Abstract
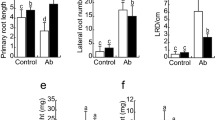
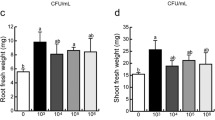
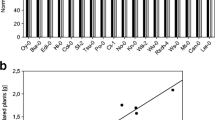
To evaluate if root architecture changes observed in Arabidopsis thaliana inoculated with Azospirillum argentinense Az39 depend exclusively on the bacterial capacity to produce indole-3-acetic acid (IAA) and plant ability to sense IAA levels. Azospirillum argentinense Az39, A. argentinense Az39 ipdC–, flagellin from A. argentinense Az39, and pure IAA were applied to A. thaliana Col-0 (wild-type) and tir1.1 (a lateral root deficient mutant) seedlings. Inoculation with heat-inactivated A. argentinense Az39 cells and a non-PGPR bacterium (Escherichia coli DH5α) was also tested. The primary root (PR) length, lateral roots (LR) number, and root hair (HR) density were assessed, and the root transcriptome was sequenced (Illumina HiSeq), followed by DEGs and GO term enrichment analyses. Inoculation with both A. argentinense strains resulted in a shorter PR and an increased number of LR and RH. IAA application (0.1 µM) led to a similar root phenotype than inoculation with Az39 (108 CFU mL−1). The addition of 1 µM flagellin, as well as plant exposure to non-lysed A. argentinense Az39 or E. coli DH5α cells, enhanced RH formation. Genes related to auxin signaling were highly expressed in the roots of Az39-inoculated seedlings; genes related to jasmonate and salicylic acid metabolism were highly expressed in the roots of plants inoculated with ipdC − . Root architecture changes in A. thaliana inoculated with A. argentinense Az39 do not depend exclusively on root IAA levels/IAA plant perception. This PGPR induces root morphological changes through both IAA-dependent and IAA-independent mechanisms. Flagellin may be a key molecule involved in IAA-independent mechanisms.





Similar content being viewed by others
Data Availability
All RNA sequencing data have been deposited at the Gene Expression Omnibus (GEO): GSE192383. All other relevant data can be found within the manuscript and its supporting materials.
References
Adu MO, Asare PA, Yawson DO, Ackah FK, Amoah KK, Nyarko MA, Andoh DA (2017) Quantifying variations in rhizosheath and root system phenotypes of landraces and improved varieties of juvenile maize. Rhizosphere 3:29–39. https://doi.org/10.1016/j.rhisph.2016.12.004
Alexeyev MF (1999) The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of Gram-negative bacteria. Biotechniques 26:824–826. https://doi.org/10.2144/99265bm05
Asari S, Tarkowská D, Rolčík J, Novák O, Palmero DV, Bejai S, Meijer J (2017) Analysis of plant growth-promoting properties of Bacillus amyloliquefaciens UCMB5113 using Arabidopsis thaliana as host plant. Planta 245:15–30. https://doi.org/10.1007/s00425-016-2580-9
Cai XT, Xu P, Zhao PX et al (2014) Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat Commun 5(1):5833. https://doi.org/10.1038/ncomms6833
Carrillo-Flores E, Arreola-Rivera J, Pazos-Solís DM, Bocanegra-Mondragón M, Fierros-Romero G, Mellado-Rojas ME, Beltrán-Peña E (2022) TOR participation on the root system changes of Arabidopsis during its interaction with Azospirillum. J Appl Biotechnol Bioeng 9:18–23. https://doi.org/10.15406/jabb.2022.09.00280
Cassán F, Coniglio A, López G, Molina R, Nievas S, de Carlan CLN, Donadio F, Torres D, Rosas S, Pedrosa FO, de Souza E, Zorita MD, de-Bashan L, Mora V (2020) Everything you must know about Azospirillum and its impact on agriculture and beyond. Biol Fert Soils 56:461–479. https://doi.org/10.1007/s00374-020-01463-y
Chávez-Herrera E, Hernández-Esquivel AA, Castro-Mercado E, García-Pineda E (2018) Effect of Azospirillum brasilense Sp245 lipopolysaccharides on wheat plant development. J Plant Growth Reg 37:859–866. https://doi.org/10.1007/s00344-018-9782-2
Cheng Z, Sun L, Qi T et al (2011) The bHLH transcription factor MYC3 interacts with the jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol Plant 4(2):279–288. https://doi.org/10.1093/mp/ssq073
Chu TN, Bui LV, Hoang MTT (2020) Pseudomonas PS01 isolated from maize rhizosphere alters root system architecture and promotes plant growth. Microorganisms 8:471. https://doi.org/10.3390/microorganisms8040471
Coniglio A, Mora V, Puente M, Cassán F (2019) Azospirillum as Biofertilizer for Sustainable Agriculture: Azospirillum brasilense AZ39 as a Model of PGPR and Field Traceability. In: Zúñiga-Dávila, D., González-Andrés, F., Ormeño-Orrillo, E. (eds) Microbial Probiotics for Agricultural Systems. Sustainability in Plant and Crop Protection. Springer, Cham. https://doi.org/10.1007/978-3-030-17597-9_4
Contesto C, Milesi S, Mantelin S, Zancarini A, Desbrosses G, Varoquaux F, Bellini C, Kowalczyk M, Touraine B (2010) The auxin-signaling pathway is required for the lateral root response of Arabidopsis to the rhizobacterium Phyllobacterium brassicacearum. Planta 232:1455–1470. https://doi.org/10.1007/s00425-010-1264-0
Costacurta A, Keijers V, Vanderleyden J (1994) Molecular cloning and sequence analysis of an Azospirillum brasilense indole-3-pyruvate decarboxylase gene. Mol Gen Genet 243:463–472. https://doi.org/10.1007/BF00280477
Creus CM, Graziano M, Casanovas EM, Pereyra MA, Simontacchi M, Puntarulo S, Barassi CA, Lamattina L (2005) Nitric oxide is involved in the Azospirillum brasilense-induced lateral root formation in tomato. Planta 221:297–303. https://doi.org/10.1007/s00425-005-1523-7
Croes C, Moens S, Bastelaere E, Vanderleyden J, Michiels KW (1993) The polar flagellum mediates Azospirillum brasilense adsorption to wheat roots. J Gen Microbiol 139:2261–2269. https://doi.org/10.1099/00221287-139-9-2261
Deng K, Yu L, Zheng X, Zhang K, Wang W, Dong P, Zhang J, Ren M (2016) Target of rapamycin is a key player for auxin signaling transduction in Arabidopsis. Front Plant Sci 7:291. https://doi.org/10.3389/fpls.2016.00291
Desrut A, Moumen B, Thibault F, Le Hir R, Coutos-Thévenot P, Vriet C (2020) Beneficial rhizobacteria Pseudomonas simiae WCS417 induce major transcriptional changes in plant sugar transport. J Exp Bot 71:7301–7315. https://doi.org/10.1093/jxb/eraa396
Di Rienzo JA, Macchiavelli RE, Casanoves F (2012) Modelos lineales mixtos. Aplicaciones en InfoStat. Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina. https://www.infostat.com.ar
dos Santos Ferreira N, Hayashi Sant’ Anna F, Massena Reis V, Ambrosini A, Gazolla Volpiano C, Rothballer M, Schwab S, Baura VA, Balsanelli E, Pedrosa FO, Pereira Passaglia LM, Maltempi de Souza E, Hartmann A, Cassán F, Zilli JE (2020) Genome-based reclassification of Azospirillum brasilense Sp245 as the type strain of Azospirillum baldaniorum sp. nov. Int J Syst Evol Microbiol 70:6203–6212. https://doi.org/10.1099/ijsem.0.004517
dos Santos Ferreira N, Coniglio A, Puente M, Hayashi Sant’ Anna F, Maroniche G, García J, Molina R, Nievas S, Gazolla Volpiano C, Ambrosini A, Pereira Passaglia LM, Pedraza RO, Massena Reis V, Zilli JE, Cassán F (2022) Genome-based reclassification of Azospirillum brasilense Az39 as the type strain of Azospirillum argentinense sp. nov. Int J Syst Evol Microbiol 72(10):1099. https://doi.org/10.1099/ijsem.0.005475
Dubrovsky JG, Puente ME, Bashan Y (1994) Arabidopsis thaliana as a model system for the study of the effect of inoculation by Azospirillum brasilense Sp245 on root hair growth. Soil Biol Biochem 26:1657–1664. https://doi.org/10.1016/0038-0717(94)90318-2
Elías JM, Ramírez-Mata A, Albornóz PL, Baca BE, Diaz-Ricci JC, Pedraza RO (2021) The polar flagellin of Azospirillum brasilense REC3 induces a defense response in strawberry plants against the fungus Macrophomina phaseolina. J Plant Growth Regul 41:2992–3008. https://doi.org/10.1007/s00344-021-10490-4
García-Gómez P, Bahaji A, Gámez-Arcas S, Muñoz FJ, Sánchez-López ÁM, Almagro G, Baroja-Fernández E, Ameztoy K, De Diego N, Ugena L, Spíchal L, Doležal K, Hajirezaei MR, Romero LC, García I, Pozueta-Romero J (2020) Volatiles from the fungal phytopathogen Penicillium aurantiogriseum modulate root metabolism and architecture through proteome resetting. Plant Cell Environ 43(10):2551–2570. https://doi.org/10.1111/pce.13817. Epub 2020 Aug 16
Gómez L, Felix G, Boller T (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18:277–284. https://doi-org.uml.idm.oclc.org/10.1046/j.1365-313X.1999.00451.x
Grover M, Bodhankar S, Sharma A, Sharma P, Singh J, Nain L (2021) PGPR mediated alterations in root traits: way toward sustainable crop production. Front Sust Food Syst 4:618230. https://doi.org/10.3389/fsufs.2020.618230
Gullett J, O’Neal L, Mukherjee T, Alexandre G (2017) Azospirillum brasilense: laboratory maintenance and genetic manipulation. Curr Protoc Microbiol 47:3E.2.1-3E.2.17. https://doi.org/10.1002/cpmc.39
Hernández-Esquivel AA, Castro-Mercado E, García-Pineda E (2020) Comparative effects of Azospirillum brasilense Sp245 and Pseudomonas aeruginosa PAO1 lipopolysaccharides on wheat seedling growth and peroxidase activity. J Plant Growth Regul 40:1903–1911. https://doi.org/10.1007/s00344-020-10241-x
Jiménez-Vázquez KR, García-Cárdenas E, Barrera-Ortiz S, Ortiz-Castro R, Ruiz-Herrera LF, Ramos-Acosta BP, Coria-Arellano JL, Sáenz-Mata J, López-Bucio J (2020) The plant beneficial rhizobacterium Achromobacter sp. 5B1 influences root development through auxin signaling and redistribution. Plant J 103:1639–1654. https://doi.org/10.1111/tpj.14853
Karimi N, Goltapeh EM, Amini J, Mehnaz S (2021) Effect of Azospirillum zeae and seed priming with zinc, manganese and auxin on growth and yield parameters of wheat, under dryland farming. Agric Res 10:44–55. https://doi.org/10.1007/s40003-020-00480-5
Khan A, Hossain MT, Park HC, Yun DJ, Shim SH, Chung YR (2016) Development of root system architecture of Arabidopsis thaliana in response to colonization by Martelella endophytica YC6887 depends on auxin signaling. Plant Soil 405:81–96. https://doi.org/10.1007/s11104-015-2775-z
Koul V, Tripathi C, Adholeya A, Kochar M (2015) Nitric oxide metabolism and indole acetic acid biosynthesis cross-talk in Azospirillum brasilense SM. Res Microbiol 166:174–185. https://doi.org/10.1016/j.resmic.2015.02.003
Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin in Plant Biol 5:325–331. https://doi.org/10.1016/S1369-5266(02)00275-3
Li Y, Shao J, Xie Y, Jia L, Fu Y, Xu Z, Zhang N, Feng H, Xun W, Liu Y, Shen Q, Xuan W, Zhang R (2021) Volatile compounds from beneficial rhizobacteria Bacillus spp. promote periodic lateral root development in Arabidopsis. Plant Cell Environ 44:1663–1678. https://doi.org/10.1111/pce.14021
van Loon, LC, Bakker P A, van der Heijdt WH, Wendehenne D, Pugin A (2008) Early responses of tobacco suspension cells to rhizobacterial elicitors of induced systemic resistance. Mol Plant Microbe Interact 21:1609–1621. https://doi-org.uml.idm.oclc.org/10.1094/MPMI-21-12-1609
López-Bucio J, Campos-Cuevas JC, Hernández-Calderón E, Velásquez-Becerra C, Farías-Rodríguez R, Macías-Rodríguez LI, Valencia-Cantero E (2007) Bacillus megaterium rhizobacteria promote growth and alter root-system architecture through an auxin- and ethylene-independent signaling mechanism in Arabidopsis thaliana. Mol Plant-Microbe Interact 20:207–217. https://doi.org/10.1094/MPMI-20-2-0207
Malhotra M, Srivastava S (2008) An ipdC gene knock-out of Azospirillum brasilense strain SM and its implications on indole-3-acetic acid biosynthesis and plant growth promotion. Antonie Van Leeuwenhoek 93:425–433. https://doi.org/10.1007/s10482-007-9207-x
Matsuda F, Miyazawa H, Wakasa K, Miyagawa H (2005) Quantification of indole-3-acetic acid and amino acid conjugates in rice by liquid chromatography-electrospray ionization-tandem mass spectrometry. Biosci Biotechnol Biochem 69:778–783. https://doi.org/10.1271/bbb.69.778
Meldau DG, Meldau S, Hoang LH, Underberg S, Wünsche H, Baldwin IT (2013) Dimethyl disulfide produced by the naturally associated bacterium Bacillus sp. B55 promotes Nicotiana attenuata growth by enhancing sulfur nutrition. Plant Cell 25:2731–2747. https://doi.org/10.1105/tpc.113.114744
Méndez-Gómez M, Castro-Mercado E, Peña-Uribe CA, Reyes-de la Cruz H, López-Bucio J, García-Pineda E (2020) Target of rapamycin signaling plays a role in Arabidopsis growth promotion by Azospirillum brasilense Sp245. Plant Sci 293:110416. https://doi.org/10.1016/j.plantsci.2020.110416
Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, Ausubel, FM (2010) Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. The Plant Cell 22:973–990. https://doi-org.uml.idm.oclc.org/10.1105/tpc.109.069658
Moens S, Michiels K, Vanderleyden J (1995) Glycosylation of the flagellin of the polar flagellum of Azospirillum brasilense, a gram-negative nitrogen-fixing bacterium. Microbiology 141:2651–2657. https://doi.org/10.1099/13500872-141-10-2651
Molina R, Obando D, Torres D, Rivera D, Cassán F (2014) Utilización de un medio de cultivo para la cuantificación y la diferenciación de bacterias presentes en la misma formulación. In: Presentación oral. XV Jornadas Argentinas de Microbiología., Córdoba, Argentina, p. 249
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 13:473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Ortíz-Castro R, Díaz-Pérez C, Martínez-Trujillo M, Del Río RE, Campos-García J, López-Bucio J (2011) Transkingdom signaling based on bacterial cyclodipeptides with auxin activity in plants. PNAS 108:7253–7258. https://doi.org/10.1073/pnas.1006740108
Pečenková T, Janda M, Ortmannová J, Hajná V, Stehlíková Z, Žárský V (2017) Early Arabidopsis root hair growth stimulation by pathogenic strains of Pseudomonas syringae. Ann Bot 120:437–446. https://doi.org/10.1093/aob/mcx073
Pound MP, French AP, Atkinson JA, Wells DM, Bennett MJ, Pridmore T (2003) RootNav: navigating images of complex root architectures. Plant Physiol 162(4):1802–14. https://doi.org/10.1104/pp.113.221531. Epub 2013 Jun 13
Prinsen E, Costacurta A, Michiels K, Vanderleyden J, Van Onckelen H (1993) Azospirillum brasilense indole-3-acetic acid biosynthesis: evidence for a non-tryptophan dependent pathway. Mol Plant Microbe Interact 6:609–615. https://doi.org/10.1094/MPMI-6-609
Puente ML, Gualpa JL, Lopez GA, Molina RM, Carletti SM, Cassán FD (2018) The benefits of foliar inoculation with Azospirillum brasilense in soybean are explained by an auxin signaling model. Symbiosis 76:41–49. https://doi.org/10.1007/s13199-017-0536-x
Ribaudo CM, Krumpholz EM, Cassán FD, Bottini R, Cantore ML, Curá JA (2006) Azospirillum sp. promotes root hair development in tomato plants through a mechanism that involves ethylene. J Plant Growth Regul 25:175–185. https://doi.org/10.1007/s00344-005-0128-5
Robertson SM, Sakariyahu SK, Bolaji A, Belmonte MF, Wilkins O (2022) Growth-limiting drought stress induces time-of-day-dependent transcriptome and physiological responses in hybrid poplar. AoB Plants 14:plac040. https://doi.org/10.1093/aobpla/plac040
Rondina ABL, dos Santos Sanzovo AW, Guimarães GS, Wendling JR, Nogueira MA, Mariangela Hungria M (2020) Changes in root morphological traits in soybean co-inoculated with Bradyrhizobium spp. and Azospirillum brasilense or treated with A. brasilense exudates. Biol Fertil Soils 56:537–549. https://doi.org/10.1007/s00374-020-01453-0
Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev 12:198–207. https://doi.org/10.1101/gad.12.2.198
Růžička K, Ljung K, Vanneste S et al (2007) Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19:2197–2212. https://doi.org/10.1105/tpc.107.052126
Ryu CM, Farag MA, Hu CH, Reddy MS, Kloepper JW, Paré PW (2004) Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol 134:1017–1026. https://doi.org/10.1104/pp.103.026583
Shi CL, Park HB, Lee JS, Ryu S, Ryu CM (2010) Inhibition of primary roots and stimulation of lateral root development in Arabidopsis thaliana by the rhizobacterium Serratia marcescens 90–166 is through both auxin-dependent and -independent signaling pathways. Mol Cells 29:251–258. https://doi.org/10.1007/s10059-010-0032-0
Shirokov A, Budanova A, Burygin G, Evseeva N, Matora L, Shchyogolev S (2020) Flagellin of polar flagellum from Azospirillum brasilense Sp245: isolation, structure, and biological activity. Int J Biol Macromol 147:1221–1227. https://doi.org/10.1016/j.ijbiomac.2019.10.092
Spaepen S, Bossuyt S, Engelen K, Marchal K, Vanderleyden J (2014) Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense. New Phytol 201:850–861. https://doi.org/10.1111/nph.12590
Steenhoudt O, Vanderleyden J (2000) Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects. FEMS Microbiol Rev 24:487–506. https://doi.org/10.1016/S0168-6445(00)00036-X
Stringlis IA, Proietti S, Hickman R, Van Verk MC, Zamioudis C, Pieterse CMJ (2018) Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant J 93:166–180. https://doi.org/10.1111/tpj.13741
Tahir HAS, Gu Q, Wu H, Raza W, Hanif A, Wu L, Colman MV, Gao X (2017) Plant growth promotion by volatile organic compounds produced by Bacillus subtilis SYST2. Front Microbiol 8:171. https://doi.org/10.3389/fmicb.2017.00171
Torres D, Benavidez I, Donadio F, Mongiardini E, Rosas S, Spaepen S, Vanderleyden J, Pěnčík A, Novák O, Strnad M, Frébortová J, Cassán F (2018) New insights into auxin metabolism in Bradyrhizobium japonicum. Res Microbiol 169:313–323. https://doi.org/10.1016/j.resmic.2018.04.002
Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F (2009) Network properties of robust immunity in plants. PLoS Genet 5:e1000772. https://doi.org/10.1371/journal.pgen.1000772
Vanstockem M, Michiels K, Vanderleyden J, Van Gool AP (1987) Transposon mutagenesis of Azospirillum brasilense and Azospirillum lipoferum: physical analysis of Tn5 and Tn5-Mob insertion mutants. Appl Environ Microbiol 53:410–415. https://doi.org/10.1128/aem.53.2.410-415.1987
Viruega-Góngora VI, Acatitla-Jácome IS, Reyes-Carmona SR, Baca BE, Ramírez-Mata A (2020) Spatio-temporal formation of biofilms and extracellular matrix analysis in Azospirillum brasilense. FEMS Microbiol Lett 367:fnaa037. https://doi.org/10.1093/femsle/fnaa037
Xun Q, Wu Y, Li H et al (2020) Two receptor-like protein kinases, MUSTACHES and MUSTACHES-LIKE, regulate lateral root development in Arabidopsis thaliana. New Phytol 227:1157–1173. https://doi.org/10.1111/nph.16599
Zamioudis C, Pieterse CM (2012) Modulation of host immunity by beneficial microbes. Mol Plant-Microbe Interact 25:139–150. https://doi-org.uml.idm.oclc.org/10.1094/MPMI-06-11-0179
Zhang H, Kim MS, Krishnamachari V, Payton P, Sun Y, Grimson M, Farag MA, Ryu CM, Allen R, Melo IS, Paré PW (2007) Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 226:839–851. https://doi.org/10.1007/s00425-007-0530-2
Zhao CZ, Huang J, Gyaneshwar P, Zhao D (2018) Rhizobium sp. IRBG74 alters Arabidopsis root development by affecting auxin signaling. Front Microbiol 8:2556. https://doi.org/10.3389/fmicb.2017.02556
Zipfel C, Robatzek S, Navarro L, Oakeley E, Jones J, Feliz G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428:764–767. https://doi.org/10.1038/nature02485
Acknowledgements
We thank Instituto de Investigaciones Agrobiotecnológicas-Consejo Nacional de Investigaciones Científicas y Tecnológicas (INIAB-CONICET) and Universidad Nacional de Río Cuarto (UNRC). We thank Dr. Federico Ariel from Instituto de Agrobiotecnología de Santa Fe (IAL-CONICET) for Col-0 seeds and Prof. Mark Estelle (Department of Cell and Developmental Biology, UC, San Diego, USA) for tir1.1 seeds. We also sincerely thank Dra. Myriam Zawoznik for her valuable comments and suggestions.
Funding
This work was supported by grants from ANPCyT (PICT-2017–0572 to CF; PICT-2018–2271 to MV, PICT-2019–2199 to PR and PICT-2019–0015 to EJM). The work was funded by the project “Plants as a tool for sustainable global development” (CZ.02.1.01/0.0/0.0/16_019/0000827) within the program Research, Development, and Education (OP RDE). This work was supported by ANID–Programa Iniciativa Científica Milenio ICN17_022, NCN2021_010 and Fondo Nacional de Desarrollo Científico y Tecnológico (1200010) to EJM.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mora, V., López, G., Molina, R. et al. Azospirillum argentinense Modifies Arabidopsis Root Architecture Through Auxin-dependent Pathway and Flagellin. J Soil Sci Plant Nutr 23, 4543–4557 (2023). https://doi.org/10.1007/s42729-023-01371-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42729-023-01371-8