Abstract
Introduction
Inaccurate, untimely diagnoses of fundus diseases leads to vision-threatening complications and even blindness. We built a deep learning platform (DLP) for automatic detection of 30 fundus diseases using ultra-widefield fluorescein angiography (UWFFA) with deep experts aggregation.
Methods
This retrospective and cross-sectional database study included a total of 61,609 UWFFA images dating from 2016 to 2021, involving more than 3364 subjects in multiple centers across China. All subjects were divided into 30 different groups. The state-of-the-art convolutional neural network architecture, ConvNeXt, was chosen as the backbone to train and test the receiver operating characteristic curve (ROC) of the proposed system on test data and external test date. We compared the classification performance of the proposed system with that of ophthalmologists, including two retinal specialists.
Results
We built a DLP to analyze UWFFA, which can detect up to 30 fundus diseases, with a frequency-weighted average area under the receiver operating characteristic curve (AUC) of 0.940 in the primary test dataset and 0.954 in the external multi-hospital test dataset. The tool shows comparable accuracy with retina specialists in diagnosis and evaluation.
Conclusions
This is the first study on a large-scale UWFFA dataset for multi-retina disease classification. We believe that our UWFFA DLP advances the diagnosis by artificial intelligence (AI) in various retinal diseases and would contribute to labor-saving and precision medicine especially in remote areas.
Similar content being viewed by others
Why carry out this study? |
Inaccurate and untimely diagnoses of retinal fundus diseases can lead to vision-threatening complications and even blindness, which decreases the quality of life and aggravates economic burden. |
Compared with conventional images, ultra-widefield fluorescein angiography (UWFFA) has the advantages of a wide imaging range and fast acquisition, which leads to a great improvement in accurate diagnosis and evaluation. |
We developed a clinically applicable deep learning model to facilitate diagnosis and evaluation in 30 fundus diseases, making it the first study on a large-scale UWFFA dataset for multi-retina disease classification. |
What was learned from the study? |
Our deep learning platform (DLP) could achieve remarkable accuracy on both test [0.940 area under the receiver operating characteristic curve (AUC)] and external test datasets (0.954 AUC), which was comparable with retinal specialists. |
We believe that our UWFFA DLP enhanced the auxiliary diagnosis in 30 types of common retinopathies immediately following examination, and this promising tool will fill the gap in current artificial intelligence (AI) tools for UWFFA image diagnosing, and contribute to labor-saving and precision medicine, especially in remote areas. |
Introduction
Millions of people in the world suffer from ocular fundus diseases such as diabetic retinopathy (DR) [1], retinal vein occlusion (RVO) [2], age-related macular degeneration (AMD) [3], retinal detachment (RD) [4], optic neuropathy, and fundus tumors [5]. Without accurate diagnoses and timely appropriate treatment, these diseases cause irreversible blurred vision, metamorphopsia, visual field defects, or even blindness, which decrease the quality of life and aggravate economic burden.
Fundus fluorescein angiography (FFA) is an examination method with routine clinical application [6]. In recent years, ultra-widefield fluorescein angiography (UWFFA) has emerged with the advantage of capturing nearly 200° and has been used to image a larger retinal area. The application of UWFFA is of great significance for the diagnosis and evaluation of a variety of vitreoretinal diseases and allowing for more accurate severity grading [7]. However, the learning curve of effectively reading either FFA or UWFFA images is long and training a professional requires significant time and cost. Especially in some rural and remote regions, characterized by insufficient ophthalmic services and a shortage of ophthalmologists, artificial intelligence (AI) and telemedicine can help physicians function more effectively. An examination using either FFA or UWFFA can be administered by nonprofessionals using an AI system and selectively delivered online to major ophthalmic institutions for diagnosis.
Although AI has already achieved impressive performance on color photography and optical coherent tomography (OCT), the studies of UWFFA are either still rare or only involve a single disease or feature, due to the lack of data. Moreover, UWFFA images provide varying dynamic information that cannot be simply analyzed one-by-one as in static OCT and color photography. Previous studies [8, 9] also ignored the difference in distribution between training and external test data, resulting in a significant performance drop on out-of-distribution (o.o.d.) testing data.
In this study, we collected a total of 61,609 UWFFA images dating from 2016 to 2021, involving more than 3364 subjects in multiple centers across China. Based on this large-scale UWFFA dataset, we have developed a multi-disease automatic detection platform by a multi-expert classification model and a test-time aggregation method that could classify 30 types of common fundus diseases and conditions based on UWFFA images. It demonstrated a competitive performance compared with retina specialists. Furthermore, our system also demonstrates high efficiency in the external multi-hospital test particularly for DR, RVO, and so on. In general, our study represents an important step in the application of AI using UWFFA images.
Methods
Data Acquisition and Preprocessing
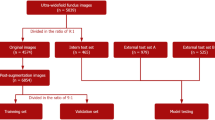
A total of 56,327 UWFFA images were collected from Eye Center of the Renmin Hospital of Wuhan University. All images were captured from February 2016 to September 2021 using Optos 200Tx or California (Optos plc, Dunfermline, UK) with 200° fields of view. The subjects were examined after mydriasis and ultra-widefield fundus (UWF) pseudocolor images were obtained. After intravenous injection of fluorescein dye, UWFFA images were captured during the early (within 1 min) and late (5–10 min) phases. A total of 2689 images were excluded due to poor quality, 12,826 images were excluded because subjects were treated before capturing images, and 230 images were excluded due to each disease only containing images from a single subject. Finally, 40,582 images were split totally randomly into three sets: the training set contained 27,156 images from 2055 subjects, the test set contained 8538 images from 671 subjects, and the external test set contained 4888 images from 379 subjects. There was no overlap in subjects between training, test, and external test sets. Furthermore, 4550 UWFFA images from 196 subjects collected from Shaanxi Eye Hospital, Xi’an People’s Hospital (Xi’an Fourth Hospital) and 732 UWFFA images from 63 subjects collected from Tianjin Medical University Eye Hospital were added to the external test set to verify the generalizability of the model. All images were preliminary annotated by two graduate students, and then reviewed by senior doctors with more than 3 years of working experience. When the images classified by the two senior doctors were inconsistent, it would be judged by a superior doctor with more than 20 years of working experience. All images were classified into 30 diseases: DR, normal, uveitis, central retinal vein occlusion (CRVO), branch retinal vein occlusion (BRVO), myopia, optic neuropathy, familial exudative retinopathy (FEVR), floaters, macular disorders without AMD, ocular ischemia syndrome (OIS), retinal degeneration, central serous chorioretinopathy (CSC), RD, AMD, choroiditis, retinal arterial obstruction (RAO), Eales disease, Coat’s disease, Stargardt disease, retinitis pigmentosa (RP), retinal vasculitis, asteroid hyalosis, acute retinal necrosis (ARN), retinal tear, retina capillary hemangioma (RCH), retinal arterial macroaneurysms (RAMs), drusen, retinal medullated nerve fibers (RMNF), and prepapillary vascular loops. After annotation, the training set and test set contained all 30 diseases, the external test set only contained 24 diseases with RAO, ARN, RCH, RAMs, drusen, and prepapillary vascular loops not included. All of the UWF images were anonymized before being utilized by research investigators. This study was approved by the Clinical Research Ethics Committee of Renmin Hospital of Wuhan University (ethics number WDRY2021-K034) and conducted in accordance with the tenets of the Declaration of Helsinki. Informed consent was waived by the ethics committee as none of the images contained personal information about the patients. We have permission to access the database.
Model Architecture
Due to the large number of disease labels and the extremely unbalanced data, as shown in Fig. 1a, the conventional deep learning strategies and architectures leveraged in [10,11,12,13] are not sufficient for our task. As the distribution of samples in the real world and in the test dataset might not be the same as the distribution of training samples, the trained model would perform poorly under that situation. Moreover, UWFFA images combined consecutive information, therefore it is hard to diagnose a case with a single image because the information contained in one image is insufficient.
The distribution of datasets. a The distribution of the training data. b The distribution of the test data. c–e The distribution of the external test data from WH, XA, and TJ, respectively. WH: Renmin Hospital of Wuhan University; XA: Shaanxi Eye Hospital, Xi’an People’s Hospital (Xi’an Fourth Hospital); TJ: Tianjin Medical University Eye Hospital
To overcome these problems, we proposed (i) a multi-expert model with shared shallow layers and independent expert networks to learn feature extraction and (ii) a test-time aggregation method to combine knowledge from multi-experts for classification. The schematics of the proposed method is shown in Fig. 2.
The structure of proposed method. \({f}_{\theta }\): shared shallow layers. \({E}_{i}\): expert subnets. \({C}_{i}\): classifier of each expert. \({\mathcal{L}}_{CE}\): cross-entropy loss. \({\mathcal{L}}_{CB}\): class balanced loss. \({\mathcal{L}}_{INV}\): inversely weighted softmax loss. \({E}_{i}^{j}\): expert subnets with jth dropout method. \({\mathcal{L}}_{DKL}\): symmetrical Kullback–Leibler divergence. \({w}_{i}^{j}\): the weight of softmax output from ith expert subnet with jth dropout method. \({P}_{i}\): the weighted prediction. a The scheme of the framework with multi-experts Sect. “Multi-expert Framework”: The framework trains multiple experts with a shared backbone to learn different knowledge. b Test-time experts aggregation Sect. “Test-time Experts Aggregation”: Data are fed into a network with different dropout methods learning the weight of each expert to fully utilize the diverse knowledge from all experts. Best viewed in color. Zoom in for details
Backbone
The state-of-the-art convolutional neural network architecture, ConvNeXt [14, 15] was chosen as the backbone, which combined the advantages of ConvNet and Transformer [16, 17], and achieves competitive performance with Transformer-based methods on image classification tasks. The reason of using ConvNeXt rather than Transformer-based methods was that Transformer-based methods are data hungry so the number of training samples of most diseases were not enough to train that type of method. A similar observation was also found in other studies [18].
Multi-Expert Framework
As shown in Fig. 2, the proposed model includes two parts: (1) shared two shallow layers of the backbone \({f}_{\theta }\) that could extract general low-level features [19]; (2) individual expert networks \({E}_{1}, {E}_{2}\) and \({E}_{3}\), each containing two deep layers of the backbone that learn objective-specific features and a classifier for different tasks [20]. The prediction of the framework is the average of the logits of all experts.
Each expert was assigned to learn different objectives and constrained by different losses. \({E}_{1}\) was designed to be good at the distribution of training data, which would perform well in many group classes. It is constrained by the traditional cross-entropy loss [21]:
where \({E}_{1}(\bullet )\) is the softmax probability calculated by the expert network \({E}_{1}\), \(N\) is the number of data; \({E}_{2}\) learns features and the classifier from balanced distribution. It is guided by the class balanced loss [22]:
where \({E}_{2}(\bullet )\) is the softmax probability produced by the expert network \({E}_{2}\), \(\beta\) is the hyperparameter, and \({N}_{y}\) is the number of samples in the data labeled as \(y\); \({E}_{3}\) aims to excel on the inverse distribution of training data, which would perform well in few group classes. It is supervised by a modified weighted softmax loss:
where \({E}_{3}(\bullet )\) is the softmax probability of the expert network \({E}_{3}\), \({\pi }_{y}\) denotes the frequency of the class y to compensate for the long-tailed distribution. To guide the whole model, the final loss is calculated as:
Test-Time Experts Aggregation
All experts are skilled in different groups and distributions, so it is important to aggregate them on unknown test distributions. The rationale of our test-time aggregation is that the expert should play a more important role and be robust in their area of expertise. To measure the robustness of experts, we introduced a self-supervised method that aggregates experts with normalized weights. As shown in Fig. 2, different from other self-supervised methods that create different views for a given sample, our method utilizes different subnets \({f}_{1}(\bullet )\) and \({f}_{2}(\bullet )\) generated by dropout proposed in R-drop [23] to find out the learnable weight \(W=[{w}_{1},{w}_{2},{w}_{3}]\in {R}^{3}\) that minimizes the symmetrical Kullback–Leibler divergence [24]
During test-time aggregation, only the learnable weight \(W\) was updated, all parameters in the networks and classifiers were frozen. Since an expert would give similar predictions from different subnets on skilled classes, minimizing Kullback–Leibler divergence would learn higher weights for the stronger expert, even though the test data distribution is unknown.
Consecutive Information Aggregation
Different from single-image classification, UWFFA images were captured in a sequence so that each image provides different information. As discussed previously, only utilizing a single UWFFA image for classification was insufficient for retinal specialists. To fully use the serial UWFFA images, we proposed to aggregate information with information entropy. At test time, the softmax of each image was calculated using test-time experts aggregation and the information entropy of the softmax from each sample was computed during aggregation. After all images from a single case were processed, the information entropy of all images were normalized, and their reciprocal were used as the weight, adding all softmax together to get the final softmax output for classification.
Results
Implementation Details
All input images were resized to 512 × 512 pixels, followed by random cropping with padding, random flipping, random erasing [25], and RandAugment [26] as data augmentation. All models were trained for 200 epochs on 4 Nvidia RTX3090 GPUs with PyTorch. During training, the optimizer AdamW [27] was applied with a learning rate of 4 × 10–3. Subsequently, there was a 10 epoch linear warmup and a cosine decaying schedule. The networks were regularized with stochastic depth [28] and label smoothing [29]. The backbone was pretrained on ImageNet-1 K [30]. The \(\beta\) was set to 0.99.
Data Characteristics
UWFFA images were collected for deep learning algorithm development and validation. Training and test data were collected from Renmin Hospital of Wuhan University, denoted as WH in the following sections. External test data were collected from Shaanxi Eye Hospital, Xi’an People’s Hospital (Xi’an Fourth Hospital), denoted as XA in the following sections and Tianjin Medical University Eye Hospital denoted as TJ in the following sections. Table 1 presents an overview of the training and test datasets, Table 2 provides a summary of the external test dataset, and Table 3 illustrates the distribution of age and gender. Among them, the training data were extremely biased (Fig. 1a), and the distributions of the test data (Fig. 1b) and the external test data (Fig. 1c–e) were different from the training data.
System Architecture
A system was developed to classify 30 types of disease from UWFFA images. A multi-experts model was trained to learn feature extraction and a test-time aggregation method was introduced to utilize knowledge from multi-experts and the sequential information from UWFFA images. The system was then evaluated under two different settings. One was similar to the real-world common clinical diagnosis methods, that is, doctors read multiple images from a single subject continuously to diagnose the disease comprehensively, the other was based on single-image classification. The technical details of the system and its implementation are explained in further detail below.
Performance of the Proposed System
Due to the biased distribution, we split data in the test dataset into three groups: Many (training samples > 1000), medium (med) (100 < training samples < 1000), and few (training samples < 100). Table 4 and Fig. 3 show that the proposed framework performed well in most diseases on test dataset. As shown in Fig. 4 for the classification accuracy of each disease, the darker color on the diagonal of the matrix indicates a better classification result. This demonstrated the effectiveness of our design. To be specific, we achieved a referable frequency-weighted average F1 score of 0.792, sensitivity of 0.778, specificity of 0.970, and area under the receiver operating characteristic curve (AUC) of 0.940. The system performs relatively well on all diseases in the “many” group. In the many group, the range of F1 scores is 0.759–0.876. The highest F1 score was achieved on normal (0.876), which has the most subjects in the training set. The second was DR (0.868), which has the most training samples. All AUC are greater than 0.9 in the “many” group. Different from the “many” group, the F1 scores in the “med” group have a wide range of 0.167–0.800. The system cannot classify OIS, Coats, and AMD well. However, the AUC of each disease in the “med” group is also greater than 0.8. In contrast, the system performs surprisingly good on the “few” group, achieving 0.512 weighted-average F1 score, and the weighted accuracy is also much higher than “med” group. The average AUC in the “few” group is almost as high as in the “many” group. The specificity of all diseases are greater than 0.95.
ROC of the proposed system on test data. a–f ROC curves and AUC for detecting every disease on test data were calculated and plotted. ROC curves of each disease and condition are listed in different colors. g–i ROC curves and AUC were calculated and plotted by “many,” “med,” and “few” groups, respectively. ROC: receiver operating characteristic curve; AUC: area under the receiver operating characteristic curve; Med: medium; DR: diabetic retinopathy; CRVO: central retinal vein occlusion; BRVO: branch retinal vein occlusion; FEVR: familial exudative vitreoretinopathy; AMD: age-related macular degeneration; OIS: ocular ischemia syndrome; CSC: central serous chorioretinopathy; RAO: retinal arterial obstruction; RP: retinitis pigmentosa; ARN: acute retinal necrosis; RCH: retina capillary hemangioma; RAMs: retinal arterial macroaneurysms; RMNF: retinal medullated nerve fibers
The confusion matrix of the proposed framework on different test dataset under different settings. a The performance of our backbone without aggregation strategy. b The performance of our system with aggregation strategy using only a single image. c The performance of our system with aggregation strategy using clinical standard diagnose. d The performance of our system with aggregation strategy using clinical standard diagnose on external test dataset. w/o: without; A: aggregation strategy; w/: with; S: single image; T: test dataset; C: clinical standard diagnose; T_ext: external test dataset
The proposed system was also evaluated on the external test dataset. The distribution of the external test dataset was different from the training and test dataset, as shown in Fig. 1, which was far more challenging. Conventional classification algorithms cannot achieve good performance on o.o.d. data. Unlike those algorithms, Table 5 and Fig. 5 indicated that our system could classify disease well, even on an o.o.d. dataset. In particular, we achieved a referable frequency-weighted average F1 score of 0.865, sensitivity of 0.831, specificity of 0.973, and AUC of 0.954. The proposed system achieved stable performance on “many” and “few” groups from o.o.d. data, and the specificity was lower than the test dataset. Unexpectedly, the system carried out better classification results in the med group from external test data. The system failed to classify RMNF, RP, Stargardt disease, choroiditis, and retinal degeneration, which could be successfully distinguished in the test dataset.
ROC of the proposed system on external test data. a–f ROC curves and AUC for detecting every disease on external test data were calculated and plotted. ROC curves of each disease and condition are listed in different colors. g–i ROC curves and AUC were calculated and plotted by “many,” “med,” and “few” groups, respectively. ROC: receiver operating characteristic curve; AUC: area under the receiver operating characteristic curve; DR: diabetic retinopathy; med: medium; CRVO: central retinal vein occlusion; BRVO: branch retinal vein occlusion; FEVR: familial exudative vitreoretinopathy; AMD: age-related macular degeneration; OIS: ocular ischemia syndrome; CSC: central serous chorioretinopathy; RP: retinitis pigmentosa; RMNF: retinal medullated nerve fibers
We also conducted ablation studies to demonstrate the effectiveness of each proposed module. Firstly, an ablation study was conducted on each loss, and the results are presented in Table 6. The table indicated that all losses are essential for our method. This necessity arises from the foundation of our method in multi-experts aggregation, enabling the utilization of knowledge acquired from various experts to enhance performance. Secondly, we also tried different number of subnets used during the test-time experts aggregation. We found the advantage of employing additional subnets was obscure. The model gained +0.008 average F1 on external test by leveraging three subnets and +0.010 average F1 by leverage four subnets. Therefore, we opted for the implementation of test-time experts aggregation using two subnets, rather than a greater number.
AI System versus Retinal Specialists
We evaluated the proposed system versus retinal specialists on two different settings: (i) The first experiment was tested on a small dataset, which contained 50 UWFFA images from 19 different diseases (DR, normal, uveitis, CRVO, BRVO, myopia, optic neuropathy, FEVR, floaters, macular disorders without AMD, CSC, RD, AMD, choroiditis, Eales disease, Coats disease, Stargardt disease, RP, retinal tear). None of these images were included in the training or testing datasets. The evaluation protocol was consistent with multiple UWFFA images from a single subject diagnosed during testing. As shown in Fig. 6a, our system performed better than retinal specialists with 3 years or less of experience, but still worse than retinal specialists with more than 6 years of experience. (ii) The second experiment was evaluated on another small dataset selected from the test dataset, which contained 50 UWFFA images from 29 diseases (similar diseases from training data except for RD), and classified with a single UWFFA image. Our system achieved much better results than retinal specialists with a single UWFFA image (Fig. 6b). This indicated that our system can automatically classify the disease from limited UWFFA images. Besides, the intergrader reliability between physicians showed substantial agreement (Cohen’s kappa statistics: graduate students 0.708, p < 0.001, senior doctors 0.802, p < 0.001), and all showed strong consistency.
The classification performance of retinal specialists verses the proposed system. a The accuracy of retinal specialists with different years of experience and our system using multi-images to diagnose. b The accuracy of retinal specialists with different years of experience and our system using single images to diagnose
Discussion
At present, the rapid development of AI technology has seen a wide range of applications throughout the years. This is especially true in computer-aided diagnosis of human diseases [31], including but not limited to Alzheimer’s disease [32], skin cancer, and lung cancer [33]. In retinal diseases, deep learning algorithms have been applied for AI-assisted diagnoses to detect DR [34], AMD [35], retinopathy of prematurity (ROP) [36], glaucoma [37], and papilledema [38]. Many of them are based on fundus color photography and OCT, while only a few AI applications are related to FFA or UWFFA. Ding et al. [39] proposed a novel pipeline to detect retinal vessels in UWFFA images using deep neural networks (DNNs). This approach reduced the effort required for generating labeled ground truth data by combining two key components: Cross-modality transfer and human-in-the-loop learning. Moreover, Pan et al. [40] also applied the model to identify the nonperfusion areas, neovascularization, laser scars, and microaneurysms in DR eyes. However, this approach did not conduct either disease classification nor grading.
Given this background, our team was the first to adopt a deep learning model based on the generative adversarial network (GAN) and convolutional neural network (CNN) models in DR. This approach allowed for DR classification across different levels of severity. Notably, the accuracy of the model was comparable to that of the diagnoses made by resident doctors [41]. Nevertheless, limited work has investigated applying AI to FFA or UWFFA. This is not because the examination is not important or clinically impractical. On the contrary, both FFA and UWFFA are very important and sometimes indispensable for the diagnosis of many retinal diseases. Rather, it is difficult to achieve AI-mediated image reading. This is because the examination is a dynamic process, and the fluorescein sodium circulates in the blood and gradually empties over time. As such, the images at different time points are different, and the characteristics of a single image are limited. Comprehensive evaluation of the entire process is required when reading films manually.
In this study, our model focused on multiple diseases instead of identifying only one disease as in most previous studies. In clinical practice, retinal disease screening of single-disease diagnostic algorithms (e.g., DR) would not recognize other fundus diseases such as AMD, glaucoma, RVO, and RAO. In reality, and especially in remote areas with an insufficient number of ophthalmologists, there is an urgent need for the ability to effectively detect various types of fundus diseases. It is very valuable to develop a multidisease detection system using fundus images to avoid missed diagnoses and delayed treatment. Additionally, the probability of occurrence of different diseases varies, which results in various data distributions at different places and biased sample data. Previous works [8, 42] have used augmentation to alleviate unbalanced samples and achieved considerable success on classification tasks. However, they have not considered o.o.d. test data, where the commonly used methods perform poorly on o.o.d. data.
To learn from biased long-tail training data and make the DLP system robust on different test distributions, we applied a multi-expert strategy to build a deep learning algorithm as described in more detail below. Each expert is constrained by different loss functions to learn diverse knowledge from the training data. A test-time multi-expert aggregation method is also introduced to fully utilize the knowledge from all experienced experts. Moreover, we utilized entropy information weighting the importance of each image in a case to aggregate consecutive information from sequential UWFFA images that normal fundus color photography and OCT do not contain. For all diseases, the high-resolution (512 × 512 pixels) images were fed into the model to extract detailed features. Figure 4 shows the confusion matrices of the proposed DLP with different testing strategies. Figure 4a demonstrates that without aggregation strategy, the backbone cannot achieve a satisfied performance; Fig. 4b tells that our system could correctly classify most disease even with only a single image, which is also proved by the results shown in Fig. 6; Fig. 4c indicates that the proposed entropy information weighting could increase the sensitivity, specificity, and F1 scores of each disease; and Fig. 4d implies that our model is robust on external test data with unknown distribution.
The deep neural network works as a black box that hinders it from being applied in clinical works [9]. To understand the framework, Grad-cam [43] is utilized to visualize the attention map of the model. The specialists could examine whether the model makes a right decision based on the correct captured features on the images. From Fig. 7 we can find that our model could locate most of the lesion correctly. Moreover, we also analyzed other samples from the test dataset, which indicated that the model paid more attention to disease-related areas that were consistent with expert domain knowledge.
UWFFA images and corresponding visualized attention map. a Our model located the lesion correctly in the AMD eye. b The model missed some correct characteristics of the lesion in ARN, resulting in misclassification. c Our model located the lesion correctly in the BRVO eye. d The RAMs shares similar features and lesion location with BRVO, thus causing a misclassification event. UWFFA: ultra-widefield fluorescein angiography; AMD: age-related macular degeneration; ARN: acute retinal necrosis; BRVO: branch retinal vein occlusion; RAMs: retinal arterial macroaneurysms
As far as we know, our study is the first to realize the multidisease classification of UWFFA based on deep learning, which well fills the gap in the application of AI. If this multidisease algorithm model were to be widely used, it would bring great convenience to patients and specialists alike. For example, the algorithm can be used in intelligent UWFFA image analysis systems and hospital electronic medical report management systems. For patients, they can obtain an AI rapid diagnosis report as soon as they finish the UWFFA examination. This would provide timely information about their condition. For specialists, this tool would reduce the workload of trained professionals and allow untrained technicians to objectively screen and handle many more patients without relying on specialists. In addition, it is also convenient for specialists to manage and track patient data, so as to better grasp the patient’s condition. This would also allow for the physicians’ needs regarding learning and scientific research to be met. Finally, we hope that our model can be applied to general clinical situations, as we believe it will benefit specialists and patients in county hospitals and rural areas in particular.
This study contains several limitations, which caused a few failure classification cases. (i) Even though our system alleviated the long-tailed problem, still the dataset in some rare disease with limited data. Our system demonstrated favorable performance in the “many” group, while less than satisfactory in “med” group, as indicated in Table 4 and Table 5. This might be caused by (1) it being challenging to extract features of lesions that are too diffused or too small from limited training samples. A typical example is shown in Fig. 7b where the model missed some correct characteristics of the lesion in ARN, resulting in misclassification. Likewise, the similar problem can be observed in OIS and prepapillary vascular loops; (2) when it comes to common features in two different diseases, it is tricky to distinguish one type in the “med”/“few” group from another disease type in the “many” group. For instance, as shown in Fig. 7d, the RAMs shares similar features and lesion location with BRVO thus causing a misclassification event. However, it is not solely exists in DLPs, until now we are facing the same issue with experienced human specialists; and (3) capturing small lesions such as leaking points and prepapillary vascular loops is subtle even in high-resolution images. The original resolution of the UWFFA is around 4 K, which was resized to 512 × 512 for our system. With a lower resolution, some lesions were lost during compression, and this will affect learning robust representations of the lesions from training data. (ii) Currently, our system lacks of the ability to distinguish the quality of the image. In this study, low-quality data were manually excluded from our datasets. However in the real world, the image quality is not always ideally high enough for our DLP, resulting in performance degradation. For decades, tremendous effort has been made in the standardization of imaging and examination, and auto-selection algorithms are also under developing; together we believe that we could fix UWFFA image quality issue in the DLP system. These common misclassifications are excusable to some extent. The main advantage of UWFFA lies in retinal vascular diseases and peripheral lesions, meanwhile it is difficult to accurately classify some tiny and occult lesions of macula and optic disc. In fact, specialists face the similar problems in reading UWFFA images. As we mentioned above, several aspects need to be further improved in our DLP system. As a next step, we will optimize this model by collecting a larger number of UWFFA images. In addition, we will conduct real-world study in the future, and continue to verify the performance of our model with real-world data.
Conclusions
To our knowledge, this is the first report to show up to 30 types of retina diseases and conditions that can be detected by deep learning algorithms in UWFFA at an accuracy level comparable to that of retinal specialists. This is also the largest UWFFA image dataset at present. Such a UWFFA DLP could be applied to telemedicine systems and would contribute to labor saving and precision medicine especially in remote areas. In the future, we will continue to accumulate larger scale of sample sizes and optimize the classification model with higher accuracy and more types of diseases, while achieving automatic classification and grading of common diseases [e.g., the Early Treatment for Diabetic Retinopathy Study (ETDRS) automatic grading in DR], to provide more comprehensive and prognostic information of certain diseases.
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Ting DS, Cheung GC, Wong TY. Diabetic retinopathy: global prevalence, major risk factors, screening practices and public health challenges: a review. Clin Exp Ophthalmol. 2016;44:260–77.
Song P, Xu Y, Zha M, Zhang Y, Rudan I. Global epidemiology of retinal vein occlusion: a systematic review and meta-analysis of prevalence, incidence, and risk factors. J Glob Health. 2019;9: 010427.
Chen YM, Huang WT, Ho WH, Tsai JT. Classification of age-related macular degeneration using convolutional-neural-network-based transfer learning. BMC Bioinformatics. 2021;22:99.
Kwok JM, Yu CW, Christakis PG. Retinal detachment. CMAJ. 2020;192:E312.
Andreoli MT, Chau FY, Shapiro MJ, Leiderman YI. Epidemiological trends in 1452 cases of retinoblastoma from the Surveillance, Epidemiology, and End Results (SEER) registry. Can J Ophthalmol. 2017;52:592–8.
Hill DW. Fluorescein angiography in fundus diagnosis. Br Med Bull. 1970;26:161–5.
Patel M, Kiss S. Ultra-wide-field fluorescein angiography in retinal disease. Curr Opin Ophthalmol. 2014;25:213–20.
Dai L, Wu L, Li H, et al. A deep learning system for detecting diabetic retinopathy across the disease spectrum. Nat Commun. 2021;12:3242.
Wong TY, Bressler NM. Artificial intelligence with deep learning technology looks into diabetic retinopathy screening. JAMA. 2016;316:2366–7.
Li Z, Guo C, Nie D, et al. Automated detection of retinal exudates and drusen in ultra-widefield fundus images based on deep learning. Eye (Lond). 2022;36:1681–6.
Bi Q, Yu S, Ji W, et al. Local-global dual perception based deep multiple instance learning for retinal disease classification. Cham: Springer International Publishing; 2021. p. 55–64.
Karthikeyan SK, Sanjay P, Madhusudan RJ, Sundaramoorthy SK, Namboori PKK. Detection of multi-class retinal diseases using artificial intelligence: an expeditious learning using deep CNN with minimal data. Biomed Pharmacol J. 2019;12:1577–86.
Arunkumar R, Karthigaikumar P. Multi-retinal disease classification by reduced deep learning features. Neural Comput Appl. 2017;28:329–34.
He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2016. pp 770–8.
Simonyan K, Zisserman A. Very deep convolutional networks for large-scale Image recognition. arXiv:14091556 2015.
Dosovitskiy A, Beyer L, Kolesnikov A, et al. An image is worth 16 × 16 words: transformers for image recognition at scale. arXiv:201011929 2021.
Touvron H, Cord M, Douze M, Massa F, Sablayrolles A, Jegou H. Training data-efficient image transformers & distillation through attention. In: Marina M, Tong Z (eds) Proceedings of the 38th International Conference on Machine Learning. Proceedings of Machine Learning Research: PMLR; 2021. p. 10347–57.
Khan S, Naseer M, Hayat M, Zamir SW, Khan FS, Shah M. Transformers in vision: a survey. arXiv:210101169. 2021.
Yosinski J, Clune J, Bengio Y, Lipson H. How transferable are features in deep neural networks? arXiv:14111792. 2014.
Wang XD, Lian L, Miao ZQ, Liu ZW, Yu SX. Long-tailed recognition by routing diverse distribution-aware experts. arXiv:201001809 2020.
Zheng Z, Zheng L, Yang Y. A discriminatively learned CNN embedding for person reidentification. ACM Trans Multimed Comput Commun Appl. 2017;14:13.
Cui Y, Jia M, Lin TY, Song Y, Belongie S. Class-balanced loss based on effective number of samples. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2019. pp 9260–9.
Liang XB, Wu LJ, Li JT, et al. R-drop: regularized dropout for neural networks. arXiv:210614448 2021.
Kullback S, Leibler RA. On information and sufficiency. Ann Math Stat. 1951;22:79–86.
Zhong Z, Zheng L, Kang GL, Li SZ, Yang Y. Random erasing data augmentation. Proc AAAI Conf Artif Intell. 2020;34:13001–8.
Ekin D, Cubuk BZ, Shlens J, Quoc V. Le. RandAugment: Practical automated data augmentation with a reduced search space. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops. 2020, pp 702–3.
Loshchilov L, Hutter F. Decoupled weight decay regularization. arXiv:171105101. 2019.
Huang G, Sun Y, Liu Z, Sedra D, Weinberger KQ. Deep networks with stochastic depth. In: Leibe B, Matas J, Sebe N, Welling M, editors. Computer vision—ECCV 2016. Cham: Springer International Publishing; 2016. p. 646–61.
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2016. pp 2818–26.
Deng J, Dong W, Socher R, Li L-J, Li K, Li F-F. ImageNet: a large-scale hierarchical image database 2009.
Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25:44–56.
Duc NT, Ryu S, Qureshi MNI, Choi M, Lee KH, Lee B. 3D-deep learning based automatic diagnosis of Alzheimer’s disease with joint MMSE prediction using resting-state fMRI. Neuroinformatics. 2020;18:71–86.
Ardila D, Kiraly AP, Bharadwaj S, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med. 2019;25:954–61.
Jin K, Pan X, You K, et al. Automatic detection of non-perfusion areas in diabetic macular edema from fundus fluorescein angiography for decision making using deep learning. Sci Rep. 2020;10:15138.
Grassmann F, Mengelkamp J, Brandl C, et al. A deep learning algorithm for prediction of age-related eye disease study severity scale for age-related macular degeneration from color fundus photography. Ophthalmology. 2018;125:1410–20.
Gensure RH, Chiang MF, Campbell JP. Artificial intelligence for retinopathy of prematurity. Curr Opin Ophthalmol. 2020;31:312–7.
Chang J, Lee J, Ha A, et al. Explaining the rationale of deep learning glaucoma decisions with adversarial examples. Ophthalmology. 2021;128:78–88.
Milea D, Najjar RP, Zhubo J, et al. Artificial intelligence to detect papilledema from ocular fundus photographs. N Engl J Med. 2020;382:1687–95.
Ding L, Bawany MH, Kuriyan AE, Ramchandran RS, Wykoff CC, Sharma G. A novel deep learning pipeline for retinal vessel detection in fluorescein angiography. IEEE Trans Image Process. 2020. https://doi.org/10.1109/TIP.2020.2991530.
Pan X, Jin K, Cao J, et al. Multi-label classification of retinal lesions in diabetic retinopathy for automatic analysis of fundus fluorescein angiography based on deep learning. Graefes Arch Clin Exp Ophthalmol. 2020;258:779–85.
Wang X, Ji Z, Ma X, et al. Automated grading of diabetic retinopathy with ultra-widefield fluorescein angiography and deep learning. J Diabetes Res. 2021;2021:2611250.
Cen L-P, Ji J, Lin J-W, et al. Automatic detection of 39 fundus diseases and conditions in retinal photographs using deep neural networks. Nat Commun. 2021;12:4828.
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. In: 2017 IEEE International Conference on Computer Vision (ICCV) 2017, pp 618–26.
Acknowledgements
The authors would like to thank all of the patients who participated in this study.
Authorship
All authors conform to the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, account for the integrity of the whole work, and have approved the version to be published.
Funding
No funding or sponsorship was received for this study or publication of this article. The Rapid Service Fee was funded by the authors.
Author information
Authors and Affiliations
Contributions
Xiaoling Wang, Changzheng Chen and Hongmei Zheng initiated the project and the collaboration. He Li developed the network architectures, training, and testing setup. Changzheng Chen, Xiaoling Wang, Hongmei Zheng and Gongpeng Sun designed the clinical setup. Xiaoling Wang and Gongpeng Sun defined the clinical labels. He Li, Mang Ye, and Bo Du contributed to software engineering. Xiaoling Wang, Hongmei Zheng, Gongpeng Sun, Wenyu Wang, Zuohuizi Yi, A’min Xu, Lu He, Haiyan Wang, Wei Jia, Zhiqing Li, and Chang Li labeled the databases. Haiyan Wang, Wei Jia, Zhiqing Li, and Chang Li contributed to external test dataset. Xiaoling Wang, Changzheng Chen, and Hongmei Zheng contributed to clinical assessments. Xiaoling Wang and He Li analyzed the data. Xiaoling Wang, Changzheng Chen, Mang Ye, and He Li interpreted the data. Xiaoling Wang and He Li wrote the paper. Changzheng Chen and Mang Ye critically revised the manuscript for important intellectual contents. Xiaoling Wang, He Li, and Hongmei Zheng contributed equally.
Corresponding authors
Ethics declarations
Conflict of Interest
Xiaoling Wang, He Li, Hongmei Zheng, Gongpeng Sun, Wenyu Wang, Zuohuizi Yi, A’min Xu, Lu He, Haiyan Wang, Wei Jia, Zhiqing Li, Chang Li, Mang Ye, Bo Du, and Changzheng Chen have nothing to disclose.
Ethical Approval
This study was approved by the Clinical Research Ethics Committee of Renmin Hospital of Wuhan University (ethics number WDRY2021- K034) and conducted in accordance with the tenets of the Declaration of Helsinki. Informed consent was waived by the ethics committee as none of the images contained personal information about the patients. We have permission to access the database.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Wang, X., Li, H., Zheng, H. et al. Automatic Detection of 30 Fundus Diseases Using Ultra-Widefield Fluorescein Angiography with Deep Experts Aggregation. Ophthalmol Ther 13, 1125–1144 (2024). https://doi.org/10.1007/s40123-024-00900-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40123-024-00900-7