Abstract
Purpose
RAS mutations represent common driver alterations in thyroid cancer. They can be found in benign, low-risk and malignant thyroid tumors with follicular architecture, which are often diagnosed as indeterminate nodules on preoperative cytology. Therefore, the detection of RAS mutations in preoperative setting has a suboptimal predictive value for malignancy. In this study, we investigated differentially expressed microRNA (miRNA) in benign and malignant thyroid tumors with follicular architecture carrying mutations in RAS genes.
Methods
Total RNA was purified from 60 RAS-mutant follicular-patterned thyroid tumors, including follicular adenoma (FA), noninvasive follicular thyroid neoplasm with papillary-like nuclear features (NIFTP), papillary and follicular thyroid carcinoma cases (PTC, FTC); 22 RAS-negative FAs were used as controls. The expression analysis of 798 miRNAs was performed by digital counting (nCounter nanoString platform).
Results
Comparing RAS-mutant and RAS-negative FAs, 12 miRNAs showed significant deregulation, which was likely related to the oncogenic effects of RAS mutations. Twenty-two miRNAs were differentially expressed in RAS-mutant benign versus malignant tumors. Considering the tumor type, 24 miRNAs were deregulated in PTC, 19 in NIFTP, and seven in FTC and compared to FA group; among these, miR-146b-5p, miR-144-3p, and miR-451a showed consistent deregulation in all the comparisons with the highest fold change.
Conclusions
The miRNA expression analysis of follicular-patterned thyroid tumors demonstrated that RAS mutations influences miRNA profile in benign tumors. In addition, several miRNAs showed a histotype-specific deregulation and could discriminate between RAS-mutant benign and RAS-mutant malignant thyroid lesions, thus deserving further investigation as potential diagnostic markers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Thyroid tumors with follicular architecture include benign lesions, low-risk neoplasms and malignant neoplasms with different treatment needs, namely follicular adenoma (FA), noninvasive follicular neoplasm with papillary-like nuclear features (NIFTP), follicular variant papillary thyroid carcinoma (FV-PTC) and follicular thyroid carcinoma (FTC) [1]. These tumors are extremely challenging to identify on fine-needle aspiration (FNA) cytology, and they are often diagnosed as indeterminate nodules [2]. Molecular analysis can be performed on FNA to refine the risk of malignancy of indeterminate nodules. However, both benign and malignant follicular-architecture thyroid neoplasms share the same class of molecular alterations, i.e., RAS-like mutations [3]. Alterations in RAS genes in thyroid tumors are well-recognized as driver genetic events and include almost exclusively activating point mutations occurring in codons 12, 13, and 61 of NRAS, HRAS, and KRAS genes. These mutations cause a constitutive activation of Ras proteins, leading to a pro-proliferative downstream signaling [4, 5]. The prevalence of RAS mutations in follicular-architecture thyroid lesions varies according to the tumor type: the reported frequency ranges from 20 to 30% in FA, and from 40 to 70% in NIFTP; RAS mutations are found in up to 70% FV-PTC and in up to 50% FTC cases [3, 6,7,8]. Therefore, when detected in indeterminate thyroid nodules, RAS mutations indicate the presence of a neoplasm, but cannot unequivocally identify a malignant lesion.
The diagnostic utility of miRNA expression analysis has been extensively explored in thyroid tumors. MiRNAs influence gene expression by inhibiting translation or promoting degradation of mRNA transcripts. MiRNAs can behave both as oncomiR and tumor suppressors, according to their mRNA targets [9]. Over the past 20 years, several miRNAs have been proposed as effective markers for the differential diagnosis of indeterminate thyroid nodules and for the prognostic stratification of tumors; also, miRNAs differentially expressed in a histotype-specific manner have been reported [10]. Across the considerable amount of data on miRNA expression analysis in thyroid tumors, there are few miRNAs widely and consistently reported as deregulated in thyroid cancer, including the overexpression of miR-146, miR-221, miR-222, and miR-181 [11,12,13]. To date, only one miRNA-based diagnostic test is available to integrate the cyto-morphological analysis of indeterminate thyroid nodules: the ThyraMIR, a commercial test offered by a private company in the USA (Interpace Diagnostics, Parsippany, NJ, USA). It is performed in combination with ThyGeNEXT test for the screening for oncogenic mutations and fusions. Specifically, only when the ThyGeNEXT is negative or alterations with relatively weak positive predictive value, including RAS mutations, are detected, the ThyraMIR test is performed to refine the risk of malignancy of the nodule [14]. The test, which has been recently improved (ThyraMIR v.2), is based on the analysis of expression levels of 11 miRNAs, measured by qRT-PCR. Although in a retrospective study the combination testing showed a high diagnostic accuracy (93%) [15], the ThyraMIR test is not part of the clinical practice of thyroid nodules.
In fact, no miRNA targets have been introduced in the daily management of indeterminate thyroid nodules. In case of RAS-positive nodules, the risk of malignancy is highly variable, and there is the need of reliable markers to resolve the uncertainty around these nodules. For this reason, this study aims to investigate miRNAs able to refine the risk of malignancy of RAS-mutant thyroid neoplasms.
Materials and methods
This study has been conducted on retrospectively selected formalin-fixed, paraffin-embedded tissue samples. The study cohort was composed by 60 RAS-mutant tumors and 22 RAS-negative FAs. RAS-mutant thyroid tumors included: 16 FAs, 15 NIFTPs, 19 PTCs (16 follicular variant and three solid variant PTCs), and ten FTCs. The selection of cases was guided by the following criteria: benign and malignant follicular-patterned thyroid lesions positive for RAS mutations and available tumor tissue for miRNA analysis. Moreover, 22 RAS-negative FAs were retrieved to serve as controls. The characteristics of the sample series are shown in Table 1.
All the available hematoxylin and eosin slides were reviewed to confirm the histological diagnosis following the current guidelines [1]. The presence of RAS mutations was assessed in routine practice by real-time PCR with a commercial kit validated for diagnostic use (EasyPGX ready Thyroid, Diatech Pharmacogenetics, Jesi, AN, Italy). The panel includes probes to identify mutations in the following hotspots: BRAF p.K601E; BRAF p.V600E; KRAS, NRAS, and HRAS codons 12/13 and 61, with no identification of the exact nucleotide change. TERT promoter mutation status was also available for 12 out of 19 PTCs, all negative, and for seven out of ten FTCs, four mutated (C228T) and three negative for mutations.
From the same tissue block used for mutation profiling, three 10 µm sections were obtained for miRNA extraction. First, tumor tissue enrichment was performed by manual dissection of areas containing tumor cells; then, total RNA, including miRNA, was purified with a specific kit, following the manufacturer’s instruction (miRNeasy FFPE kit, Qiagen, Hilden, Germany). Total RNA quantity and quality were assessed by the spectrophotometer XPose (Trinean, Gentbrugge, Belgium).
The miRNA expression analysis was performed on 82 samples, 60 RAS-mutant and 22 RAS-wild-type, by using the nCounter Human v3 miRNA Expression Assay (nanoString Technologies, Seattle, WA, USA) on the nCounter platform (nanoString Technologies), by using 100 ng RNA, as recommended by the manufacturer. Due to an assay upgrade made by the manufacturer, part of cases was analyzed by v3a panel version (n = 22), and part by v3b version (n = 60); both versions contain probe pairs targeting 798 endogenous miRNAs. Raw data obtained after digital counting from v3a and v3b panels were separately analyzed with the nSolver analysis software (v4.0, nanoString Technologies); data were normalized by using the mean expression of the 100 most expressed miRNAs in the dataset. To select miRNA highly expressed among samples, the mean plus two standard deviations of negative control probes was used as threshold, and targets detected in less than 50% of samples were filtered out. Batch effects due to different version of the panel were removed using the ComBat function of the sva Bioconductor package v.3.20.0. Principal component analysis (PCA) was carried out using the PCAtools Bioconductor package v.2.8.0. Hierarchical clustering was performed using Euclidean distance and Ward clustering method following the procedures of the heatmap3 R package v.1.1.9. Differentially expressed miRNA were computed using the moderated t statistics and following the procedures of the limma Bioconductor package v. 3.52.2. Age and sex were used as confounders, and p values were adjusted using the Benjamini–Hochberg method. Adjusted p values ≤ 0.05 were considered significant. Receiver operating characteristics (ROC) curves analysis was carried out using the pROC R package v.1.18.0. Confidence intervals were estimated by 2000 bootstrap resampling. For data analysis, NIFTP class has been considered as malignant, as it requires surgery.
The set of significantly deregulated miRNAs in benign versus malignant RAS-mutant tumors has been entered on DIANA-miRPath v3.0 database (https://dianalab.e-ce.uth.gr/html/mirpathv3, last accessed August 22, 2022), a freely available online tool, which uses both validated and predicted targets to identify pathways controlled by multiple miRNAs, by using Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations [17]. Results were combined by genes union; the options of conservative statistics and correction for multiple comparisons (Benjamini–Hochberg’s FDR) were selected; significance was set as ≤ 0.01.
Statistical analyses (except for pathway enrichment analysis) were performed in R environment (https://www.r-project.org, v.4.1.2, last accessed August 8, 2022).
Results
miRNA expression profile of thyroid tumors
After filtering and merging miRNAs from v3a and v3b panels, 128 miRNAs were selected for further analyses and adjusted for batch effect (Suppl. Fig. 1a, b).
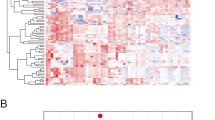
The heatmap shown in Fig. 1a represents the unsupervised clustering of all samples, including the RAS-negative FAs, based on the expression of the selected 128 miRNAs. Figure 1b shows the same heatmap after removal of RAS-negative samples. The distribution of samples according to RAS status and histotype indicates that these factors do have an influence on miRNA expression. This is confirmed by the PCA shown in Fig. 2. In fact, when RAS-negative FAs are included, they are grouped together, separately from RAS-mutant samples (Fig. 2a). It is interesting to note that RAS-mutant FAs are closer to the group of malignant tumors, but still occupy a circumscribed portion of the graph (Fig. 2b). When RAS-negative FAs are excluded, samples do not assume a well-defined grouping looking at RAS mutation type (Fig. 2c) nor at the tumor type, though FAs occupy the bottom right part of the graph (Fig. 2d).
Heatmaps representing the unsupervised clustering of miRNAs and samples. a All samples were included (n = 82), b only RAS-mutant samples were included (n = 60). Each row represents one miRNA, each column one sample, the colors indicate high (red) and low (blue) gene expression level. Colored sidebars represent mutational status, histological diagnosis, and benign or malignant neoplasm, as it is indicated in the legend
a, b Principal component analysis of all samples, including RAS-negative FAs (n = 82), labeled for mutational status and histological class. c, d Principal component analysis of the 60 RAS-mutant tumors, tumors are shown with different colors according to the involved RAS gene (NRAS, HRAS, KRAS) and to the histological class
miRNAs deregulated in RAS-mutant versus RAS-negative benign tumors
To identify miRNAs whose deregulation is likely related to RAS mutations, a comparison between RAS-positive FAs and RAS-negative FAs was performed; as shown in Table 2, 12 miRNAs were significantly deregulated.
miRNAs deregulated in benign versus malignant RAS-mutant thyroid tumors
To investigate subtle differences among RAS-mutant tumor types, differentially expressed miRNA between benign and malignant cases and among histotypes have been investigated. Comparing all the malignant cases versus RAS-mutant FAs, 22 miRNAs were significantly deregulated (12 up- and 10 down-regulated). In addition, 24 miRNAs were deregulated in PTCs, seven in FTCs and 19 in NIFTPs compared to FAs (Table 3). Several miRNAs were deregulated in more than one comparison, as shown in Fig. 3a.
a Venn diagram representing the number of significantly deregulated miRNAs in different histological classes. b–d Volcano plots showing the relative expression (logFC) of miRNAs in PTC, NIFTP, and FTC classes compared to FA group. MiRNAs have been highlighted based on their expression level (green for logFC >|1|), significance level (light blue for P value ≤ 0.05), or both (orange for miRNAs showing logFC >|1| and P value ≤ 0.05). FTC follicular thyroid carcinoma, NIFTP noninvasive follicular thyroid neoplasm with papillary-like nuclear features, PTC papillary thyroid carcinoma, FA follicular adenoma, NS not significant, Log2 FC base 2 logarithm of the fold change
The 22 deregulated miRNAs in benign versus malignant cases have been used for pathway enrichment analysis on DIANA-miRPath v3.0 tool, and 48 pathways resulted significantly enriched. In Table 4, the top ten pathways showing significant enrichment, ranked by ascending p values, are shown.
Three miRNAs showed significant deregulation in all the comparisons with an absolute logFC value >|1|; miR-146b-5p, miR-144-3p, and miR-451a (volcano plots in Fig. 3b–d). The expression levels of these miRNAs in all the histological classes are illustrated in Fig. 4a–c; RAS-negative FAs have been also included in the graph for a direct comparison. The up-regulation of miR-146b-5p appears as a sort of continuum, from RAS-negative benign tumors to RAS-positive benign and malignant tumors; the expression of miR-144-3p and miR-451a does not show evident differences between RAS-negative and RAS-positive benign cases. MiR-146b-5p, miR-144-3p, and miR-451a have been tested by ROC analysis to investigate their potential usefulness in discriminating malignant versus benign RAS-positive tumors (Fig. 4d; Table 5). Area under the curve (AUC) was 0.83 (95% CI 0.70–0.93), 0.79 (95% CI 0.64–0.91) and 0.79 (95% CI 0.67–0.90) for miR-146b-5p, miR-144-3p, and miR-451a, respectively.
a–c Relative expression levels of miR-146b-5p, miR-144-3p, and miR-451a (expressed in log2FC) in all the histological classes. d ROC analysis showing the diagnostic performance of the three selected miRNAs. FA WT wild-type follicular adenoma, FA follicular adenoma, NIFTP noninvasive follicular thyroid neoplasm with papillary-like nuclear features, FTC follicular thyroid carcinoma, PTC papillary thyroid carcinoma
Discussion
Follicular-patterned well-differentiated thyroid tumors represent a morphologically and molecularly homogeneous class of lesions, with a highly heterogeneous clinical course. FA is a benign neoplastic lesion, with clonal development and a well-circumscribed capsule. NIFTP is a low-risk lesion, with cyto-architectural characteristics of FV-PTC but an intact tumor capsule. FV-PTC is often considered a low-risk tumor, at least compared with more aggressive PTC variants, such as tall cell and hobnail variants, and especially when encapsulated [1, 6]. Nevertheless, in a small proportion of cases, FV-PTC can show dedifferentiation, and can cause recurrence and distant metastases [18]. Finally, FTCs show an increasingly aggressive clinical course according to their degree of invasiveness, from minimally invasive to angioinvasive to widely invasive types [6]. All these follicular-patterned tumors, besides the follicular growth pattern, share the same class of molecular alterations, i.e., RAS-like mutations. The different level of aggressiveness can be only in part explained by the presence of co-occurring molecular alterations, such as PIK3CA, TP53, and TERT promoter mutations, found in less than 10% of FV-PTCs and in 10–20% of FTCs [7, 19, 20]. In fact, there is controversy about the diagnostic and prognostic role of RAS mutations. NRAS, KRAS and HRAS are oncogenes: their activating mutations are known to promote cell growth and proliferation. However, if detected preoperatively on thyroid nodules, RAS mutations show a suboptimal positive predictive value for malignancy [21]. Therefore, in this study we investigated miRNA expression profiles in benign and malignant RAS-mutant thyroid neoplasms with the aim of identifying miRNAs that could serve as diagnostic markers in presurgical setting, specifically in RAS-mutant thyroid nodules, thus helping refine their risk of malignancy.
As a first step, we compared miRNA expression levels in RAS-negative and RAS-mutant FAs; in this way, we sought to identify miRNAs selectively altered in RAS-positive tumors. In fact, it is known that RAS mutations influence miRNA expression profiles in thyroid tumors [22, 23]. However, authors mostly focused on differential miRNA expression between malignant and benign tumors, without considering the presence of RAS mutations as a confounding factor [24,25,26]; on the contrary, several miRNAs are known to be specifically deregulated in BRAF mutant thyroid tumors compared to wild-type cases [27, 28]. In previous studies from our group, we demonstrated that wild-type NIFTPs show a benign-like miRNA phenotype, while RAS-mutant NIFTPs show a malignant-like miRNA phenotype [29, 30]; in detail, 12 miRNAs were significantly deregulated in mutant versus wild-type NIFTPs (miR-221-5p, miR-221-3p, miR-222-3p, miR-146b-5p, miR-181a-3p, miR-28-5p, miR-363-3p, miR-342-3p, miR-1285-5p, miR-152-3p, miR-25-3p, miR-30e-3p) [30]. Likewise, comparing RAS-negative and RAS-mutant FAs, herein we report 12 differentially expressed miRNAs, whose deregulation is likely related to RAS mutations (miR-222-3p, miR-7-5p, miR-221-3p, miR-221-5p, miR-19a-3p, miR-135a-5p, miR-146b-5p, let-7a-5p, miR-92a-3p, miR-142-3p, miR-296-5p, miR-26a-5p). Interestingly, among these, miR-221, miR-222 and miR-146b-5p, well-known to be cancer-related biomarkers [11,12,13], were significantly up-regulated in mutant FAs. Therefore, activating mutations in RAS genes are likely related to the expression of oncogenic miRNAs also in benign thyroid tumors; this could affect the diagnostic performance of molecular classifiers based on miRNA expression profile, resulting in false positive calls. On the other hand, whether these oncogenic miRNAs contribute to promote tumor invasiveness in conjunction with RAS mutations is yet to be clarified.
Then, to identify miRNAs with diagnostic value, we analyzed the miRNA expression profile of FAs, NIFTPs, PTCs and FTCs harboring RAS mutations. Comparing the FAs versus all the malignant lesions, 22 miRNAs showed significant deregulation. Among these, the most widely described as up-regulated in thyroid cancer are miR-146b-5b, miR-221, miR-181, and miR99b [10, 11, 13]; on the other hand, the most well-known downregulated miRNAs are miR144, miR-451a, miR-497, miR-195, miR-143, and miR-152 [13]. The analysis of enriched pathways showed that the 22 miRNAs deregulated in malignant RAS-positive thyroid tumors play a key role in several cancer-related pathways. It is interesting to note that, in the hypothesis of a diagnostic setting, only one out of 22 differentially expressed miRNAs is currently included in the commercially available miRNA-based diagnostic panel (i.e., miR-146b-5p) [15].
The three miRNAs showing significant deregulation in all the comparisons with the highest logFC, i.e., miR-146b-5p, miR-144-3p, and miR-451a, demonstrated good accuracy in distinguishing benign from malignant lesions, with AUC of 0.83, 0.79, and 0.79, and positive predictive value of 0.97, 0.93, and 0.94, respectively. MiR-146b is a well-recognized oncomiR in several solid tumors, and it has been often described as a PTC-specific marker. There are even studies exploring therapeutic strategies based on miR-146b silencing [31, 32]. Biologically, it is known that miR-146b overexpression promotes thyroid cell survival, proliferation, migration, and reduces radioiodine sensitivity through the downregulation of genes such as ZNRF3, SMAD4, NIS, IRAK1 [30]. In thyroid cancer, the up-regulation of miR-146b has been correlated to BRAF p.V600E mutation, increased tumor stage and tumor aggressiveness, and higher rate of lymph node metastases [13, 28]. Herein, we demonstrated that up-regulation of miR-146b-5p occurs also in benign tumors harboring RAS mutations: these findings offer interesting cues on miR-146b biological role, but raise also new questions around its diagnostic utility, since it is difficult to establish an absolute cutoff able to reliably predict malignancy. MiR-144-3p and miR-451a, both downregulated in RAS mutant thyroid malignancies in the present study, have been previously described as tumor suppressor miRNAs [23, 33, 34]. The physio-pathological role of miR-451a has been investigated in several tumors. In vitro, miR-451a inhibits thyroid cancer cells growth, proliferation, epithelial-mesenchymal transition, and induces apoptosis [35, 36]. In thyroid cancer, mainly PTC, miR-451a is downregulated, and significant correlation with aggressive clinical-pathological features (tall cell variant, advanced stage, extra-thyroidal extension) has been described [23, 33]. Similarly to miR-146b, also miR-451a expression seems modulated by the presence of BRAF mutations. In fact, significant downregulation of miR-451a has been reported in PTCs compared to normal thyroid tissue irrespective of mutation status, but the lower miR-451a levels were observed in BRAF p.V600E PTCs [23]. In our study, miR-451a was not deregulated in wildtype versus RAS mutant FAs, but it was significantly downregulated in RAS-mutant FAs compared to malignant tumors, thus representing a promising diagnostic marker irrespective of RAS mutations. Considering miR-144-3p, its overexpression inhibits proliferation, migration, and invasion of thyroid cancer cells in vitro [37]. Although the majority of reports found that miR-144-3p is downregulated [10, 13, 23, 34], there are few studies reporting that miR-144-3p is overexpressed in thyroid cancer [38, 39]; in one study, the overexpression of miR-144-3p was inversely correlated with the transcript quantity of the tumor suppressor gene PTEN [38]. In our study, miR-144-3p was significantly down-regulated in all the RAS-mutant malignant tumors compared to RAS-mutant FAs, therefore our report supports the evidence that miR-144-3p acts as a tumor suppressor miRNA.
Interestingly, in a study conducted by Knyazeva and colleagues, it was reported that the combination of miR-146b-5p and miR-451a, which showed reciprocal deregulation, was able to distinguish FA from FTC with an AUC of 0.92 [24]. Using prediction algorithms, the authors hypothesized that the concomitant up-regulation of miR-146b-5p and downregulation of miR-451a led to the activation of downstream pathways that ultimately promote an invasive behavior in follicular thyroid tumors. Unfortunately, the RAS genes status of these tumors was unknown.
As regards miRNA expression profiles among tumor types, the number of deregulated miRNAs was higher in the comparisons between PTCs and FAs (n = 24) and between NIFTPs and FAs (n = 19), while only seven miRNAs were significantly deregulated in FTCs; some miRNAs showed deregulation in more than one comparison. The fact that NIFTPs and PTCs shared 12 deregulated miRNAs is not surprising, since FV-PTCs and NIFTPs are morphologically similar, with the main difference standing in their degree of invasiveness; on the other hand, the 12 remaining miRNAs deregulated in PTCs and not in NIFTPs might be related to an invasive tumor behavior. In particular, miR-324-5p and miR-22-3p showed significant up-regulation and down-regulation, respectively, in both PTCs and FTCs versus FAs. Studies conducted in various cancer types, including thyroid cancer, reported that miR-324-5p is generally up-regulated [40]. Several authors investigated the biological role of miR-22-3p, and described it as a tumor suppressor miRNA [41,42,43]. In cervical squamous cell carcinoma, miR-22-3p downregulation has been associated with metastatic tumor progression [44]; the biological functions of miR-22-3p in the context of RAS-mutant thyroid tumors has not been investigated.
Of the seven miRNAs deregulated in FTCs compared to FAs, six were in common with PTCs and NIFTPs, and only one was significant in FTCs only, i.e., miR-145-5p. The utility of this miRNA in thyroid cancer has been poorly explored. In PTCs, miR-145-5p showed downregulation compared to non-tumor tissue, and negative correlation with tumor progression and metastatic spread [45]. However, there is no specific investigation regarding FTCs, thus the biological and clinical role of miR-145-5p should be further elucidated.
Our results present some potential limitations. First, it was conducted on a limited set of thyroid cancer types. The cases were retrieved upon careful histological and molecular selection, but it would have been interesting to investigate and compare miRNA expression profiles of aggressive histotypes (poorly differentiated and anaplastic thyroid cancer), and also RAS-driven medullary thyroid cancer. Second, only a small set of genetic alterations were screened, and wild-type cases could have harbored uncommon molecular alterations with a potential influence on miRNA expression profiles. Finally, the study was conducted on tumor tissue samples, thus the significance of these results cannot be directly extended to thyroid cytology, and an independent cohort is necessary to validate the diagnostic value of the identified miRNAs.
In conclusion, comparing RAS-mutant and RAS-negative benign thyroid tumors, we found that RAS mutations increase the expression of miRNA with a recognized oncogenic role; therefore, when evaluating miRNA expression profile of thyroid tumors, RAS status should be considered as a confounding factor. Moreover, we demonstrated that, besides the morphological similarities and the shared presence of RAS mutations, there are miRNAs able to distinguish benign from malignant follicular-patterned neoplasms, with the most promising miRNAs being miR-146b-5p, miR-451a, and miR-144-3p. Moreover, we highlighted that some miRNAs, whose utility in thyroid cancer has been poorly investigated until now, could serve as histotype-specific miRNAs. Our results could help improving the diagnostic performance of already existing molecular classifiers and could support in the selection of miRNA targets for setting up new diagnostic panels.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Lloyd R, Osamura R, Kloppel G, Rosai J (2017) WHO classification of tumours of endocrine organs, 4th edn. IARC Press, Lyon
LiVolsi VA, Baloch ZW (2011) Follicular-patterned tumors of the thyroid: the battle of benign vs malignant vs so-called uncertain. Endocr Pathol 22:184–189. https://doi.org/10.1007/s12022-011-9183-6
Macerola E, Poma AM, Vignali P, Basolo A, Ugolini C, Torregrossa L et al (2021) Molecular genetics of follicular-derived thyroid cancer. Cancers 13(5):1139. https://doi.org/10.3390/cancers13051139
Gimple RC, Wang X (2019) RAS: Striking at the core of the oncogenic circuitry. Front Oncol 9:965. https://doi.org/10.3389/fonc.2019.00965
Prior IA, Hood FE, Hartley JL (2020) The frequency of Ras mutations in cancer. Cancer Res 80(14):2969–2974. https://doi.org/10.1158/0008-5472.CAN-19-3682
Baloch ZW, Asa SL, Barletta JA, Ghossein RA, Juhlin CC, Jung CK et al (2022) Overview of the 2022 WHO classification of thyroid neoplasms. Endocr Pathol 33(1):27–63. https://doi.org/10.1007/s12022-022-09707-3
Cancer Genome Atlas Research Network (2014) Integrated genomic characterization of papillary thyroid carcinoma. Cell 159(3):676–690. https://doi.org/10.1016/j.cell.2014.09.050
Marotta V, Bifulco M, Vitale M (2021) Significance of RAS mutations in thyroid benign nodules and non-medullary thyroid cancer. Cancers 13(15):3785. https://doi.org/10.3390/cancers13153785
Peng Y, Croce CM (2016) The role of MicroRNAs in human cancer. Signal Transduct Target Ther 1:15004. https://doi.org/10.1038/sigtrans.2015.4
De Martino M, Esposito F, Capone M, Pallante P, Fusco A (2022) Noncoding RNAs in thyroid-follicular-cell-derived carcinomas. Cancers 14(13):3079. https://doi.org/10.3390/cancers14133079
Santiago K, Chen Wongworawat Y, Khan S (2020) Differential MicroRNA-signatures in thyroid cancer subtypes. J Oncol 2020:1–14. https://doi.org/10.1155/2020/2052396
Pishkari S, Paryan M, Hashemi M, Baldini E, Mohammadi-Yeganeh S (2018) The role of MicroRNAs in different types of thyroid carcinoma: a comprehensive analysis to find new MiRNA supplementary therapies. J Endocrinol Invest 41(3):269–283. https://doi.org/10.1007/s40618-017-0735-6
Papaioannou M, Chorti AG, Chatzikyriakidou A, Giannoulis K, Bakkar S, Papavramidis TS (2022) MicroRNAs in papillary thyroid cancer: what is new in diagnosis and treatment. Front Oncol 11:755097. https://doi.org/10.3389/fonc.2021.755097
Lupo MA, Walts AE, Sistrunk JW, Giordano TJ, Sadow PM, Massoll N et al (2020) Multiplatform molecular test performance in indeterminate thyroid nodules. Diagn Cytopathol 48(12):1254–1264. https://doi.org/10.1002/dc.24564
Finkelstein SD, Sistrunk JW, Malchoff C, Thompson DV, Kumar G, Timmaraju VA et al (2022) A retrospective evaluation of the diagnostic performance of an interdependent pairwise MicroRNA expression analysis with a mutation panel in indeterminate thyroid nodules. Thyroid. https://doi.org/10.1089/thy.2022.0124
Nardi F, Basolo F, Crescenzi A, Fadda G, Frasoldati A, Orlandi F et al (2014) Italian consensus for the classification and reporting of thyroid cytology. J Endocrinol Invest 37(6):593–599. https://doi.org/10.1007/s40618-014-0062-0
Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T et al (2015) DIANA-MiRPath v3.0: deciphering MicroRNA function with experimental support. Nucleic Acids Res 43(W1):W460–W466. https://doi.org/10.1093/nar/gkv403
Condello V, Torregrossa L, Sartori C, Denaro M, Poma AM, Piaggi P et al (2019) MRNA and MiRNA expression profiling of follicular variant of papillary thyroid carcinoma with and without distant metastases. Mol Cell Endocrinol 479:93–102. https://doi.org/10.1016/j.mce.2018.09.005
Duan H, Liu X, Ren X, Zhang H, Wu H, Liang Z (2019) Mutation profiles of follicular thyroid tumors by targeted sequencing. Diagn Pathol 14(1):39. https://doi.org/10.1186/s13000-019-0817-1
Yoo SK, Song YS, Park YJ, Seo JS (2020) Recent improvements in genomic and transcriptomic understanding of anaplastic and poorly differentiated thyroid cancers. Endocrinol Metab 35(1):44–54. https://doi.org/10.3803/EnM.2020.35.1.44
Torregrossa L, Poma AM, Macerola E, Rago T, Vignali P, Romani R et al (2022) The Italian consensus for the classification and reporting of thyroid cytology: cytohistologic and molecular correlations on 37,371 nodules from a single institution. Cancer Cytopathol. https://doi.org/10.1002/cncy.22618
Nikiforova MN, Chiosea SI, Nikiforov YE (2009) MicroRNA expression profiles in thyroid tumors. Endocr Pathol 20(2):85–91. https://doi.org/10.1007/s12022-009-9069-z
Minna E, Romeo P, Dugo M, De Cecco L, Todoerti K, Pilotti S et al (2016) MiR-451a is underexpressed and targets AKT/MTOR pathway in papillary thyroid carcinoma. Oncotarget 7(11):12731–12747. https://doi.org/10.18632/oncotarget.7262
Knyazeva M, Korobkina E, Karizky A, Sorokin M, Buzdin A, Vorobyev S et al (2020) Reciprocal dysregulation of MiR-146b and MiR-451 contributes in malignant phenotype of follicular thyroid tumor. Int J Mol Sci 21(17):5950. https://doi.org/10.3390/ijms21175950
Jahanbani I, Al-Abdallah A, Ali RH, Al-Brahim N, Mojiminiyi O (2018) Discriminatory MiRNAs for the management of papillary thyroid carcinoma and noninvasive follicular thyroid neoplasms with papillary-like nuclear features. Thyroid 28(3):319–327. https://doi.org/10.1089/thy.2017.0127
Rogucki M, Sidorkiewicz I, Niemira M, Dzięcioł JB, Buczyńska A, Adamska A et al (2022) Expression profile and diagnostic significance of MicroRNAs in papillary thyroid cancer. Cancers 14(11):2679. https://doi.org/10.3390/cancers14112679
Galuppini F, Censi S, MeranteBoschin I, Fassan M, Sbaraglia M, Valeri N et al (2022) Papillary thyroid carcinoma: molecular distinction by MicroRNA profiling. Front Endocrinol 13:834075. https://doi.org/10.3389/fendo.2022.834075
Celakovsky P, Kovarikova H, Chrobok V, Mejzlik J, Laco J, Vosmikova H et al (2021) MicroRNA deregulation in papillary thyroid cancer and its relationship with BRAF V600E mutation. In Vivo 35(1):319–323. https://doi.org/10.21873/invivo.12262
Macerola E, Poma AM, Proietti A, Romani R, Torregrossa L, Ugolini C et al (2020) Digital gene expression analysis on cytology smears can rule out malignancy in follicular-patterned thyroid tumors. J Mol Diagn 22(2):179–187. https://doi.org/10.1016/j.jmoldx.2019.09.008
Denaro M, Ugolini C, Poma AM, Borrelli N, Materazzi G, Piaggi P et al (2017) Differences in MiRNA expression profiles between wild-type and mutated NIFTPs. Endocr Relat Cancer 24(10):543–553. https://doi.org/10.1530/ERC-17-0167
Chou CK, Liu RT, Kang HY (2017) MicroRNA-146b: a novel biomarker and therapeutic target for human papillary thyroid cancer. Int J Mol Sci 18(3):636. https://doi.org/10.3390/ijms18030636
de Santa-Inez DC, Fuziwara CS, Saito KC, Kimura ET (2021) Targeting the highly expressed MicroRNA MiR-146b with CRISPR/Cas9n gene editing system in thyroid cancer. Int J Mol Sci 22(15):7992. https://doi.org/10.3390/ijms22157992
Liu B, Shi H, Qiu W, Wu X, Li L, Wu W (2021) A two-MicroRNA signature predicts the progression of male thyroid cancer. Open Life Sci 16(1):981–991. https://doi.org/10.1515/biol-2021-0099
Zhou M, Wu Y, Li H, Zha X (2020) MicroRNA-144: a novel biological marker and potential therapeutic target in human solid cancers. J Cancer 11(22):6716–6726. https://doi.org/10.7150/jca.46293
Fan X, Zhao Y (2019) MiR-451a inhibits cancer growth, epithelial-mesenchymal transition and induces apoptosis in papillary thyroid cancer by targeting PSMB8. J Cell Mol Med 23(12):8067–8075. https://doi.org/10.1111/jcmm.14673
Wang Q, Shang J, Zhang Y, Zhou Y, Tang L (2020) MiR-451a restrains the Growth and metastatic phenotypes of papillary thyroid carcinoma cells via inhibiting ZEB1. Biomed Pharmacother 127:109901. https://doi.org/10.1016/j.biopha.2020.109901
Yi D, Zhang D, Zeng Z, Zhang S, Li M, Zhang Y (2022) MicroRNA-144–3p Represses the growth and EMT of thyroid cancer via the E2F2/TNIK axis in cells and male BALB/c nude mice. Endocrinology 163(7):bqac071. https://doi.org/10.1210/endocr/bqac071
Cao HL, Gu MQ, Sun Z, Chen ZJ (2020) MiR-144-3p contributes to the development of thyroid tumors through the PTEN/PI3K/AKT pathway. Cancer Manag Res 12:9845–9855. https://doi.org/10.2147/CMAR.S265196
Liu C, Su C, Chen Y, Li G (2018) MiR-144–3p promotes the tumor growth and metastasis of papillary thyroid carcinoma by targeting paired box gene 8. Cancer Cell Int 18:54. https://doi.org/10.1186/s12935-018-0550-y
Kuo WT, Yu SY, Li SC, Lam HC, Chang HT, Chen WS et al (2016) MicroRNA-324 in human cancer: MiR-324–5p and MiR-324–3p have distinct biological functions in human cancer. Anticancer Res 36:5189–5196. https://doi.org/10.21873/anticanres.11089
Li C, Li X, Wang H, Guo X, Xue J, Wang X et al (2022) MicroRNA-22–3p and MicroRNA-149–5p inhibit human hepatocellular carcinoma cell growth and metastasis properties by regulating methylenetetrahydrofolate reductase. Curr Issues Mol Biol 44:952–962. https://doi.org/10.3390/cimb44020063
Wang X, Yao Z, Fang L (2021) MiR-22-3p/PGC1β suppresses breast cancer cell tumorigenesis via PPARγ. PPAR Res 2021:6661828. https://doi.org/10.1155/2021/6661828
Yang X, Su W, Li Y, Zhou Z, Zhou Y, Shan H et al (2021) MiR-22-3p suppresses cell growth via MET/STAT3 signaling in lung cancer. Am J Transl Res 13(3):1221–1232
Kwon AY, Jeong JY, Park H, Hwang S, Kim G, Kang H et al (2022) MiR-22-3p and MiR-30e-5p are associated with prognosis in cervical squamous cell carcinoma. Int J Mol Sci 23(10):5623. https://doi.org/10.3390/ijms23105623
Zhang W, Ji W, Li T, Liu T, Zhao X (2020) MiR-145 functions as a tumor suppressor in papillary thyroid cancer by inhibiting RAB5C. Int J Med Sci 17(13):1992–2001. https://doi.org/10.7150/ijms.44723
Acknowledgements
The authors would like to thank Cristina Niccoli and Serena Pelliccioni for their technical support.
Funding
Open access funding provided by Università di Pisa within the CRUI-CARE Agreement. Reagents have been acquired partly with funds of the University of Pisa (recipient F.B.; no specific grant number), and partly with funds from Tuscany Region (recipient F.B.; PRE-MED2, Bando Ricerca Salute 2018).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Ethical approval
The study was approved by the Institutional Ethical Committee (CEAVNO, protocol number 9989/2019).
Informed consent
Patient consent was waived because it was conducted on anonymous archive samples. All the experimental procedures have been performed in accordance with the Declaration of Helsinki.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Macerola, E., Poma, A.M., Vignali, P. et al. MicroRNA expression profiling of RAS-mutant thyroid tumors with follicular architecture: microRNA signatures to discriminate benign from malignant lesions. J Endocrinol Invest 46, 1651–1662 (2023). https://doi.org/10.1007/s40618-023-02023-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40618-023-02023-5