of human tumors. These include: “sustaining proliferative signaling, evading growth suppressors, resisting cell death, enabling replicative immortality, inducing angiogenesis, and activating invasion and metastasis.” They also suggested that genome instability and inflammation are underlying these hallmarks (Hanahan and Weinberg 2000). Genome instability generates genetic diversity, due to acquired or inherited defects in DNA repair or in cell-cycle checkpoints, thus expediting the acquisition of hallmark capabilities. Inflammation is a known intrinsic defense mechanism activated by the immune system against infection or injury that fosters multiple hallmark functions (Hanahan and Weinberg 2000).
A few years after the publication of this position paper, it has also become increasingly evident that mutated cells on their way leading to a tumor manifestation have to acquire further capabilities such as, how to thrive in a chronically inflamed microenvironment, evade immune recognition, and suppress immune reactivity (Cavallo et al. 2011). The recognition of these capabilities have verified that evading immune destruction is an essential hallmark of cancer (Hanahan and Weinberg 2011). Reprogramming of cellular metabolism, has also been added, a decade later (Hanahan and Weinberg 2011). Importantly, it is shown that most of the hallmarks of cancer are interconnected with an altered cancer cell-intrinsic metabolism (Kroemer and Pouyssegur 2008). As knowledge of cancer mechanisms has progressed due to applying more advanced computational tools, genomic technologies and "big data" approaches, other potential hallmark features of cancer have been identified (Hanahan 2022). Out of these both disrupted differentiation and unlocking phenotypic plasticity have recently been added as a “discrete hallmark capability”, while both nonmutational epigenetic reprogramming (Baylin et al. 2000; Hanahan 2022) and polymorphic microbiomes (Senga and Grose 2021; Hanahan 2022) have been added as “distinctive enabling characteristics” that enhance the procurement of hallmark capabilities (Hanahan 2022). Altered neuronal signaling has also been proposed for considering as distinctive enabling characteristics (Senga and Grose 2021). Recent studies have also demonstrated that splicing regulatory factors play an important role in nearly all hallmarks of cancer (Koedoot et al. 2019). Notably, the role of alternative splicing in breast tumorigenesis was proposed for BRCA1 gene even in the absence of deleterious inherited mutations (Orbán and Oláh 2003).
In addition, it is now increasingly clear that tumor microenvironment—which is made up of heterogeneous and interactive populations of cancer cells, cancer stem cells and various stromal cells—plays an integral role in tumorigenesis and malignant progression (Beck and Blanpain 2013; Hanahan 2022).
The lucid conceptualization of complex cancer biology by Hanahan and Weinberg (2000, 2011) is widely accepted and has also inspired development of advanced cancer therapeutics and therapies by targeting the various hallmarks.
In summary, Fig. 1 shows major hallmarks of cancer.
Hallmark flower of cancer, 2022. Other emerging hallmarks and "enabling characteristics" that facilitate the acquisition of hallmark capabilities are described in the session of "Biology of cancer development." After: Hanahan and Weinberg (2000), Hanahan and Weinberg (2011), Senga and Grose (2021), Hanahan (2022). Asterisk denotes those hallmarks where interplay is demonstrated between cancer and COVID-19
COVID-19 and cancer
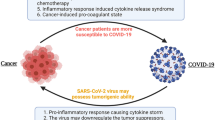
Numerous studies have shown that older adults and individuals with certain preexisting health conditions, such as patients with cancer, are at a higher risk of SARS-CoV-2 infection (e.g., Liang et al. 2020) and are more likely to experience severe disease, including mortality, from COVID-19 (Bakouny et al. 2020). Several studies have reported a poor prognosis for the patients with cancer and COVID-19, especially those who if they are affected those with hematological malignancies, and male patients who have lung cancer have a higher risk of death (e.g., Yang et al. 2020). Furthermore, therapeutic interventions, including surgery, chemotherapy, radiotherapy, and immunotherapy, have been associated with worse COVID-19 outcomes among cancer patients (Lee et al. 2020a; Derosa et al. 2020).
The interplay between COVID-19 and cancer biology
Early in the pandemic, it became clear that there were several risk factors shared by COVID-19 and cancer, which increase the risk for both diseases. These include comorbidities (such as cardiovascular and respiratory diseases), advanced age and smoking, obesity, or cancer type and staging (e.g., Lee et al. 2020; Pinato et al. 2020). These risk factors may significantly weaken the host immune system, making cancer patients more vulnerable to infectious agents, including SARS-CoV-2 (e.g., Swann and Smyth 2007; van Dam et al. 2020).
Recent studies on genetic risk factors for severe COVID-19, such as the OAS gene family, have also shown potential overlap with cancer research (Pairo-Castineira et al. 2021).
A recent study by Zhang and Yu (2020) shed new light on the prognostic roles of OAS in breast cancer. The study found that high mRNA expression of OAS1 and OAS3 was associated with a worse prognosis for all breast cancer patients, while OAS2 was linked to a favorable prognosis. Furthermore, a subgroup of interferon-stimulated genes, including the OAS family, STAT1, IRF7, and BST2, collectively known as the interferon-related DNA damage resistant signature (IRDS), were found to be upregulated in various cancer types. The upregulation of these genes promotes resistance to DNA damaging chemotherapy and radiotherapy (Padariya et al. 2021). Targeting the IRDS genes has been suggested as a potentially effective strategy to resensitize tumor cells and improve the outcome of anticancer treatment (Padariya et al. 2021).
While cancer and COVID-19 have different origins, development, and effects on the body, there are some shared manifestations between the two diseases. Although much remains to be learned about the interplay between cancer and COVID-19, researchers have already leveraged knowledge from cancer biology in the first two years of the pandemic to gain insight into the effects of COVID-19. Multiple lines of evidence have emerged, revealing clinical and molecular similarities between cancer and COVID-19 (e.g., Bakouny et al. 2020; Zong et al. 2021; American Association for Cancer Research 2022).
Entry factors of SARS-CoV-2 internalization
The first biological link between COVID-19 and cancer was reported on entry factors of SARS-CoV-2 internalization (Hoffmann et al. 2020). Based on the correlations seen between age-related elevation of ACE2 protein in human lung tissue and the most common incidence of cancer in the same age group (65 and older), it is suggested that the abundance of lung ACE2 may contribute to severe COVID-19 among elder patients with lung cancer (Sinha et al. 2021). Moreover, it has also reported that the highly elevated ACE2 levels seen in the lungs of individuals who are regular smokers may explain why smokers, or patients with smoking-related chronic obstructive pulmonary disease (COPD) or lung cancer, are especially sensitive to adverse outcomes from COVID-19 (e.g., Bakouny et al. 2020; Sinha et al. 2021).
Another potential point of connection between cancer and COVID-19 involves TMPRSS2 protein, the other entry factor of SARS-CoV-2 internalization (Hoffmann et al. 2020). Insights into this link have been derived from research in prostate cancer, where TMPRSS2 protein is present in high levels. Research has revealed that TMPRSS2 is not only highly altered in prostate cancer cells, as demonstrated by Stopsack et al. (2020), but it is also regulated by androgen—a hormone that signals the cell to grow and develop male characteristics. As androgen-targeting therapies are already in use for treating prostate cancer, TMPRSS2 has become an attractive target for potential COVID-19 therapies (Montopoli et al. 2020). Therefore, there are ongoing clinical studies to assess whether therapeutics that lower TMPRSS2 levels or inhibit its function could alleviate COVID-19 symptoms (Zong et al. 2021).
Moreover, researchers have gained critical insights into the interplay between the biology of cancer and COVID-19 from other active research areas, including immune interactions, arterial and venous thrombotic events, and reprogrammed metabolism, which are all associated with the currently accepted hallmarks of cancer (discussed in the next sessions and see Fig. 1).
Immune interactions between cancer and COVID-19
Important research discoveries in cancer immunology, such as the immune system's response to cancer cells and the mechanisms used by tumors to evade the immune system, have provided crucial insights into the immune response to COVID-19 (Costello et al. 1999; Seliger 2005; Derosa et al. 2020). These findings have uncovered significant dysregulation in multiple components of the immune system and provided further data supporting the biological link between COVID-19 and cancer, as recently reviewed (e.g., Bakouny et al. 2020; Zong et al. 2021; American Association for Cancer Research 2022).
As mentioned earlier, evading immune destruction is a crucial hallmark of cancer (Hanahan and Weinberg 2011). Unfortunately, both cancer cells and SARS-CoV-2 can avoid the immune system by suppressing the host's defense mechanisms. The immune system can be suppressed by lowering levels of major histocompatibility complex (MHC) proteins, which are required to flag virus-infected cells or cancer cells for recognition and elimination by immune cells, among other factors (Derosa et al. 2020). Another mechanism that is also able to suppress the immune system is known as “T-cell exhaustion,” which results in reduced numbers of T-cells. This can occur when T cells are persistently stimulated due to chronic inflammation, which is often associated with cancer or long-term infection (Coussens and Werb 2002). These exhausted T-cells are also seen in patients with severe COVID-19. Furthermore, overlapping hyper-inflammatory reactions and arterial and venous thrombotic events have been linked to immune system dysfunction, as outlined below.
Overlapping hyperinflammatory reactions in severe COVID-19 and in cancer patients treated with CAR T-cell therapy:
Cytokine release syndrome (CRS) is an immune system dysfunction with uncontrolled production of pro-inflammatory cytokines and chemokines and manifest as “cytokine storm.” This serious life-threating condition is frequently seen in patients with severe COVID-19. CRS is similar to the known adverse event seen for certain cancer patients treated with CAR T-cell therapy (Turnquist et al. 2020; Moore and June 2020). Recently, shared inflammatory pathways (Iovino et al. 2021) and common biomarkers have been identified that are characteristic for both COVID-19 and CAR T-cell therapy-induced CRS, such as IL-6 (e.g., Turnquist et al. 2020; Zong et al. 2021). These similarities between the mechanisms of CRS in cancer patients treated with CAR T-cell therapy and severe COVID-19 led to repurposing of “immunotherapy-inspired” agents to reduce the cytokine release in COVID-19 (e.g., Agarwal and June 2020).
Arterial and venous thrombotic events in the interplay between SARS-CoV-2 and cancer biology:
It is demonstrated that development of thrombotic events, at least in part, may be related to inflammation and the activation of the innate immune system, which can activate systemic coagulation pathways (Engelmann and Massberg 2013; Connors and Levy 2020; Bakouny et al. 2020). Understanding the effects of COVID-19 on blood vessels, blood, and immune system is another important area of research because an overactive inflammatory response and abnormal blood clotting (a prominent coagulation disorder) are important factors in severe COVID-19. Moreover, these coagulation disorders are associated with the formation of new blood vessels in the patient’s lungs. Discussed before that abnormal vasculature known as pathological neoangiogenesis is another hallmark of solid tumors (Hanahan and Weinberg 2011). It is well documented that patients with cancer are at somewhat elevated risk of thrombotic complications, particularly those patients who are complicated with both cancer and COVID-19 (Lee and Levine 2003). Recently, it has been shown that several proteins that are related to tumor angiogenesis, such as VEGF, HIF and others are also elevated in the plasma or lungs of patients with COVID-19. These data are suggested to be considered in utilization of antiangiogenic cancer therapeutics for the treatment of COVID-19 (Smadja et al. 2021).
Parallels and intersections in cancer cell metabolism with COVID-19
Complexity of malignant phenotype includes metabolic reprogramming of cancer cells,—either as a consequence or as a cause what is an essential hallmark feature of cancer (Kroemer and Pouyssegur 2008; Hanahan and Weinberg 2011). It is demonstrated that multiple alterations in signal transduction pathways and/or enzymatic machineries are responsible for metabolic reprogramming of cancer cells that serve to the advantage of these transformed cells (Kroemer and Pouyssegur 2008). Recently, evidence was provided for the direct link of SARS‑CoV-2 infection with Warburg effect/elevated glucose levels in association with inflammatory response, exploring that SARS‑CoV‑2 infection can elicit metabolic reprogramming in tumor cells and perhaps also tumor progression (Icard et al. 2021; Codo et al. 2020).
The elevated activity of guanylate synthetic pathways of cancer cells is largely due to increased activity of inositide-5’-monophosphate dehydrogenase (EC 1.1.1.205) (IMPDH), the rate-limiting enzyme of GTP biosynthesis. It was suggested and was proven by Weber that IMPDH is a sensitive and selective target for enzyme-pattern-targeted chemotherapy (Weber 1983). Here, we briefly present the impact of three clinically significant inhibitors of IMPDH (Tiazofurin, Ribavirin and Mycophenolic acid), on metabolic reprogramming and its links with other capabilities (later nominated as hallmarks) of cancer. It is demonstrated that targeting of the deregulated de novo purine biosynthesis leads to a strong anti-proliferative effect with restoring of normal cell differentiation and apoptosis program through inhibiting of IMPDH activity/depleting GTP production, and down-regulation of gene expression programs important for proliferation and adaptive survival of cancer cells (Oláh et al. 1988, 2006). Early studies with Tiazofurin provided the first evidence that the proliferative arrest, the differentiation toward the erythroid pathway and the apoptosis of BCR-ABL positive, K562 human leukemia cells, induced by strong inhibition of IMPDH, are mediated, at least in parts, by the down-regulation of the K-RAS and by the profound down-regulation of c-MYC oncogenes (Oláh et al. 1990). Tiazofurin also exhibited remarkable anti-metastatic activity (a hallmark of cancer progression) in different human and experimental tumors (Timar et al. 1996). Moreover, it has demonstrable anti-leukemia effects, when used to treat patients with BCR-ABL positive acute myelogenous leukemias, which are resistant to standard anti-leukemia chemotherapy (Tricot et al. 1990). Ribavirin, a potent anti-viral drug with a wide anti-viral spectrum (Sidwell et al. 1972) that is clinically used for hepatitis C-related liver diseases, such as hepatocellular carcinoma. Some data on its anti-cancer activity, were also reported, in human cancer cell lines (Kökény et al. 2009). It was demonstrated, that treatment with Ribavirin—similarly to Tiazofurin, caused a significant growth inhibition via activating the differentiation and more profoundly, the apoptosis pathway, in response to sub-toxic and toxic doses of the drug, respectively. Mycophenolic acid, by inhibiting de novo purine biosynthesis, functions as an antifungal, antibacterial, antiviral, and immunosuppressive agent and used to prevent rejection following organ transplantation and to treat autoimmune conditions (Bentley 2000). Interestingly, but not surprisingly, HL-60 promyelocytic leukemia cells treated with sub-toxic and toxic doses of mycophenolic acid, respectively, also undergo terminal differentiation and apoptosis (Görtz et al. 1997).
Quite early in the pandemic, IMPDH inhibitors—among many other anticancer/antiviral agents—were repurposed to assess their impact against SARS-CoV-2. Preliminary data showed that Ribavirin can suppress the replication of SARS‑CoV‑2 at a low molar concentration (Elfiky 2020). However, the efficacy of various agents, including ribavirin, hydroxychloroquine, lopinavir/ritonavir that were initially proclaimed to be effective against SARS-CoV-2, was later disproved (Drożdżal et al. 2021). Also, there are data for the activity of mycophenolic acid against SARS-CoV-2 (Kato et al. 2020; Han et al. 2021), however, further in-depth studies would be needed to assess the real impact of the inhibitors of IMPDH against SARS-CoV-2.
It is demonstrated above, that targeting deregulated cellular metabolism of cancer with clinically significant selective inhibitors offers an efficient strategy to uncover all cancer hallmarks that are intertwined with altered cancer cell-intrinsic metabolism.
Recent studies also demonstrate that cancer cell metabolism links epigenetic modifications to transcriptional regulation through modulation of gene expression programs important for proliferation and adaptive survival of cancer cells (Morrison et al. 2022). The close link of metabolic and epigenetic pathways in cancer cells may provide further opportunities to therapeutically target malignant growth via the hallmark features of cancer (Morrison et al. 2022).
Transposable elements in cancer and COVID-19
Transposable elements (TEs) are mobile DNA sequences that constitute about 45% of the human genome. TEs are important for genome plasticity and evolution as they can create new genes and functions. However, TE activation can contribute to cancer and other human diseases (Solyom and Kazazian 2012; Grundy et al. 2022). TE insertion in human genomes may cause genetic dysfunction and alteration of gene expression, contributing to plasticity, a hallmark of cancer (Grundy et al. 2022). A recent report suggests that TEs (transposases) may play a role in SARS-CoV-2’s ability to evade the host immune system, indicating that TEs could be potential drug targets for COVID-19 in gene therapies (Sorek et al. 2022). These findings suggest that TEs, the understudied residents of our genome, could uncover further interactions between COVID-19 and cancer, serving as potential drug targets for both diseases.
Messenger RNA (mRNA): from a highly immunogenic molecule to mRNA-based vaccines and therapies
mRNA was first discovered in 1961 (Brenner et al 1961). A further 60 years passed until finally the first mRNA became a FDA-approved product in the form of COVID-19 mRNA vaccine (Food and Drug Administration FDA 2021). During those years, however, particularly during the past more than three decades, a lot of progress has been made toward mRNA-based therapies and vaccination by hundreds of scientists, including cancer researchers. Due to the extensive literature, the last part of this review briefly summarizes the key discoveries and breakthrough events that contributed to the rise of mRNA technology in vaccinology and cancer therapy, giving the milestones in Table 1.
In 1984, the first successful in vitro synthesis of mRNA and its subsequent translation into functional proteins were accomplished, enabling the production of large amounts of protein from any cDNA clone (Melton et al. 1984; Krieg and Melton 1984). Initially, in the early 1990s, in vitro-transcribed (IVT) mRNA was mainly used for vaccines by ex vivo pulsing of dendritic cells. In 1990, reporter gene mRNAs were injected into mice, and protein production was successfully detected, marking the first successful use of IVT mRNA to produce protein in animals (Wolff et al. 1990). RNA technology was initially motivated by the fight against cancer, and the first RNA cancer vaccines were published more than 25 years ago (Conry et al. 1995; Zhou et al. 1995; Boczkowski et al. 1996).
IVT mRNA encoding therapeutic proteins or viral antigens had great potential for treating or preventing various diseases, but for years the innate immunogenicity and instability of mRNA hampered its medical use.
Development of non-inflammatory, nucleoside-modified mRNA for therapy and vaccine
Katalin Karikó Hungarian biochemist began her pioneering work with mRNA in the mid-1980s, and generated functional, highly processed proteins from IVT mRNA transfected into cultured cells, with the goal of developing mRNA encoding therapeutic proteins that has the potential for treating various diseases (Karikó et al. 1998, 1999).
In the late 1990s, Karikó continued studying IVT mRNA and began uncovering the mechanisms of its inflammatory activity with Drew Weissman, American immunologist at the Perelman School of Medicine, University of Pennsylvania. Over the following years, they identified innate immune receptors, including TLR3, TLR7 and TLR8 that responded to the transfected mRNA (Karikó et al. 2004).
Based on their interesting findings that tRNAs (known to contain many modified nucleosides) were non-immunogenic, Karikó and Weissman developed and tested a hypothesis that nucleoside modification of mRNA avoids activation of RNA sensors and inflammation. They achieved a monumental breakthrough when they successfully warded off the inherent immunogenic response of mRNA and improved translational efficiency, by incorporating naturally-occurring modified nucleosides, including pseudouridine into the mRNA, so thus preventing activation of RNA sensors (TLR7 and TLR8) (Karikó et al. 2005). In parallel studies, Karikó and colleagues demonstrated that uridine in mRNA was responsible for triggering the immune reaction and most importantly, also demonstrated that exchanging the uridine to pseudouridine in the mRNA resulted in an optimal transcript that is non-inflammatory, more stable and translates very efficiently, thus making pseudouridine-containing mRNA an ideal platform for delivering therapeutic proteins (Karikó et al. 2008). They also invented a purification procedure to remove contaminating double-stranded byproducts from the IVT mRNA that further increased the translational capacity of the mRNA and eliminated its remaining immunogenicity by avoiding activation of TLR3, showing that the nucleoside-modified mRNA encoded proteins were functional, thus enabling the use of mRNA for therapy (Karikó et al. 2011). Moreover, utilizing the discovery, that lipid nanoparticles (LNPs) are appropriate carriers for mRNA as RNA vaccines (Geall et al. 2012), they formulated a potential mRNA vaccine with LNP (Pardi et al. 2015).
Anti-SARS-CoV-2 mRNA vaccines
Patent coinvented by Katalin Karikó and Drew Weissman on nucleoside-modified mRNA (Karikó K and Weissman D Issued patents 2012–2019) were used to create the anti-SARS-CoV-2 mRNA vaccines, BNT162b2 by BioNTech/Pfizer and mRNA-1273 by Moderna/NIH. When the SARS-CoV-2 pandemic emerged, biotechnology companies BioNTech and Moderna immediately jumped to action, testing nucleoside-modified mRNA vaccine against the virus in mice (Laczko et al. 2020) and soon after in clinical trials (Sahin et al. 2020).
While the COVID mRNA vaccines have rolled out in an unprecedentedly short time: researchers have gone from identification of SARSCoV-2, the causative agent of disease to mass production of multiple vaccines, in less than 1 calendar year, but the achievements began decades earlier, in the devoted work of hundreds of scientists—including that of cancer researchers on mRNA (e.g., Weissman 2015; Kwok et al. 2021), (Table 1). mRNA experts now consider pseudouridine an essential component of the mRNA technology, as the chemical modification of selected nucleotides was the groundwork that solved the two most concerning issues in mRNA, immunogenicity and stability (Sahin et al. 2014; Weissman 2015).
Many different vaccine platforms against SARS- CoV-2 have been developed, but the mRNA vaccines developed with the nucleoside-modified mRNA technology of Karikó and Weissman (Karikó et al. 2005) formulated with LNPs rivals the best available conventional vaccine manufacture methods, as it is more safety, more efficient and much more reproducible vaccine as compared to the use of other (subunit, killed and live attenuated virus, as well as DNA-based) vaccines (Pardi et al. 2018). Priority COVID-19 vaccination began in patients with cancer, the largest risk population, during the pandemic (Ribas et al. 2021), confirming the results of the early studies that COVID-19 vaccines are effective in most patients with cancer, with few to no side effects (Sahin et al. 2020).
Applications of mRNA technology
The rapid success of mRNA-based COVID-19 vaccines has highlighted the vast potential of mRNA technology (Polack et al. 2020; Baden et al. 2021). Accumulated knowledge and technical advancements have led to the emergence of various mRNA-based applications in many branches of medicine, including cancer, infectious diseases, autoimmune diseases, and regenerative medicine. These applications encompass viral vaccines, protein replacement and supplementation therapies, cancer immunotherapies, cell-based therapies, and reprogramming strategies (e.g., Weissman 2015; Chabanovska et al. 2021). Protein replacement and supplementation therapies and vaccination approaches have been developed primarily for cancer and infectious diseases.
mRNA cancer vaccines and other cancer immunotherapies
Vaccination is a highly effective therapeutic modality that has saved countless lives every year, with preventive vaccines being particularly successful (World Health Organization 2022). Most cancer vaccines are therapeutic and aim to stimulate cell-mediated responses capable of clearing or reducing tumor burden (Coulie et al. 2014). The goals of cancer and antiviral vaccines are similar, as they aim to elicit potent antigen-specific immunity. However, while antiviral vaccines generate neutralizing antibodies against exposed virion proteins, antitumor immunity depends on T-cell reactivity against HLA-restricted tumor antigens, making their identification critical (Coulie et al. 2014; Kwok et al. 2021). Earlier research on RNA vaccines was primarily focused on cancer treatment, and the response to the COVID-19 pandemic has leveraged data from years of preclinical and clinical trials exploring mRNA vaccines for cancer treatment (Dolgin 2021; see Table 1).
Despite the formidable barriers presented by the genomic and phenotypic heterogeneity of cancer, as well as the immune-dampening effects of cancer cells, significant progress has been made in the development of cancer vaccines, including mRNA-based cancer vaccines, over the past few years (e.g., Pardi et al. 2018; Kwok et al. 2021).
Advanced immunotherapies, such as immune checkpoint inhibitors and CAR T-cell therapy, have led to remarkable treatments for previously untreatable or refractory cancers (Ribas and Wolchok 2018; Schuster et al. 2017). The success of mRNA-based COVID-19 vaccines has reignited interest in mRNA-based cancer immunotherapies. Recent advancements in next-generation sequencing (NGS), such as whole-exome and transcriptome sequencing, have facilitated the precise identification of immunogenic epitopes for each tumor by integrating mutational and gene expression data obtained from NGS (Wells et al. 2020). This has significantly improved the rational design of neoepitope cancer vaccines in a patient-specific manner. Enhancing cell-mediated immunity through the use of cancer vaccines is considered one of the most promising options for modern cancer immunotherapy, nevertheless, several challenges in mRNA vaccine immunogenicity and efficacy still need to be addressed (Bidram et al. 2021).
In recent years, mRNA-based drug discovery in cancer immunotherapy has advanced significantly (Di Trani et al. 2022), leading to the enrollment of patients in various clinical trials exploring the efficacy of mRNA-based cancer vaccine therapies. Early results from these trials, both as monotherapy and in combination with checkpoint inhibitors, have been encouraging (Lorentzen et al. 2022; ClinicalTrials.gov). Importantly, these mRNA-based immunotherapies have shown promise in treating previously untreatable diseases, including stage IV non-small cell lung cancer (Papachristofilou et al. 2019), unresectable/advanced solid tumors (Haanen et al. 2022), and glioblastomas (Meister et al. 2022). Promising results have also been seen with advanced melanoma (De Keersmaecker et al. 2020), highlighting the potential of mRNA-based cancer vaccines in treating various types of cancer.
Concluding remarks
Karikó’s and Weissman’s discovery in the field of mRNA vaccines and therapeutics was a groundbreaking achievement that has revolutionized the delivery of effective and safe vaccines, therapeutics, and gene therapies across the spectrum of medicine. Katalin Karikó has already received numerous prestigious prizes, including the Breakthrough Prizes and the Lasker award, for her seminal contributions in mRNA field. The limitless opportunities of the mRNA platform and the immense interest in it from both academic communities and mRNA companies, together predict that the novel mRNA-based therapeutics, including vaccines, are poised to become a key strategy in cancer treatment in the near future.
References
Agarwal S, June CH (2020) Harnessing CAR T-cell insights to develop treatments for hyperinflammatory responses in patients with COVID-19. Cancer Discov 10:775–778. https://doi.org/10.1158/2159-8290.CD-20-0473
Alberer M, Gnad-Vogt U, Hong HS, Mehr KT, Backert L, Finak G et al (2017) Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet 390(10101):1511–1520. https://doi.org/10.1016/S0140-6736(17)31665-3
American Association for Cancer Research (2022). AACR Report on the Impact of COVID-19 on Cancer Research and Patient Care. https://www.AACR.org/COVIDReport
Anderson JL, May HT, Knight S, Bair TL, Muhlestein JB, Knowlton KU, Horne BD (2021) Association of sociodemographic factors and blood group type with risk of COVID-19 in a US population. JAMA Netw Open 4:e217429. https://doi.org/10.1001/jamanetworkopen.2021.7429
Andreakos E, Abel L, Vinh DC, Kaja E, Drolet BA, Zhang Q, Human Genetic Effort COVID, Su HC, Casanova JL, Spaan AN et al (2022) A global effort to dissect the human genetic basis of resistance to SARS-CoV-2 infection. Nat Immunol 23(2):159–164. https://doi.org/10.1038/s41590-021-01030-z
Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R et al (2021) Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med 384:403–416. https://doi.org/10.1056/NEJMoa2035389
Bakouny Z, Hawley Jessica E, Choueiri TK, Peters S, Rini B, Warner JL, Painter CA (2020) COVID-19 and Cancer: current challenges and perspectives. Cancer Cell 38:629–646. https://doi.org/10.1016/j.ccell.2020.09.018
Barnkob MB, Pottegard A, Stovring H, Haunstrup TM, Homburg K, Larsen R et al (2020) Reduced prevalence of SARS-CoV-2 infection in ABO blood group O. Blood Adv 4:4990–4993. https://doi.org/10.1182/bloodadvances.2020002657
Baylin SB, Belinsky SA, Herman JG (2000) DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet 16(4):168–174. https://doi.org/10.1016/s0168-9525(99)01971-x
Bentley R (2000) Mycophenolic Acid: a one hundred year odyssey from antibiotic to immunosuppressant. Chem Rev 100:3801–3826. https://doi.org/10.1021/cr990097b
Bidram M, Zhao Y, Shebardina NG, Baldin AV, Bazhin AV, Ganjalikhany MR et al (2021) mRNA-based cancer vaccines: a therapeutic strategy for the treatment of melanoma patients. Vaccines (basel) 9:1060. https://doi.org/10.3390/vaccines9101060
Boczkowski D, Nair SK, Snyder D, Gilboa E (1996) Dendritic cells pulsed with RNA are potent antigen-presenting cells in vitro and in vivo. J Exp Med 184:465–472. https://doi.org/10.1084/jem.184.2.465
Breast Cancer Association Consortium, Dorling L, Carvalho S, Allen J, González-Neira A, Luccarini C, Wahlström C et al (2021) Breast cancer risk genes—association analysis in more than 113,000 women. N Engl J Med 384(5):428–439. https://doi.org/10.1056/NEJMoa1913948
Brenner S, Jacob F, Meselson M (1961) An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature 190:576–581. https://doi.org/10.1038/190576a0
Buitrago-Garcia D, Egli-Gany D, Counotte MJ, Hossmann S, Imeri H, Ipekci AM et al (2020) Occurrence and transmission potential of asymptomatic and presymptomatic SARS-CoV-2 infections: a living systematic review and meta-analysis. PLoS Med 17:e1003346. https://doi.org/10.1371/journal.pmed
Callaway E (2021) The quest to find genes that drive severe COVID. Nature 595:346–348. https://doi.org/10.1038/d41586-021-01827-w
Cavallo F, De Giovanni G, Nanni P, Forni G, Lollini P-L (2011) The immune hallmarks of cancer. Cancer Immunol Immunother 60:319–326. https://doi.org/10.1007/s00262-010-0968-0
Centers for Disease Control and Prevention (2022) People with certain medical conditions [updated Nov 24, 2021]. https://www.cdc.gov/coronavirus/2019-ncov/needextra-precautions/people-with-medical-conditions.html/. Accessed 24 July 2022.
Chabanovska O, Galow AM, David R, Lemcke H (2021) mRNA—a game changer in regenerative medicine, cell-based therapy and reprogramming strategies. Adv Drug Deliv Rev 179:114002. https://doi.org/10.1016/j.addr.2021.114002
Chenevix-Trench G, Milne RL, Antoniou AC, Couch FJ, Easton DF, Goldgar DE (2007) CIMBA. An international initiative to identify genetic modifiers of BRCA1 and BRCA2: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA). Breast Cancer Res 9(2):104. https://doi.org/10.1186/bcr1670
ClinicalTrials.gov: https://clinicaltrials.gov
Codo AC, Davanzo GG, Monteiro LB, de Souza GF, Muraro SP et al (2020) Elevated glucose levels favor SARS-CoV-2 infection and monocyte response through a HIF-1α/glycolysis-dependent axis. Cell Metab 32(3):437–446. https://doi.org/10.1016/j.cmet.2020.07.007
Connors JM, Levy JH (2020) COVID-19 and its implications for thrombosis and anticoagulation. Blood 135:2033–2040. https://doi.org/10.1182/blood.2020006000
Conry RM, LoBuglio AF, Wright M, Sumerel L, Pike MJ, Johanning F et al (1995) Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res 55:1397–1400
Costello RT, Gastaut JA, Olive D (1999) Tumour escape from immune surveillance. Arch Immunol Ther Exp (warsz) 47(2):83–88
Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T (2014) Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer 14:135–146. https://doi.org/10.1038/nrc3670
Coussens LM, Werb Z (2002) Inflammation and cancer. Nature 420:860–867. https://doi.org/10.1038/nature01322
Couzin-Frankel J (2013) Breakthrough of the year 2013. Cancer Immunother Sci 342(6165):1432–1433. https://doi.org/10.1126/science.342.6165.1432
Cullis PR, Hope MJ (2017) Lipid nanoparticle systems for enabling gene therapies. Mol Ther 25(7):1467–1475. https://doi.org/10.1016/j.ymthe.2017.03.013
Dai M, Liu D, Liu M, Zhou F, Li G, Chen Z et al (2020) Patients with cancer appear more vulnerable to SARS‑CoV‑2. A multicenter study during the COVID‑19 outbreak. Cancer Discov 10:783–791. https://doi.org/10.1158/2159-8290.CD-20-0422
De Keersmaecker B, Claerhout S, Carrasco J, Bar I, Corthals J, Wilgenhof S et al (2020) TriMix and tumor antigen mRNA electroporated dendritic cell vaccination plus ipilimumab: link between T-cell activation and clinical responses in advanced melanoma. J Immunother Cancer 8:e000329. https://doi.org/10.1136/jitc-2019-000329
Derosa L, Melenotte C, Griscelli F, Gachot B, Marabelle A, Kroemer G, Zitvogel L (2020) The immuno-oncological challenge of COVID-19. Nat Cancer 1:946–964. https://doi.org/10.1038/s43018-020-00122-3
Di Trani CA, Fernandez-Sendin M, Cirella A, Segués A, Olivera I, Bolaños E et al (2022) Advances in mRNA-based drug discovery in cancer immunotherapy. Expert Opin Drug Discov 17:41–53. https://doi.org/10.1080/17460441.2021.1978972
Dolgin E (2021) The tangled history of mRNA vaccines. Nature 597:318–324. https://doi.org/10.1038/d41586-021-02483-w
Drożdżal S, Rosik J, Lechowicz K, Machaj F, Szostak B, Przybyciński J et al (2021) An update on drugs with therapeutic potential for SARS-CoV-2 (COVID-19) treatment. Drug Resist Update 59:100794. https://doi.org/10.1016/j.drup.2021.100794
Elfiky AA (2020) Ribavirin, remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci 253:117592. https://doi.org/10.1016/j.lfs.2020.117592
Elkrief A, Wu JT, Jani C, Enriquez KT, Glover M, Shah MR et al (2022) Learning through a pandemic: the current state of knowledge on COVID-19 and cancer. Cancer Discov 12(2):303–330. https://doi.org/10.1158/2159-8290.CD-21-1368
Engelmann B, Massberg S (2013) Thrombosis as an intravascular effector of innate immunity. Nat Rev Immunol 13:34–45. https://doi.org/10.1038/nri3345
Fearon ER, Vogelstein B (1990) A genetic model for colorectal tumorigenesis. Cell 61:759–767. https://doi.org/10.1016/0092-8674(90)90186-i
Ferreira MA (2021) Genome-wide analysis in 756,646 individuals provides first genetic evidence that ACE2 expression influences COVID-19 risk and yields genetic risk scores predictive of severe disease. medRxiv. 2020.12.14.20248176. https://doi.org/10.1101/2020.12.14.20248176. Preprint.
Food and Drug Administration (FDA). News release (2021) FDA approves first COVID-19 vaccine. Approval signifies key achievement for public health. https://www.fda.gov/news-events/press-announcements/fda-approves-first-covid-19-vaccine
Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR et al (2004) A census of human cancer genes. Nat Rev Cancer 4(3):177–183. https://doi.org/10.1038/nrc1299
Geall AJ, Verma A, Otten GR, Shaw CA, Hekele A, Banerjee K et al (2012) Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci USA 109:14604–14609. https://doi.org/10.1073/pnas.1209367109
Global Cancer Observatory: Cancer Today. Lyon: International Agency for Research on Cancer. Ferlay J, Ervik M, Lam F, Colombet M, Mery L, Piñeros M, et al. (2020) https://gco.iarc.fr/today. Accessed February 2021
Görtz A, Franklin TJ, Dive C, Hickman JA (1997) Cell cycle specific induction of HL-60 cell differentiation and apoptosis by mycophenolic acid. Cell Death Differ 4:787–795. https://doi.org/10.1038/sj.cdd.4400300
Grundy EE, Diab N, Chiappinelli KB (2022) Transposable element regulation and expression in cancer. FEBS J 289:1160–1179. https://doi.org/10.1111/febs.15722
Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS et al (2020) Extrapulmonary manifestations of COVID-19. Nat Med 26(7):1017–1032. https://doi.org/10.1038/s41591-020-0968-3
Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, King MC (1990) Linkage of early-onset familial breast cancer to chromosome 17q21. Science 250(4988):1684–1689. https://doi.org/10.1126/science.2270482
Haanen J, Mackensen A, Koenecke C, Alsdorf W, Wagner-Drouet E, Heudobler D et al (2022) BNT211: a phase I trial to evaluate safety and efficacy of CLDN6 CAR-T cells and CARVac-mediated in vivo expansion in patients with CLDN6-positive advanced solid tumors. Cancer Res Proc Am Assoc Cancer Res 82(12):CT002
Han Y, Duan X, Yang L, Nilsson-Payant BE, Wang P, Duan F, Tang X, Yaron TM et al (2021) Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589:270–275. https://doi.org/10.1038/s41586-020-2901-9
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100:57–70. https://doi.org/10.1016/S0092-8674(00)81683-9
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674. https://doi.org/10.1016/j.cell.2011.02.013
Hanahan D (2022) Hallmarks of cancer: new dimensions. Cancer Discov 12:31–46. https://doi.org/10.1158/2159-8290.CD-21-1059
Hartenian E, Nandakumar D, Lari A, Ly M, Tucker JM, Glaunsinger BA (2020) The molecular virology of coronaviruses. J Biol Chem 295(37):12910–12934. https://doi.org/10.1074/jbc.REV120.013930
Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S et al (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181(2):271-280.e8. https://doi.org/10.1016/j.cell.2020.02.052
Hou X, Zaks T, Langer R, Dong Y (2021) Lipid nanoparticles for mRNA delivery. Nat Rev Mater 6(12):1078–1094. https://doi.org/10.1038/s41578-021-00358-0
Hou Y, Zhao J, Martin W, Kallianpur A, Chung MK, Jehi L et al (2020) New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med 18(1):216. https://doi.org/10.1186/s12916-020-01673-z
Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD et al (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31(3):227–229. https://doi.org/10.1038/nbt.2501
Icard P, Lincet H, Wu Z, Coquerel A, Forgez P, Alifano M, Fournel L (2021) The key role of Warburg effect in SARS-CoV-2 replication and associated inflammatory response. Biochimie 180:169–177. https://doi.org/10.1016/j.biochi.2020.11.010
International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437(7063):1299–1320. https://doi.org/10.1038/nature04226
International Agency for Research on Cancer (IARC): http://monographs.iarc.fr
International Human Genome Sequencing Consortium (2004) Finishing the euchromatic sequence of the human genome. Nature 431(7011):931–945. https://doi.org/10.1038/nature03001
Iovino L, Thur LA, Gnjatic S, Chapuis A, Milano F, Hill JA (2021) Shared inflammatory pathways and therapeutic strategies in COVID-19 and cancer immunotherapy. J Immunother Cancer 9:e002392. https://doi.org/10.1136/jitc-2021-002392
Karikó K (2022) Lasker-APSA Lecture https://laskerfoundation.org/katalin-kariko-developing-mrna-for-therapy/
Karikó K, Kuo A, Barnathan ES, Langer DJ (1998) Phosphate-enhanced transfection of cationic lipid-complexed mRNA and plasmid DNA. Biochim Biophys Acta 1369:320–334. https://doi.org/10.1016/s0005-2736(97)00238-1
Karikó K, Kuo A, Barnathan E (1999) Overexpression of urokinase receptor in mammalian cells following administration of the in vitro transcribed encoding mRNA. Gene Ther 6(6):1092–1100. https://doi.org/10.1038/sj.gt.3300930
Karikó K, Weissman D Issued patents (2012–2019) RNA Containing modified nucleosides and methods of use thereof U.S. Patent No. 8,278,036 (2012); 8,691,966 (2014); 8,748,089 (2014); 8,835,108 (2014); 9,750,824 (2017); 10,232,055 (2019)
Karikó K, Ni H, Capodici J, Lamphier M, Weissman D (2004) mRNA is an endogenous ligand for Toll-like receptor 3. J Biol Chem 279:12542–12550. https://doi.org/10.1074/jbc.M310175200
Karikó K, Buckstein M, Ni H, Weissman D (2005) Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23(2):165–175. https://doi.org/10.1016/j.immuni.2005.06.008
Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D (2008) Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther 16(11):1833–1840. https://doi.org/10.1038/mt.2008.200
Karikó K, Muramatsu H, Ludwig J, Weissman D (2011) Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res 39:e142. https://doi.org/10.1093/nar/gkr695
Kato F, Matsuyama S, Kawase M, Hishiki T, Katoh H, Takeda M (2020) Antiviral activities of mycophenolic acid and IMD-0354 against SARS-CoV-2. Microbiol Immunol 64:635–639. https://doi.org/10.1111/1348-0421.12828
Koedoot E, Wolters L, van de Water B, Le Dévédec SE (2019) Splicing regulatory factors in breast cancer hallmarks and disease progression. Oncotarget 10:6021–6037. https://doi.org/10.18632/oncotarget.27215
Kökény S, Papp J, Weber G, Vaszko T, Carmona-Saez P, Olah E (2009) Ribavirin acts via multiple pathways in inhibition of leukemic cell proliferation. Anticancer Res 29:1971–1980
Kovács ME, Papp J, Szentirmay Z, Ottó S, Oláh E (2009) Deletions removing the last exon of TACSTD1 constitute a distinct class of mutations predisposing to Lynch syndrome. Hum Mutat 30(2):197–203. https://doi.org/10.1002/humu.20942
Kousathanas A, Pairo-Castineira E, Rawlik K, Stuckey A, Odhams CA, Walker S et al (2022) Whole-genome sequencing reveals host factors underlying critical COVID-19. Nature 607(7917):97–103. https://doi.org/10.1038/s41586-022-04576-6
Kreiter S, Selmi A, Diken M, Koslowski M, Britten CM, Huber C, Türeci O, Sahin U (2010) Intranodal vaccination with naked antigen-encoding RNA elicits potent prophylactic and therapeutic antitumoral immunity. Cancer Res 70(22):9031–9040. https://doi.org/10.1158/0008-5472.CAN-10-0699
Krieg PA, Melton DA (1984) Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res 12:7057–7070. https://doi.org/10.1093/nar/12.18.7057
Kroemer G, Pouyssegur J (2008) Tumor cell metabolis. Cancer’s Achilles’ Heel j ccr. https://doi.org/10.1016/j.ccr.2008.05.005
Kuba K, Imai Y, Rao S, Gao H, Guo F, Guan B et al (2005) A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med 11:875–879. https://doi.org/10.1038/nm1267
Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips KA, Mooij TM, Roos-Blom MJ et al (2017) Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 317(23):2402–2416. https://doi.org/10.1001/jama.2017.7112
Kuenzi BM, Ideker T (2020) A census of pathway maps in cancer systems biology. Nat Rev Cancer 20(4):233–246. https://doi.org/10.1038/s41568-020-0240-7
Kwok M, Fritsch EF, Wu CJ (2021) Cancer and COVID-19: on the quest for effective vaccines. Blood Cancer Discov 2:13–18. https://doi.org/10.1158/2643-3230.BCD-20-0205
Laczko D, Hogan MJ, Toulmin SA, Hicks P, Lederer K, Gaudette BT et al (2020) A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in mice. Immunity 53:724–732. https://doi.org/10.1016/j.immuni.2020.07.019
Lakeman IMM, Hilbers FS, Rodríguez-Girondo M, Lee A, Vreeswijk MPG, Hollestelle A et al (2019) Addition of a 161-SNP polygenic risk score to family history-based risk prediction: impact on clinical management in non-BRCA1/2 breast cancer families. J Med Genet 56(9):581–589. https://doi.org/10.1136/jmedgenet-2019-106072
Lee AYY, Levine MN (2003) Venous thromboembolism and cancer: risks and outcomes. Circulation 107:17I – 21. https://doi.org/10.1161/01.CIR.0000078466.72504.AC
Lee LY, Cazier JB, Angelis V, Arnold R, Bisht V, Campton NA (2020) COVID-19 mortality in patients with cancer on chemotherapy or other anticancer treatments: a prospective cohort study. Lancet 395:1919–1926. https://doi.org/10.1016/S0140-6736(20)31173-9
Liang W, Guan W, Chen R, Wang W, Li J, Xu K et al (2020) Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol 21:335–337. https://doi.org/10.1016/S1470-2045(20)30096-6
Lorentzen CL, Haanen JB, Met Ö, Svane IM (2022) Clinical advances and ongoing trials on mRNA vaccines for cancer treatment. Lancet Oncol 23(10):e450–e458. https://doi.org/10.1016/S1470-2045(22)00372-2
Mardis ER (2008) Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet 9:387–402. https://doi.org/10.1146/annurev.genom.9.081307.164359
Mavaddat N, Pharoah PD, Michailidou K, Tyrer J, Brook MN, Bolla MK et al (2015) Prediction of breast cancer risk based on profiling with common genetic variants. J Natl Cancer Inst 107(5):djv036. https://doi.org/10.1093/jnci/djv036
Meister H, Look T, Roth P, Pascolo S, Sahin U, Lee S (2022) Multifunctional mRNA-Based CAR T cells display promising antitumor activity against glioblastoma. Clin Cancer Res 28:4747–4756. https://doi.org/10.1158/1078-0432.CCR-21-4384
Melton DA, Krieg PA, Rebagliati MR, Maniatis T, Zinn K, Green MR (1984) Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res 12:7035–7056. https://doi.org/10.1093/nar/12.18.7035
Meric-Bernstam F, Farhangfar C, Mendelsohn J, Mills GB (2013) Building a personalized medicine infrastructure at a major cancer center. J Clin Oncol 31(15):1849–1857. https://doi.org/10.1200/JCO.2012.45.3043
Michailidou K, Lindström S, Dennis J, Beesley J, Hui S, Kar S et al (2017) Association analysis identifies 65 new breast cancer risk loci. Nature 551(7678):92–94. https://doi.org/10.1038/nature24284
Montopoli M, Zumerle S, Vettor R, Rugge M, Zorzi M, Catapano CV et al (2020) Androgen-deprivation therapies for prostate cancer and risk of infection by SARS-CoV-2: a population-based study (N = 4532). Ann Oncol 31:1040–1045. https://doi.org/10.1016/j.annonc.2020.04.479
Moore JB, June CH (2020) Cytokine release syndrome in severe COVID-19. Science 368:473–474. https://doi.org/10.1126/science.abb8925
Morrison AJ (2022) Cancer cell metabolism connects epigenetic modifications to transcriptional regulation. FEBS J 289:1302–1314. https://doi.org/10.1111/febs.16032
Nowell PC (1974) The clonal evolution of tumour cell populations. Science 194:23–28. https://doi.org/10.1126/science.959840
Oláh E (2021) A new era in understanding the genetic background of cancer susceptibility. Hungarian Academy of Sciences, Budapest, (Based on Inaugural Lecture, 2013). https://doi.org/10.36820/szekfoglalo.2021.olah (in Hungarian)
Oláh E, Natsumeda Y, Ikegami T, Kote Z, Horanyi M, Szelenyi J et al (1988) Induction of erythroid differentiation and modulation of gene expression by tiazofurin in K-562 leukemia cells. Proc Natl Acad Sci U S A 85(17):6533–6537. https://doi.org/10.1073/pnas.85.17.6533
Oláh E, Kote Z, Natsumeda Y, Yamaji Y, Jarai G, Lapis E, Financsek I, Weber G (1990) Down-regulation of c-myc and c-Ha-ras gene expression by tiazofurin in rat hepatoma cells. Cancer Biochem Biophys 11(2):107–117
Oláh E, Kökény S, Papp J, Bozsik A, Keszei M (2006) Modulation of cancer pathways by inhibitors of guanylate metabolism. Adv Enzyme Regul 46:176–190. https://doi.org/10.1016/j.advenzreg.2006.01.002
Online Mendelian Inheritance in Man®, OMIM®: An Online Catalog of Human Genes and Genetic Disorders http://www.ncbi.nlm.nih.gov/omim/ Accessed 4 May 2022
Orbán TI, Oláh E (2003) Emerging roles of BRCA1 alternative splicing. Mol Pathol 56(4):191–197. https://doi.org/10.1136/mp.56.4.191
Padariya M, Sznarkowska A, Kote S, Gómez-Herranz M, Mikac S, Pilch M et al (2021) Functional interfaces, biological pathways, and regulations of interferon-related DNA damage resistance signature (IRDS) genes. Biomolecules 11(5):622. https://doi.org/10.3390/biom11050622
Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D et al (2021) Genetic mechanisms of critical illness in COVID-19. Nature 591:92–98. https://doi.org/10.1038/s41586-02003065-y
Papachristofilou A, Hipp MM, Klinkhardt U, Früh M, Sebastian M, Weiss C et al (2019) Phase Ib evaluation of a self-adjuvanted protamine formulated mRNA-based active cancer immunotherapy, BI1361849 (CV9202), combined with local radiation treatment in patients with stage IV non-small cell lung cancer. J Immunother Cancer 7:38. https://doi.org/10.1186/s40425-019-0520-5
Pardi N, Tuyishime S, Muramatsu H, Kariko K, Mui BL, Tam YK et al (2015) Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J Control Release 217:345–351. https://doi.org/10.1016/j.jconrel.2015.08.007
Pardi N, Hogan MJ, Porter FW, Weissman D (2018) mRNA vaccines—a new era in vaccinology. Nat Rev Drug Discov 17(4):261–279. https://doi.org/10.1038/nrd.2017.243
Pinato DJ, Zambelli A, Aguilar-Company J, Bower M, Sng C, Salazar R et al (2020) Clinical portrait of the SARS-CoV-2 epidemic in European cancer patients. Cancer Discov 10(10):1465–1474. https://doi.org/10.1158/2159-8290.CD-20-0773
Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S et al (2020) Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 383:2603–2615. https://doi.org/10.1056/NEJMoa2034577
Ponder BA (1990) Inherited predisposition to cancer. Trends Genet 6(7):213–218. https://doi.org/10.1016/0168-9525(90)90181-5
Ponder BAJ (1997) Genetic testing for cancer risk. Science 278:1050–1054. https://doi.org/10.1126/science.278.5340.1050
Ribas A, Wolchok JD (2018) Cancer immunotherapy using checkpoint blockade. Science 359:1350–1355. https://doi.org/10.1126/science.aar4060
Ribas A, Sengupta R, Locke T, Zaidi SK, Campbell KM, Carethers JM et al (2021) Priority COVID-19 vaccination for patients with cancer while vaccine supply is limited. AACR COVID-19 and cancer task force. Cancer Discov 11(2):233–236. https://doi.org/10.1158/2159-8290.CD-20-1817
Rivera DR, Peters S, Panagiotou OA, Shah DP, Kuderer NM, Hsu CY et al (2020) Utilization of COVID-19 treatments and clinical outcomes among patients with cancer: a COVID-19 and Cancer Consortium (CCC19) Cohort Study. Cancer Discov 10:1514–1527. https://doi.org/10.1158/2159-8290.CD-20-0941
Sahin U, Karikó K, Türeci Ö (2014) mRNA-based therapeutics–developing a new class of drugs. Nat Rev Drug Discov 13(10):759–780. https://doi.org/10.1038/nrd4278
Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, Vormehr M et al (2020) COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 586:594–599. https://doi.org/10.1038/s41586-020-2814-7
Schuster SJ, Svoboda J, Chong EA, Nasta SD, Mato AR, Anak Ö et al (2017) Chimeric antigen receptor T cells in refractory B-cell lymphomas. N Engl J Med 377:2545–2554. https://doi.org/10.1056/NEJMoa1708566
Seliger B (2005) Strategies of tumour immune evasion. BioDrugs 19:347–354. https://doi.org/10.2165/00063030-200519060-00002
Senga SS, Grose RP (2021) Hallmarks of cancer—the new testament. Open Biol 11:200358. https://doi.org/10.1098/rsob.20.0358
Severe Covid-19 GWAS Group, Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P (2020) Genome wide association study of severe Covid-19 with respiratory failure. N Engl J Med 383:1522–1534. https://doi.org/10.1056/NEJMoa2020283
Sidwell RW, Huffman JH, Khare GP, Allen LB, Witkowski JT, Robins RK (1972) Broad-spectrum antiviral activity of Virazole: 1-beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science 177(4050):705–706. https://doi.org/10.1126/science.177.4050.705
Singer CF, Balmaña J, Bürki N, Delaloge S, Filieri ME, Gerdes AM et al (2019) Genetic counselling and testing of susceptibility genes for therapeutic decision-making in breast cancer-an European consensus statement and expert recommendations. Eur J Cancer 106:54–60. https://doi.org/10.1016/j.ejca.2018.10.007
Sinha S, Kundu CN (2021) Cancer and COVID-19: why are cancer patients more susceptible to COVID-19? Med Oncol 38:101. https://doi.org/10.1007/s12032-021-01553-3
Smadja DM, Mentzer SJ, Fontenay M, Laffan MA, Ackermann M, Helms J et al (2021) COVID-19 is a systemic vascular hemopathy: insight for mechanistic and clinical aspects. Angiogenesis 24:755–788. https://doi.org/10.1007/s10456-021-09805-6/
Solyom S, Kazazian HH Jr (2012) Mobile elements in the human genome: implications for disease. Genome Med 4:12. https://doi.org/10.1186/gm311
Sorek M, Meshorer E, Schlesinger S (2022) Impaired activation of transposable elements in SARS-CoV-2 infection. EMBO Rep 23(9):e55101. https://doi.org/10.15252/embr.202255101
Soriano JB, Murthy S, Marshall JC, Relan P, Diaz JV (2022) A clinical case definition of post-COVID-19 condition by a Delphi consensus. WHO Clinical Case Definition Working Group on Post-COVID-19 Condition. Lancet Infect Dis 22:e102–e107. https://doi.org/10.1016/S1473-3099(21)00703-9
Stopsack KH, Mucci LA, Antonarakis ES, Nelson PS, Kantoff PW (2020) TMPRSS2 and COVID-19: serendipity or opportunity for intervention? Cancer Discov 10:779–782. https://doi.org/10.1158/2159-8290.CD-20-0451
Tangye SG, Al-Herz W, Bousfiha A, Cunningham-Rundles C, Franco JL, Holland SM et al (2021) The ever-increasing array of novel inborn errors of immunity: an interim update by the IUIS committee. J Clin Immunol 41(3):666–679. https://doi.org/10.1007/s10875-021-00980-1.Review
Tímár J, Tóvári J, Pogány G, Ladányi A, Paku S, Rásó E, Bocsi J, Jeney A, Lapis K (1996) The antimetabolite Tiazofurin (TR) inhibits glycoconjugate biosynthesis and invasiveness of tumour cells. Eur J Cancer 32A(1):152–159. https://doi.org/10.1016/0959-8049(95)00544-7
Tricot G, Jayaram HN, Weber G, Hoffman R (1990) Tiazofurin: biological effects and clinical uses. Int J Cell Cloning 8(3):161–170. https://doi.org/10.1002/stem.5530080303
Turnquist C, Ryan BM, Horikawa I, Harris BT, Harris CC (2020) Cytokine storms in cancer and COVID-19. Cancer Cell 38:598–601. https://doi.org/10.1016/j.ccell.2020.09.019
van Dam PA, Huizing M, Mestach G, Dierckxsens S, Tjalma W, Trinh XB et al (2020) SARS-CoV-2 and cancer: are they really partners in crime? Cancer Treat Rev 89:102068. https://doi.org/10.1016/j.ctrv.2020.102068
Vrdoljak E, Sullivan R, Lawler E (2020) Cancer and coronavirus disease 2019; how do we manage cancer optimally through a public health crisis? Eur J Cancer 132:98–99. https://doi.org/10.1016/j.ejca.2020.04.001
Weber G (1983) Biochemical strategy of cancer cells and the design of chemotherapy: G. H. A. Clowes Memorial Lecture. Cancer Res 43(8):3466–3492
Weide B, Carralot JP, Reese A, Scheel B, Eigentler TK, Hoerr I et al (2008) Results of the first phase I/II clinical vaccination trial with direct injection of mRNA. J Immunother 31(2):180–188. https://doi.org/10.1097/CJI.0b013e31815ce501
Weissman D (2015) mRNA transcript therapy. Expert Rev Vaccines 14:265–281. https://doi.org/10.1586/14760584.2015.973859
Wells DK, van Buuren MM, Dang KK, Hubbard-Lucey VM, Sheehan KCF, Campbell KM et al (2020) Key parameters of tumor epitope immunogenicity revealed through a consortium approach improve neoantigen prediction. Cell 183:818–834. https://doi.org/10.1016/j.cell.2020.09.015
Wilson IA (2020) A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368:630–633. https://doi.org/10.1126/science.abb7269
Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL (1990) Direct gene transfer into mouse muscle in vivo. Science 247:1465–1468. https://doi.org/10.1126/science.1690918
World Health Organization, WHO (2020) Coronavirus disease (COVID-19) pandemic. https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed 22 Aug 2022
World Health Organization. Immunization coverage (2022) http://www.who.int/mediacentre/factsheets/fs378/en
Xu S, Yang K, Li R, Zhang L (2020) mRNA vaccine era—mechanisms, drug platform and clinical prospection. Int J Mol Sci 21:6582. https://doi.org/10.3390/ijms21186582
Yang K, Sheng Y, Huang C, Jin Y, Xiong N, Jiang K et al (2020) Clinical characteristics, outcomes, and risk factors for mortality in patients with cancer and COVID-19 in Hubei, China: a multicentre, retrospective, cohort study. Lancet Oncol 21:904–913. https://doi.org/10.1016/S1470-2045(20)30310-7
Yi C, Sun X, Ye J, Ding L, Liu M, Yang Z, Lu X et al (2020) Key residues of the receptor binding motif in the spike protein of SARS-CoV-2 that interact with ACE2 and neutralizing antibodies. Cell Mol Immunol 17:621–630. https://doi.org/10.1038/s41423-020-0458-z
Zeberg H, Pääbo S (2020) The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature 587:610–612. https://doi.org/10.1038/s41586-020-2818-3
Zeberg H, Pääbo S (2021) A genomic region associated with protection against severe COVID-19 is inherited from Neandertals. Proc Natl Acad Sci USA 118:e2026309118. https://doi.org/10.1073/pnas.2026309118
Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J et al (2020a) Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370:eabd4570. https://doi.org/10.1126/science.abd4570
Zhang YD, Hurson AN, Zhang H, Choudhury PP, Easton DF, Milne RL (2020b) Assessment of polygenic architecture and risk prediction based on common variants across fourteen cancers. Nat Commun 11(1):3353. https://doi.org/10.1038/s41467-020-16483-3
Zhang Y, Yu C (2020) Prognostic characterization of OAS1/OAS2/OAS3/OASL in breast cancer. BMC Cancer 20:575. https://doi.org/10.1186/s12885-020-07034-6
Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B (2020) Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395:1054–1062. https://doi.org/10.1016/S0140-6736(20)30566-3
Zhou WZ, Hoon DS, Huang SK, Fujii S, Morishita HKR et al (1995) RNA melanoma vaccine: induction of antitumor immunity by human glycoprotein 100 mRNA immunization. Hum Gene Ther 10:2719–2724. https://doi.org/10.1089/10430349950016762
Zhou S, Brunet-Ratnasingham E, Henry D, Kimchi N, Afrasiabi Z et al (2021) A Neanderthal OAS1 isoform protects individuals of European ancestry against COVID-19 susceptibility and severity. Nat Med 27:659–667. https://doi.org/10.1038/s41591-021-01281-1
Zong Z, Wei Y, Ren J, Zhang L, Zhou F (2021) The intersection of COVID-19 and cancer: signaling pathways and treatment implications. Mol Cancer 20(1):76. https://doi.org/10.1186/s12943-021-01363-1Review
zur Hausen H (2008) Papillomaviruses–to vaccination and beyond. Biochemistry (mosc) 73(5):498–503. https://doi.org/10.1134/s0006297908050027.Review
Acknowledgements
This work was supported in part by the Hungarian Research Grants KTIA-OTKA CK80745, NKFIH-OTKA K112228, and from the National Research, Development and Innovation Fund Hungary – Befektetés a jövőbe: NKFIH-2020-1.1.6-Jövő-2021-00008.
Funding
Open access funding provided by National Institute of Oncology.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Oláh, E. Learning from cancer to address COVID-19. BIOLOGIA FUTURA 74, 29–43 (2023). https://doi.org/10.1007/s42977-023-00156-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42977-023-00156-5