Abstract
Introduction
Rapid identification of the causal organism and antibiotic resistance is crucial for guiding targeted therapy in patients with suspected staphylococcal infection. A meta-analysis was carried out to evaluate the diagnostic relevance of Xpert™ MRSA/SA (Xpert) from clinical samples of various origins for limiting the use of unnecessary empirical methicillin-resistant Staphylococcus aureus (MRSA) therapy.
Methods
Five databases, including the Cochrane Library, Scopus, PubMed, Web of Science, and Embase, were comprehensively inspected from inception to October 12, 2021. The pooled summary estimates were evaluated using a bivariate random-effects model.
Results
Our inclusion criteria were met by 49 publications containing 68 datasets out of 735 citations. A total of 21 studies (n = 4996) examined the accuracy of Xpert in detecting methicillin-sensitive S. aureus (MSSA), while 47 studies (n = 45,430) examined the accuracy of Xpert in detecting MRSA. As compared to MRSA, Xpert’s diagnostic performance for MSSA detection was markedly higher [sensitivity: 0.97 (0.96–0.98), specificity: 0.97 (0.97–0.98), area under curve (AUC): 0.99 (0.99–1.0)]. Xpert’s pooled sensitivity and specificity differed marginally across sample types, including screening of colonization, lower respiratory tract (LRT), osteoarticular, and bloodstream samples. Notably, the Xpert pooled specificity was consistently ≥ 92% against microbiological culture across all sample types. The diagnostic efficiency heterogeneity was not explained by a meta-regression and subgroup analysis of research design, sample conditions, and sampling methods (P > 0.05).
Conclusion
Our findings suggest that Xpert could be used as the favoured screening test for the early detection of staphylococcal infection in a variety of sample types, with the goal of guiding therapeutic decisions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Rapid identification of the causal organism and antibiotic resistance is crucial for guiding treatment. |
Evaluation of Xpert accuracy in various sample settings including nasal, blood, lower respiratory tract (LRT), and osteoarticular. |
Detection of methicillin-sensitive and-methicillin resistant S. aureus to guide therapeutic decisions. |
Utilization of Xpert as the preferred early diagnostic test for staphylococcal infection to avoid empiric broad-spectrum antibiotics. |
Introduction
Staphylococcus aureus is both a commensal and an opportunistic human pathogen that may cause anything from minor skin blemishes to life-threatening conditions like sepsis [1]. Approximately 20–40% of adults carry staphylococcal strains on various body sites, which could lead to subsequent infection [2]. MRSA infection continues to be a major threat globally [3, 4], with higher morbidity and mortality than methicillin-sensitive S. aureus (MSSA) [5], leading to the recommendation of vancomycin as empirical antimicrobial therapy. Other antibiotics used as an alternative therapy for MRSA include linezolid, teicoplanin, daptomycin, and delafloxacin [6, 7]. Although there are no known immediate hazards to this method, patients may ultimately end up taking too many broad-spectrum antibiotics, which may modify the patient's microflora, expose individuals to the dangers of drug toxicity, and maximize the rate of drug-resistant bacteria [8]. Also, vancomycin is less effective than anti-staphylococcal penicillin in treating MSSA infections [9]. If the primary medications are insufficient and are changed after diagnostic procedures are accessible, the fatality rate does not significantly improve. All these conditions eventually contribute to extended hospital stays and higher treatment costs.
The current intervention of staphylococcal infections has primarily relied on gram stain and detection of pathogenic organisms using traditional culture-based methods. However, due to gram stain's low sensitivity, diagnosing staphylococcal infection remains challenging [10, 11], and a microbiological diagnosis takes approximately 48–72 h to provide a result and has reduced sensitivity compared to amplification assays [12, 13]. Poor bacterial culture detection rates may be ascribed to a combination of previous antibiotic medication before acquiring specimens [14], low bacterial concentration in fluid samples, and perhaps causative agents that are difficult to discern in the laboratory owing to rigorous criteria. The limited sensitivity of these tests hinders patient care and antibiotic selection, causing patients to lose out on the most effective treatment alternatives. Therefore, rapid identification of causative agents and prompt contact precautions are crucial for infection control and transmission prevention.
Xpert™ MRSA/SA (Xpert) (Cepheid, Sunnyvale, CA) has been shown to detect S. aureus, MRSA, and non-staphylococcal strains in a timely and efficient manner and is unaffected by prior antibiotic exposure [15, 16]. According to Brown and Paladino (2010), using the Xpert assay and implementing directed therapy based on test findings can reduce costs and mortality rates compared to empiric therapy alone [17]. The Xpert assay employs an automated real-time polymerase chain reaction (PCR)-based system that integrates sample purification and amplification and detects MSSA and MRSA in clinical specimens within 1 h [15]. This assay requires minimal training and biosafety facilities, avoids cross-contamination, and has a high responsiveness in culture negative specimens [15, 16, 18]. Xpert detects MSSA by detecting the staphylococcal protein A (spa) gene and can distinguish MSSA from MRSA given the presence of both the methicillin-resistant gene (mecA) and the staphylococcal cassette chromosome mec (SCCmec) embedded into the S. aureus chromosome attB site [19]. Spa-negative gram-positive cocci in clusters can be assumed to be coagulase-negative staphylococci because internal control detects assay failures [20]. In recent years, Xpert has been extensively documented to identify staphylococcal infection [21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69]; however, the literature on the usefulness of these tests for prompt staphylococcal infection care is widely dispersed. We therefore aimed to conduct a diagnostic test accuracy meta-analysis based on the most up-to-date research evidence to inform support in clinical decisions.
Methods
Data Sources and Searches
This study was carried out following the guidelines by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses of Diagnostic Test Accuracy (PRISMA-DTA) (see Supplementary Table 1) [70]. Five databases, including the Cochrane Library, Scopus, PubMed, Web of Science, and Embase, were methodically inspected using electronic databases from the library's inception until October 12, 2021. The search strategy was developed based on broader terms used in literature for staphylococcal infections, which include: (‘Staphylococcus aureus’ OR ‘S. aureus’ OR ‘Methicillin-resistant Staphylococcus aureus’ OR ‘MRSA’) AND (‘Xpert’ OR ‘GeneXpert’ OR ‘Roche’ OR ‘Cepheid’ OR ‘Abbott’) AND (‘Sensitivity’ OR ‘Specificity’ OR ‘Accuracy’). Furthermore, references of reviews and included papers were searched for possibly relevant research. This article is based on previously conducted studies and does not contain any new studies with human participants or animals performed by any of the authors.
Study Selection
All search results were imported into the citation manager (EndNote X9, Thomson Reuters, New York, USA), and duplicates were carefully filtered to ensure no overlapping publications. Two authors (S.C. Ojha and K. Chen) independently assessed citations by title and abstract according to preset eligibility criteria, and unrelated papers were eliminated. All studies requesting a diagnosis, whether for colonization screening or determining the causal agent of suspected staphylococcal infections, were included. The full text of all qualifying studies was reviewed for diagnostic accuracy data and that any differences of opinion were settled through mutual agreement.
Inclusion Criteria
Inclusion criteria comprised: (i) Xpert accuracy as an index test in various specimens; (ii) patients clinically or radiographically suspected of having a staphylococcal infection; (iii) implication of traditional culture as the reference standard; (iv) comprises data for specificity and sensitivity or provides adequate information to construct 2 × 2 contingency tables.
Exclusion Criteria
Exclusion criteria involved meta-analyses, conference proceedings and abstracts, reviews, letters to the editor, animal experiments, editorials, commentaries, case reports, and mechanism studies, as were studies with fewer than ten participants. Studies that failed to detect staphylococcal strains in clinically suspected patients using the index test and the microbiological reference standard were excluded.
Data Extraction
Two independent analysts (S.C. Ojha and K. Chen) piloted the data extraction form. The investigating authors independently extracted results from all selected studies using a predefined strategy. Findings were compared after data extraction, and disputes were resolved by consultation with a third investigator (C. Sun). When accuracy data or sample preparation processes were ambiguous, the authors of published research were approached. Using data from the publications, we constructed two-by-two contingency tables for Xpert performance against the microbiological culture reference standard. Investigations that included the identification of both MSSA and MRSA were regarded as distinct studies.
Quality Assessment
Independent studies were evaluated for bias using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) tool [71]. The methodological quality was evaluated independently by two investigating analysts. Molecular approaches as a reference standard were not considered. The risk of bias was judged in four QUADAS-2 domains (reference standard, flow and timing, patient selection, and index test), and applicability concerns were judged through three domains (reference standard, index test, and patient selection). The spectrum and selection biases of individuals were identified. The risk of bias in each domain was evaluated by asking signaling queries to which the answer could be "yes" or "no". The signaling questions were deemed “unclear” due to a lack of information that could not be answered as “yes” or “no”.
Statistical Analysis
Data obtained from 2 × 2 contingency tables were utilized to compute pooled summary estimates and the related confidence intervals (CIs). Missing values were substituted with 0.5 to attain a zero correction in the contingency tables. RevMan (version 5.4; Nordic Cochrane Centre, Copenhagen, Denmark) was employed to evaluate the quality of studies and generate summary plots [72]. As per random-effects model, Xpert's diagnostic accuracy with a was calculated against a culture reference standard. Meta-DiSc 1.4 (from the Cochrane Colloquium in Barcelona, Spain) was used to get pooled summary estimates of factors like the likelihood ratios, sensitivity, area under the curve (AUC), specificity, and diagnostic odds ratio (DOR) [73]. In addition, I-square (I2) statistics was used to determine the degree of data heterogeneity among the studies [74]. Meta-regression and subgroup analyses were used to examine for probable causes of heterogeneity in different research designs (prospective/other), sample conditions (fresh/frozen), and sampling methods (consecutive/ convenience). To assess publication bias, Deek's funnel asymmetry test was used [75]. Generally, a P value of < 0.05 was regarded as statistically relevant.
Results
Literature Selection
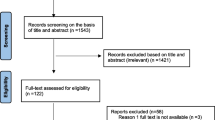
A total of 735 distinct publications were searched (PubMed, 259; Scopus, 185; the Cochrane Library, 6; Web of Science, 135; Embase, 150) (Fig. 1). Of these, 262 records were eliminated owing to database duplication. After evaluating the titles and abstracts of 473 papers, 125 studies that were judged possibly relevant were submitted for full-text review. Supplementary Table 1 summarizes the papers that were examined as well as the reasons why those studies were eliminated. Finally, future analyses included 49 papers that met all of the inclusion criteria [21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69].
Characteristics of the Included Studies
The characteristic features of the studies that were included are shown in Table 1 [21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69]. The majority of the research was carried out in high-income states, with one study being carried out in a lower-middle-income nation [53]. A single study that assessed Xpert accuracy for both MSSA and MRSA against a microbiological culture was treated as two investigations. Based on this proposition, this meta-analysis comprised 49 studies with a total of 68 datasets. Of 49 publications, 20 articles comprising 21 datasets (n = 4996) evidenced the Xpert’s pooled summary estimates for MSSA detection [24, 26, 28, 29, 31, 33, 43, 44, 49, 52, 53, 56,57,58,59,60,61,62, 67, 69], while 47 publications (n = 45,430) evidenced the Xpert’s accuracy for detection of MRSA [21,22,23,24,25,26,27,28, 30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60, 62,63,64,65,66,67,68]. For S. aureus diagnostic assessment, the median sample size per study was 182 (interquartile range: 107–333), whereas for MRSA clinical assessment, the median number of samples per study was 401 (interquartile range: 135–1074). Likewise, five publications (n = 2636) evaluated the Xpert’s accuracy for screening MSSA colonization [24, 43, 52, 62, 67], whereas 32 studies (n = 42,247) assessed MRSA colonization [21,22,23,24,25, 27, 30, 32, 34,35,36,37,38,39,40,41,42,43, 45, 46, 50,51,52, 54, 55, 62,63,64,65,66,67,68]. Four publications (n = 480) investigated the Xpert's accuracy in detecting bone and joint infection caused by MSSA, while other publications (n = 478) assessed the Xpert's accuracy in detecting MRSA in osteoarticular samples [33, 56, 57, 60]. Xpert was also evaluated for lower respiratory tract (LRT) infections. Four publications (n = 561) evaluated the Xpert’s MSSA detection accuracy [28, 29, 49, 59], while four other publications (n = 582) evaluated MRSA in LRT specimens [28, 47, 49, 59]. The remaining studies assessed Xpert's accuracy in bloodstream infections. The Xpert's accuracy to detect bloodstream MSSA was evaluated in eight publications (n = 2622) [26, 31, 44, 53, 58, 61, 67, 69], while MRSA in blood samples was evaluated in seven publications (n = 2123) [26, 31, 44, 48, 53, 58, 67]. In all cases, the research was carried out in referral hospitals or academic research laboratories. Up to October 12, 2021, all articles published in English were included.
Quality Appraisal
The quality of included studies was demonstrated using QUADAS-2 tool (see Supplementary Fig. 1A, 1B). Twenty-four publications demonstrated a high risk of bias in the patient selection domain, as the studies were unable to prevent improper sample exclusion [23,24,25,26,27,28, 32, 34, 35, 37, 38, 40, 43, 44, 46, 51,52,53,54, 56, 57, 62, 68]. Applicability concern in the patients’ selection domain was not a concern as all publications used samples from patients with possible infections, signaling a low risk of bias. Regarding index tests, all studies did not report about blinding of index tests except for three studies [65,66,67]; therefore, the risk of bias in the index test domain was considered unclear. The index test's applicability for all studies was considered unclear since there is not a globally accepted test methodology. The reference standard domain was said to have a low risk of bias due to the Xpert's use of pre-established binary dependent inquiry standards. The reference standards for all publications were performed in either referral hospitals or academic research laboratories, so we do not expect operator error bias to be an issue. Finally, because the reference standards and the index test were performed on the same samples, there was no possibility of bias in the flow and timing domains or their applicability.
Summary Estimates
Because the studies were heterogeneous, combining all of them to obtain Xpert accuracy estimates for total staphylococcal infections was not considered to be meaningful for guiding therapy choices. To restrict the spread of staphylococcal infection within clinical settings, we examined the accuracy of Xpert for MSSA and MRSA in samples from diverse sample settings. Based on the availability of data for various samples such as nasal, LRT, osteoarticular, and bloodstream samples, we sorted studies into subgroups to assess the Xpert's accuracy against a microbiological reference standard in diverse samples and obtained pooled summary estimates.
Detection of Staphylococcal Infections
A total of 20 publications (n = 4996) compared Xpert to traditional culture for MSSA detection [24, 26, 28, 29, 31, 33, 43, 44, 49, 52, 53, 56,57,58,59,60,61,62, 67, 69], while 47 publications (n = 45,430) evidenced the Xpert’s accuracy in detecting MRSA [21,22,23,24,25,26,27,28, 30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60, 62,63,64,65,66,67,68]. The Xpert’s MSSA detection sensitivity and specificity varied from 0.85 (0.72–0.94) to 1.0 (0.97–1.0) and from 0.87 (0.8–0.91) to 1.00 (0.98–1.0), respectively (see Supplementary Fig. 2A). To detect MSSA, the Xpert’s pooled estimates in overall samples were [specificity: 0.97 (0.96–0.98), sensitivity: 0.97 (0.96–0.98), negative likelihood ratio (NLR): 0.033 (0.02–0.06), positive likelihood ratio (PLR): 40.95 (23.5–71.45), DOR: 1452.6 (670.14–3148.8)]. Significant heterogeneity was seen in the I2 statistics, where the sensitivity and specificity are 84.1% and 86.8%, respectively. With an area under the curve (AUC) of 0.99 (0.99–1.0), the summary receiver-operating characteristics (SROC) were found to have high overall clinical utility (see Supplementary Fig. 3A).
Similarly, 47 publications [21,22,23,24,25,26,27,28, 30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60, 62,63,64,65,66,67,68] with a total of 45,430 samples fulfill the criteria for MRSA diagnosis in suspected patients. The Xpert’s MSSA detection sensitivity and specificity varied from 0.6 (0.47–0.73) to 1.0 (0.97–1.0) and from 0.9 (0.84–0.94) to 1.0 (0.99–1.0), respectively (see Supplementary Fig. 2B). The Xpert’s pooled estimates to detect MRSA were [specificity: 0.98 (0.97–0.98), sensitivity: 0.87 (0.86–0.88), NLR: 0.11 (0.09–0.14), PLR: 33.78 (26.8–42.57), and DOR: 352.42 (249.33–498.13)]. Significant heterogeneity was seen in the I2 statistics, where the sensitivity and specificity are 83.4% and 90.3%, respectively. With an AUC of 0.99 (0.98–1.0), the SROC were found to have high overall clinical utility (see Supplementary Fig. 3B).
Detection of Staphylococcal Colonization
A total of 2636 swabs from five publications were used to detect MSSA in clinically suspected patients [24, 43, 52, 62, 67] that met the criteria for contrasting Xpert with culture standard. The Xpert’s MSSA detection sensitivity and specificity varied from 0.87 (0.82–0.91) to 1.0 (0.94–1.0) and from 0.87 (0.80–0.91) to 0.99 (0.97–0.99), respectively (Fig. 2A). The Xpert’s pooled estimates to detect MSSA in swabs were [specificity: 0.95 (0.94–0.96), sensitivity: 0.93 (0.91–0.95), NLR: 0.06 (0.02–0.13), PLR: 19.3 (9.18–40.6), DOR: 359.64 (234.78–559.9)]. Significant heterogeneity was seen in the I2 statistics, where the specificity and sensitivity were 90.3% and 83.4%, respectively. With an AUC of 0.99 (0.98–1.0), the SROCs were found to have higher clinical utility (see Fig. 3A).
Forest plot for detection of (A) MSSA; pooled sensitivity: 0.93 (0.91–0.95), pooled specificity: 0.95 (0.94–0.96) and (B) MRSA, pooled sensitivity: 0.86 (0.84–0.87), pooled specificity: 0.98 (0.97–0.98) in nasal samples. The square stands for the sensitivity and specificity of a particular study; the black line represents its confidence interval. TP true positive, FP false positive, FN false negative, TN true negative, CI confidence interval
Correspondingly, 32 publications with 42,247 samples examined MRSA colonization in clinically suspected patients [21,22,23,24,25, 27, 30, 32, 34,35,36,37,38,39,40,41,42,43, 45, 46, 50,51,52, 54, 55, 62,63,64,65,66,67,68]. The Xpert’s MRSA detection sensitivity and specificity varied from 0.6 (0.47–0.73) to 1.0 (0.97–1.0) and from 0.91 (0.88–0.93) to 1.0 (0.97–1.0), respectively (Fig. 2B). The pooled estimates for detecting MRSA in swabs were [specificity: 0.98 (0.97–0.98), sensitivity: 0.86 (0.84–0.87), NLR: 0.13 (0.1–0.17), PLR: 30.64 (24.13–38.92), and DOR: 252.91 (181.41–352.57)]. Considerable heterogeneity was seen in the I2 statistics, where the specificity and sensitivity were 90.9% and 85.3%, respectively. With an AUC of 0.99 (95% CI: 0.98–0.99), the SROC were found to have greater clinical diagnostic value (see Fig. 3B).
Detection of Staphylococcal Infection in LRT Specimens
Four publications [28, 29, 49, 59] assessed the Xpert’s MSSA detection accuracy using 561 LRT specimens, while four publications [28, 47, 49, 59] demonstrated the Xpert’s MRSA detection accuracy using 582 specimens. The Xpert’s MSSA detection sensitivity and specificity varied from 0.96 (0.78–1.0) to 1.0 (0.94–1.00) and 0.93 (0.86–0.97) to 1.00 (0.91 to 1.00), respectively (Fig. 4A), while MRSA detection sensitivity and specificity varied from 0.83 (0.36–1.0) to 1.0 (0.90–1.00) and from 0.90 (0.84–0.94) to 1.00 (0.96–1.0), respectively (Fig. 4B). The Xpert’s pooled MSSA detection estimates were comparable [specificity: 0.97 (0.95–0.99), sensitivity: 0.99 (0.96–1.0), NLR: 0.03 (0.01–0.11), PLR: 34.15 (8.81–132.43), and DOR: 1204.6 (188.1–7715.6)] to MRSA [specificity: 0.93 (0.90–0.95), sensitivity: 0.97 (0.92–0.99), NLR: 0.05 (0.02–0.14), PLR: 13.04 (6.16–27.62), and DOR: 374.04 (76.19–1836.3)]. Low to moderate heterogeneity was seen in the I2 statistics for MSSA detection, where the specificity and sensitivity were 65.8% and 12.7%, respectively, while the I2 statistical values of Xpert sensitivity and specificity for MRSA identification were 26.8 and 82.2%, respectively, suggesting low to moderate heterogeneity. With an AUC of 0.99 (0.99–1.00), the SROCs were found to have greater clinical diagnostic value for both MSSA and MRSA (Fig. 4C, 4D).
Detection of staphylococcal strains in LRT samples. A Forest plot for MSSA; pooled sensitivity: 0.99 (0.96–1.0), pooled specificity: 0.97 (0.95–0.99). B Forest plot for MRSA; pooled sensitivity: 0.97 (0.92–0.99), pooled specificity: 0.93 (0.90–0.95). The black line shows the study's confidence interval, while the blue square reflects its sensitivity and specificity. C SROC plot for MSSA; D SROC plot for MRSA. Red circles in SROC curve represent each investigation's data point, while the solid blue line shows the SROC curve. TP true positive; FP false positive; FN false negative; TN true negative; CI confidence interval; LRT lower respiratory tract; AUC area under the curve
Detection of Staphylococcal Infection in Osteoarticular Samples
In four publications [33, 56, 57, 60], the accuracy of Xpert was tested in osteoarticular samples for both MSSA (n = 480) and MRSA (n = 478). The Xpert’s MSSA detection sensitivity and specificity varied from 0.85 (0.72–0.94) to 1.0 (0.79–1.00) and 0.98 (0.94–1.0) to 1.0 (0.94–1.0), respectively (Fig. 5A). For MRSA identification, the Xpert’s sensitivity and specificity varied from 0.82 (0.48–0.98) to 1.0 (0.66–1.0) and 1.00 (0.95–1.00) to 1.00 (0.98–1.00), respectively (Fig. 5B). The Xpert’s pooled diagnostic accuracy was comparable for both MSSA [specificity: 0.99 (0.97–1.0), sensitivity: 0.92 (0.86–0.96), NLR: 0.11 (0.06–0.18), PLR: 46.48 (21.66–99.76), DOR: 445.72 (145.19–1368.3)] and MRSA [specificity: 1.0 (0.99–1.0), sensitivity: 0.92 (0.75–0.99), NLR: 0.16 (0.07–0.39), PLR: 184.07 (45.74–740.74), DOR: 1560.1 (241.57–10,075.9)]. The I2 statistical scores for MSSA detection specificity and sensitivity were 6.0% and 53.1%, respectively, suggesting low to moderate heterogeneity, whereas the I2 MRSA statistical scores were < 18.3%, indicating low heterogeneity across studies. Both MSSA and MRSA had an AUC of 1.0 (0.99–1.00), denoting remarkably higher diagnostic value (Fig. 5C, D).
Detection of staphylococcal strains in osteoarticular samples. A Forest plot for MSSA; pooled sensitivity: 0.92 (0.86–0.96), pooled specificity: 0.99 (0.97–1.0). B Forest plot for MRSA; pooled sensitivity: 0.92 (0.75–0.99), pooled specificity: 1.0 (0.99–1.0). The black line shows the study's confidence interval, while the blue square reflects its sensitivity and specificity. C SROC plot for MSSA; D SROC plot for MRSA. Red circles in SROC curve represent each investigation's data point, while the solid blue line shows the SROC curve. TP true positive; FP false positive; FN false negative; TN true negative; CI confidence interval; AUC area under the curve
Detection of Bloodstream Staphylococcal Infection
Eight publications [26, 31, 44, 53, 58, 61, 67, 69] with a total of 2622 specimens analyzed the Xpert’s MSSA detection accuracy in blood specimens, whereas seven studies [26, 31, 44, 48, 53, 58, 67] with a total of 2123 samples evaluated the Xpert’s accuracy for MSSA detection in samples from bloodstream. The Xpert’s MSSA detection sensitivity and specificity varied from 0.99 (0.98–1.0) to 1.0 (0.99–1.00) and 0.98 (0.92–1.0) to 1.00 (0.98 to 1.00) and 0.98 (0.92–1.0) to 1.00 (0.98 to 1.00), respectively (Fig. 6A). On the other hand, Xpert’s MRSA detection sensitivity and specificity varied from 0.98 (0.93–1.00) to 1.0 (0.82–1.00) and 0.99 (0.98–1.0) to 1.00 (0.99–1.00), respectively (Fig. 6B). Xpert evidenced roughly similar diagnostic value for both MSSA [specificity: 1.0 (0.99–1.00), sensitivity: 1.0 (0.99–1.00), NLR: 0.01 (0.004–0.03), PLR: 90.1 (31.86–254.75), DOR: 11,653.5 (3311.8–41,006.4)] and MRSA [specificity: 1.0 (0.99–1.00), sensitivity: 0.99 (0.96–1.0), NLR: 0.04 (0.02–0.10), PLR: 218.05 (106.36–447.01), DOR: 7328.5 (2366.0–22,699.0)]. In the case of MSSA detection, the I2 scores for sensitivity and specificity were 0% and 48.7%, reflecting low to substantial heterogeneity, whilst the Xpert’s MRSA detection sensitivity and specificity were 0% and 3.9%, reflecting low heterogeneity. With an AUC of 1.0 (0.99–1.00), the SROCs were found to have greater clinical diagnostic value for both MSSA and MRSA (Fig. 6C, 6D).
Detection of staphylococcal strains in blood samples. A Forest plot for MSSA; pooled sensitivity: 1.0 (0.99–1.00), pooled specificity: 1.0 (0.99–1.00). B Forest plot for MRSA; pooled sensitivity: 0.99 (0.96–1.0), pooled specificity: 1.0 (0.99–1.00). The black line shows the study's confidence interval, while the blue square reflects its sensitivity and specificity. C SROC plot for MSSA; D SROC plot for MRSA. Red circles in SROC curve represent each investigation's data point, while the solid blue line shows the SROC curve. TP true positive; FP false positive; FN false negative; TN true negative; CI confidence interval; AUC area under the curve
Meta-Regression and Subgroup Analysis
Because there was high heterogeneity across studies, a meta-regression test was utilized to inspect the source of heterogeneity in designated subgroups. Meta-regression analysis demonstrated that except for the sampling methods (consecutive/convenience; P = 0.03), all other factors including research design (prospective/others) and sample conditions (fresh/frozen) were not major contributors to heterogeneity (meta-regression P = 0.86 and P = 0.6, respectively).
Publication Bias
Deek's funnel plot asymmetry analysis was conducted to examine publication bias of all included studies. In our study, we found no evidence of significant publication bias (P > 0.05).
Discussion
Timely and efficient microbiological detection of staphylococcal infection is critical in the care of both colonized and infected patients to choose suitable therapy and avoid infection transmission via the use of appropriate barrier measures. However, the various studies' use of distinct case definitions and samples makes comparing study results difficult and limits illness treatment. Staphylococcal infections are traditionally tested using culture-based approaches, which are time-consuming (48–72 h) and need further phenotypic assessments. In fact, a 24–48-h period is generally predicted between the detection of gram-positive cocci in clusters on gram-stain and the results of antibiotic susceptibility testing. According to the existing literature, prompt selection of the best suitable antibacterial may minimize mortality and expenses [76, 77]. One effective tool that may aid with this is an Xpert MRSA/SA system (Cepheid, Sunnyvale, CA), which is a unique, completely automated approach that distinguishes MRSA from MSSA in < 1 h after acquiring gram stain. This technique has been successfully examined for the identification of Clostridium difficile [78], group B Streptococci [79], Bacillus anthracis [80], Staphylococcal strains [21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69], and others from clinical specimens. Nonetheless, there is a paucity of data to decide on the accuracy of the Xpert MRSA/SA assay for the diagnosis of staphylococcal infections to facilitate the targeted administration of the most appropriate antibiotic.
Based on our findings, Xpert was better at detecting MSSA [specificity: 0.97 (0.96–0.98), sensitivity: 0.97 (0.96–0.98), AUC: 0.99 (0.99–1.0)] than MRSA [specificity: 0.98 (0.97–0.98), sensitivity: 0.87 (0.86–0.88), AUC: 0.99 (0.99–1.0)]. It should be emphasized that there was substantial diversity in the number of studies included and sample settings, which may not necessarily depict the true scenario and aid in pragmatic therapeutic advice. Therefore, we initially aimed to screen colonization of both MSSA and MRSA in carriers, who can then be subjected to contact precautions to prevent spread to other patients. We observed that a larger number of studies were conducted for screening colonization and were mostly compared to direct culture, which demonstrated sensitivity of 86% and specificity of 98%, which correlated well to the overall summary estimates of staphylococcal strains (Table 2). Low sensitivity for Xpert might be attributed to patient features, sampling error, and low bacterial load in swab samples. Conversely, Xpert's ability to correctly identify patients who did not have an infection was consistently above 95%, depicting Xpert’s ability to identify patients without infection correctly. Compared to previous studies, an investigation by Parente and coworkers [81] found that MRSA nares screening can be useful to rule out MRSA pneumonia, but the study was not unique in its ability to accurately diagnose staphylococcal colonization.
Similarly, diagnosing the causative organism and administering appropriate antibiotic therapy are critical in treating staphylococcal BJI in osteoarticular specimens. Traditional bacterial culture and gram stain tests have low to moderate sensitivity, which impedes the treatment strategy of staphylococcal BJI [12, 82], leading to delays in the administration of antibacterial medications against the causative microorganisms. Numerous recent studies have shown that Xpert may be useful for distinguishing staphylococcal BJI from other pathogenic microbes [33, 56, 57, 60]; however, the findings of these studies have not been fully examined. According to the results of this investigation, the testing of Xpert in osteoarticular samples compared to those using the culture technique demonstrated great sensitivity and specificity for distinguishing MSSA and MRSA (Table 2). It should be mentioned that in patients with suspected staphylococcal BJI, identifying staphylococcal species and resistance markers as soon as possible is critical, as a timely therapy may significantly increase overall life expectancies and minimize hospital burden.
The role of the diagnostic laboratory in the treatment of LRT infection is critical. Traditional testing method of the respiratory secretions is still the best way to look for pathogenic microorganisms [83]. However, a positive test can only be made in about 30% of cases [13], with a high rate of false negatives due to normal flora complicating the analysis and necessitating a significant amount of samples to start culture, which is not ideal. Several studies have examined the Xpert’s ability to distinguish between MSSA and MRSA in possible LRT specimens [28, 29, 47, 49, 59]; nevertheless, a clear understanding of Xpert accuracy has not been demonstrated. According to the findings of this study, the pooled estimates of Xpert’s MSSA detection [specificity: 0.97 (0.95–0.99), sensitivity: 0.99 (0.96–1.0), AUC: 1.0 (0.99–1.00)] were relatively similar to the MRSA detection [specificity: 0.93 (0.90–0.95), sensitivity: 0.97 (0.92–0.99), AUC: 1.0 (0.99–1.00)], exemplifying Xpert's impressive overall performance. In contrast to previously published systematic reviews, a publication by Chen et al. reported on the effective diagnostic relevance of NAAT for staphylococcal strains in LRT specimens [18], which is similar to the results of our investigation.
We also compared the Xpert assay to established reference standards for detecting staphylococcal strains in blood specimens, where most studies employed gram-positive cocci culture broths. The Xpert assay demonstrated statistically equivalent sensitivities and specificities for identifying MSSA and MRSA in positive blood cultures compared to the culture method (Fig. 6). The superior sensitivity of Xpert across enrichment samples, as well as the fairly low non-interpretable data, encourage the Xpert’s utility for monitoring bloodstream infections. A potential advantage of the Xpert assay is the simplified sample-to-result workflow and on-demand flexibility, which minimizes turnaround time for blood cultures. The advantages of quick diagnosis of MSSA and MRSA with the deployment of a molecular test straight from positive blood culture broths have been extensively established in terms of reduced patient numbers, time to commence treatment, duration of hospital stay, and related healthcare expenditures [20, 84]. Table 2 shows the pooled Xpert estimates for distinguishing MSSA and MRSA across all specimens. Importantly, these benefits can only be realized when molecular testing is available on demand and the findings are actively disclosed to the clinician.
Our research strengths include a thorough search approach that retrieved all relevant publications from five of the most often used databases. The searches were carried out methodically, and at least two investigators examined the titles and abstracts of all publications. Following a group discussion, the authors' collective opinion was represented in the publications included in this meta-analysis. The PRISMA-DTA criteria for systematic reviews and the QUADAS-2 tool were applied to ascertain the included publications quality. In the following analysis, publications that did not satisfy the conditions for identifying staphylococcal BJI were exempted. Furthermore, studies that compared different genera of Xpert with the same sample were aggregated rather than included as separate studies, which could have resulted in an overestimation of the clinical outcomes of index tests.
This research has certain limitations that should be considered. Despite doing a thorough search of databases, it is conceivable that we overlooked a few pertinent studies. We also could not address the impact of attributes such as specimen volume, non-standardized processing, amplification protocols, expert knowledge with the Xpert test, and research laboratory facilities on the Xpert’s accuracy because of the high level of discrepancies in reporting of these factors in the publications. Also, we did not evaluate the accuracy of Xpert among independent traditional reference standards, including direct and enrichment cultures, which could be probable reasons for the heterogeneity. Studies comparing different generations of Xpert assay with culture were averaged for their representation in our meta-analysis. Although the research design, sampling condition, and test methods were not a major source of inconsistency in the meta-regression analysis, these factors may have elevated variability and limited the Xpert’s universal applicability. It is also worth noting that most studies were conducted in high-income countries, with only one study conducted in a lower-middle income country, which may limit the generalizability of the study. Furthermore, there was also a limited availability of osteoarticular and LRT-related publications, which should be considered when interpreting the results of this study.
Conclusions
To the best of the author's knowledge, this is the first study of its kind to evaluate Xpert's diagnostic accuracy across multifaceted specimens to guide appropriate therapeutic decisions. Our findings suggest that Xpert is a reliable and robust automated assay for distinguishing staphylococcal strains from potentially infected patients in diverse sample settings. This research suggests that if a short time delay (≤ 60 min) does not influence mortality, as no literature has claimed so far, then commencing suitable medication may be more important than immediately starting empiric use of broad-spectrum antibiotics. Xpert can be utilized as the preferable first testing method for a point-of-care diagnosis of staphylococcal infection to avoid costly anti-MRSA therapy. Whenever possible, the use of Xpert in conjunction with microbiological culture should be considered, as susceptibilities must be obtained to properly guide treatment. Furthermore, Xpert results and local clinical data should be taken into account when developing new guidelines for anti-staphylococcal medication use. Given the paucity of Xpert data across osteoarticular and LRT sample types, a thorough examination involving a relatively large number of prospective studies may be helpful in determining the best sample type for future implications. It should also be noted that, compared to traditional methods, the use of molecular techniques incurs additional costs and necessitates substantial facilities, and only a limited number can be tested at any given time, which could be interesting to estimate in future studies. Furthermore, future investigations are required to thoroughly substantiate the treatment benefit associated with Xpert's cost-effectiveness, mortality prediction, and time to antibiotic discontinuation.
References
von Allmen N, Gorzelniak K, Liesenfeld O, Njoya M, Duncan J, Marlowe EM, Hartel T, Knaust A, Hoppe B, Walter M. Liquid and dry swabs for culture-and pcr-based detection of colonization with methicillin-resistant Staphylococcus aureus during admission screening. Eur J Microbiol Immunol. 2019;9:131–7.
Andersen BM, Tollefsen T, Seljordslia B, Hochlin K, Syversen G, Jonassen TO, Rasch M, Sandvik L. Rapid MRSA test in exposed persons: costs and savings in hospitals. J Infect. 2010;60:293–9.
Arcenas RC, Spadoni S, Mohammad A, Kiechle FL, Walker K, Fader RC, Perdreau-Remington F, Osiecki J, Liesenfeld O, Henrickson S, Rao A. Multicenter evaluation of the LightCycler MRSA advanced test, the Xpert MRSA Assay, and MRSASelect directly plated culture with simulated workflow comparison for the detection of methicillin-resistant Staphylococcus aureus in nasal swabs. J Mol Diagn. 2012;14:367–75.
Bebko SP, Byers P, Green DM, Awad SS. Identification of methicillin-susceptible or methicillin-resistant Staphylococcus aureus carrier status preoperatively using polymerase chain reaction in patients undergoing elective surgery with hardware implantation. Infect Cont Hosp Epidemiol. 2015;36:738–41.
Blanc DS, Nahimana I, Zanetti G, Greub G. MRSA screening by the Xpert MRSA PCR assay: pooling samples of the nose, throat, and groin increases the sensitivity of detection without increasing the laboratory costs. Eur J Clin Microbiol Infect Dis. 2013;32:565–8.
Bulliard E, Grandbastien B, Senn L, Greub G, Blanc DS. Evaluation of three consecutive versions of a commercial rapid PCR test to screen for methicillin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2019;25:1430.e1-1430.e4.
Creamer E, Dolan A, Sherlock O, Thomas T, Walsh J, Moore J, Smyth E, O’Neill E, Shore A, Sullivan D, Rossney AS, Cunney R, Coleman D, Humphreys H. The effect of rapid screening for methicillin-resistant Staphylococcus aureus (MRSA) on the identification and earlier isolation of MRSA-positive patients. Infect Control Hosp Epidemiol. 2010;2010(31):374–81.
Dewar S, Vass D, MacKenzie FM, Parcell BJ. Point-of-care testing by healthcare workers for detection of meticillin-resistant Staphylococcus aureus, Clostridioides difficile, and norovirus. J Hosp Infect. 2019;103:447–53.
Gray J, Patel M, Turner H, Reynolds F. MRSA screening on a paediatric intensive care unit. Arch Dis Child. 2012;97:243–4.
Hombach M, Pfyffer GE, Roos M, Lucke K. Detection of methicillin-resistant Staphylococcus aureus (MRSA) in specimens from various body sites: performance characteristics of the BD GeneOhm MRSA assay, the Xpert MRSA assay, and broth-enriched culture in an area with a low prevalence of MRSA infections. J Clin Microbiol. 2010;48:3882–7.
Jacqmin H, Schuermans A, Desmet S, Verhaegen J, Saegeman V. Performance of three generations of Xpert MRSA in routine practice: approaching the aim? Eur J Clin Microbiol Infect Dis. 2017;36:1363–5.
Jonckheere S, Van Vaerenbergh K, Boel A, Vankeerberghen A, Beenhouwer HD. How is the Xpert MRSA Gen 3 assay (Cepheid) performing on pooled eSwab medium? Diagn Microbiol Infect Dis. 2015;83:219–21.
Kelley PG, Grabsch EA, Howden BP, Gao W, Grayson ML. Comparison of the xpert methicillin-resistant Staphylococcus aureus (MRSA) assay, BD GeneOhm MRSA assay, and culture for detection of nasal and cutaneous groin colonization by MRSA. J Clin Microbiol. 2009;47:3769–72.
Lee S, Park YJ, Park KG, Jekarl DW, Chae H, Yoo JK, Seo SW, Choi JE, Lim JH, Heo SM, Seo JH. Comparative evaluation of three chromogenic media combined with broth enrichment and the real-time pcr-based xpert mrsa assay for screening of methicillin-resistant Staphylococcus aureus in nasal swabs. Ann Lab Med. 2013;33:255–60.
Lepainteur M, Delattre S, Cozza S, Lawrence C, Roux AL, Rottman M. Comparative evaluation of two PCR-based methods for detection of methicillin-resistant Staphylococcus aureus (MRSA): Xpert MRSA Gen 3 and BD-Max MRSA XT. J Clin Microbiol. 2015;53:1955–8.
Malhotra-Kumar S, Van Heirstraeten L, Lee A, Cortinas A, Lammens C, Vanhommerig E, Molenberghs G, Aerts M, Harbarth S, Goossens H, MOSAR WP2 Study Team. Evaluation of molecular assays for rapid detection of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2010;2010(48):4598–601.
May L, McCann C, Brooks G, Rothman R, Miller L, Jordan J. Dual-site sampling improved detection rates for MRSA colonization in patients with cutaneous abscesses. Diagn Microbiol Infect Dis. 2014;80:79–82.
May LS, Rothman RE, Miller LG, Brooks G, Zocchi M, Zatorski C, Dugas AF, Ware CE, Jordan JA. A randomized clinical trial comparing use of rapid molecular testing for Staphylococcus aureus for patients with cutaneous abscesses in the emergency department with standard of care. Infect Control Hosp Epidemiol. 2015;36:1423–30.
Nielsen XC, Madsen TV, Engberg J. Evaluation of Xpert MRSA gen 3 and BD MAX MRSA XT for meticillin-resistant Staphylococcus aureus screening in a routine diagnostic setting in a low-prevalence area. J Med Microbiol. 2017;66:90–5.
Nulens E, Descheemaeker P, Deurenberg RN, Stobberingh EE, Gordts B. Contribution of two molecular assays as compared to selective culture for MRSA screening in a low MRSA prevalence population. Infection. 2010;38:98–101.
Parcell BJ, Phillips G. Use of Xpert (R) MRSA PCR point-of-care testing beyond the laboratory. J Hosp Infect. 2014;87:119–21.
Patel PA, Robicsek A, Grayes A, Schora DM, Peterson KE, Wright MO, Peterson LR. Evaluation of multiple real-time PCR tests on nasal samples in a large MRSA surveillance program. Am J Clin Pathol. 2015;143:652–8.
Patel PA, Schora DM, Peterson KE, Grayes A, Boehm S, Peterson LR. Performance of the Cepheid Xperte (R) SA Nasal Complete PCR assay compared to culture for detection of methicillin-sensitive and methicillin-resistant Staphylococcus aureus colonization. Diagn Microbiol Infect Dis. 2014;2014(80):32–4.
Roisin S, Laurent C, Nonhoff C, Deplano A, Hallin M, Byl B, Struelens MJ, Denis O. Positive predictive value of the Xpert MRSA assay diagnostic for universal patient screening at hospital admission: influence of the local ecology. Eur J Clin Microbiol Infect Dis. 2012;31:873–80.
Rossney AS, Herra CM, Brennan GI, Morgan PM, O’Connell B. Evaluation of the Xpert methicillin-resistant Staphylococcus aureus (MRSA) assay using the GeneXpert real-time PCR platform for rapid detection of MRSA from screening specimens. J Clin Microbiol. 2008;2008(46):3285–90.
Wang H, Salamon D, Jean S, Leber AL. Evaluation of the Cepheid Xpert SA Nasal Complete for direct detection of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus in nasal swabs from pediatric patients. Diagn Microbiol Infect Dis. 2021;101:115417.
Wassenberg M, Kluytmans J, Erdkamp S, Bosboom R, Buiting A, Elzakker EV, Melchers W, Thijsen S, Troelstra A, Vandenbroucke-Grauls C, Visser C, Voss A, Wolffs P, Wulf M, van Zwet T, de Wit A, Bonten M. Costs and benefits of rapid screening of methicillin-resistant Staphylococcus aureus carriage in intensive care units: a prospective multicenter study. Crit Care. 2012;16:R22.
Wassenberg MW, Kluytmans JA, Box AT, Bosboom RW, Buiting AGM, van Elzakker EPM, Melchers WJG, van Rijen MML, Thijsen SFT, Troelstra A, Vandenbroucke-Grauls CMJE, Visser CE, Voss A, Wolffs PFG, Wulf MWH, van Zwet AA, de Wit GA, Bonten MJM. Rapid screening of methicillin-resistant Staphylococcus aureus using PCR and chromogenic agar: a prospective study to evaluate costs and effects. Clin Microbiol Infect. 2010;16:1754–61.
Wolk DM, Marx JL, Dominguez L, Driscoll D, Schifman RB. Comparison of MRSASelect Agar, CHROMagar methicillin-resistant Staphylococcus aureus (MRSA) medium, and Xpert MRSA PCR for detection of MRSA in Nares: diagnostic accuracy for surveillance samples with various bacterial densities. J Clin Microbiol. 2009;47:3933–6.
Wolk DM, Picton E, Johnson D, Davis T, Pancholi P, Ginocchio CC, Finegold S, Welch DF, de Boer M, Fuller D, Solomon MC, Rogers B, Mehta MS, Peterson LR. Multicenter evaluation of the Cepheid Xpert methicillin-resistant Staphylococcus aureus (MRSA) test as a rapid screening method for detection of MRSA in nares. J Clin Microbiol. 2009;47:758–64.
Yarbrough ML, Warren DK, Allen K, Burkholder D, Daum R, Donskey C, Knaack D, LaMarca A, May L, Miller LG, Parenti DM, Peterson L, Tan TY, Widen R, Hernandez DR, Wolk DM, Burnham CA. Multicenter evaluation of the Xpert MRSA NxG assay for detection of methicillin-resistant Staphylococcus aureus in nasal swabs. J Clin Microbiol. 2018;56:e01381-e1417.
Wolk DM, Struelens MJ, Pancholi P, Davis T, Della-Latta P, Fuller D, Picton E, Dickenson R, Denis O, Johnson D, Chapin K. Rapid detection of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) in wound specimens and blood cultures: multicenter preclinical evaluation of the Cepheid Xpert MRSA/SA skin and soft tissue and blood culture assays. J Clin Microbiol. 2009;47:823–6.
Cercenado E, Marín M, Burillo A, Martín-Rabadán P, Rivera M, Bouza E. Rapid detection of Staphylococcus aureus in lower respiratory tract secretions from patients with suspected ventilator-associated pneumonia: evaluation of the Cepheid Xpert MRSA/SA SSTI assay. J Clin Microbiol. 2012;50:4095–7.
Coppens J, Van Heirstraeten L, Ruzin A, Yu L, Timbermont L, Lammens C, Matheeussen V, McCarthy M, Jorens P, Leven M, Kumar-Singh S, Malhotra-Kumar S. Comparison of GeneXpert MRSA/SA ETA assay with semi-quantitative and quantitative cultures and nuc gene-based qPCR for detection of Staphylococcus aureus in endotracheal aspirate samples. Antimicrob Resist Infect Control. 2019;8:4.
Oh AC, Lee JK, Lee HN, Hong YJ, Chang YH, Hong S, Kim DH. Clinical utility of the Xpert MRSA assay for early detection of methicillin-resistant Staphylococcus aureus. Mol Med Rep. 2013;7:11–5.
Paonessa JR, Shah RD, Pickens CI, Lizza BD, Donnelly HK, Malczynski M, Qi C, Wunderink RG. Rapid detection of methicillin-resistant Staphylococcus aureus in BAL: a pilot randomized controlled trial. Chest. 2019;2019(155):999–1007.
Trevino SE, Pence MA, Marschall J, Kollef MH, Babcock HM, Burnham CAD. Rapid MRSA PCR on respiratory specimens from ventilated patients with suspected pneumonia: a tool to facilitate antimicrobial stewardship. Eur J Clin Microbiol Infect Dis. 2017;36:879–85.
Dubouix-Bourandy A, de Ladoucette A, Pietri V, Mehdi N, Benzaquen D, Guinand R, Gandois JM. Direct detection of Staphylococcus osteoarticular infections by use of Xpert MRSA/SA SSTI real-time PCR. J Clin Microbiol. 2011;49:4225–30.
Sambri A, Pignatti G, Romagnoli M, Donati D, Marcacci M, Cadossi M. Intraoperative diagnosis of Staphylococcus aureus and coagulase-negative Staphylococcus using Xpert MRSA/SA SSTI assay in prosthetic joint infection. New Microbiol. 2017;40:130–4.
Searns JB, Robinson CC, Wei Q, Yuan J, Hamilton S, Pretty K, Donaldson N, Parker SK, Dominguez SR. Validation of a novel molecular diagnostic panel for pediatric musculoskeletal infections: Integration of the Cepheid Xpert MRSA/SA SSTI and laboratory-developed real-time PCR assays for clindamycin resistance genes and Kingella kingae detection. J Microbiol Methods. 2019;156:60–7.
Valour F, Blanc-Pattin V, Freydière AM, Bouaziz A, Chanard E, Lustig S, Ferry T, Laurent F, Lyon Bone Joint Infection Study Group. Rapid detection of Staphylococcus aureus and methicillin resistance in bone and joint infection samples: Evaluation of the GeneXpert MRSA/SA SSTI assay. Diagn Microbiol Infect Dis. 2014;78:313–5.
Buchan BW, Allen S, Burnham CAD, Tekippe EM, Davis T, Levi M, Mayne D, Pancholi P, Relich RF, Thomson R, Ledeboer NA. Comparison of the next-generation Xpert MRSA/SA BC assay and the GeneOhm StaphSR assay to routine culture for identification of Staphylococcus aureus and methicillin-resistant S. aureus in positive-blood-culture broths. J Clin Microbiol. 2015;2015(53):804–9.
Davies J, Gordon CL, Tong SYC, Baird RW, Davis JS. Impact of results of a rapid Staphylococcus aureus diagnostic test on prescribing of antibiotics for patients with clustered gram-positive cocci in blood cultures. J Clin Microbiol. 2012;2012(50):2056–8.
McHugh MP, Parcell BJ, MacKenzie FM, Templeton KE, SMVN Molecular Diagnostics Evaluation Group. Rapid molecular testing for Staphylococcus aureus bacteraemia improves clinical management. J Med Microbiol. 2020;69:552–7.
Page A, O’Rourke S, Brennan M, Clooney L, Blanc DL, Griffin J, Eogan M, Drew RJ. Impact of Xpert MRSA/SA blood culture PCR assay on management of positive blood cultures in obstetric patients: a retrospective audit. Ir J Med Sci. 2017;2017(186):995–8.
Reddy K, Whitelaw A. Can the Xpert MRSA/SA BC assay be used as an antimicrobial stewardship tool? A prospective assay validation and descriptive impact assessment study in a South African setting. BMC Infect Dis. 2021;21:177.
Spencer DH, Sellenriek P, Burnham CAD. Validation and implementation of the GeneXpert MRSA/SA blood culture assay in a pediatric setting. Am J Clin Pathol. 2011;136:690–4.
Wade-Cummings M, Mailman JF, Degelman ML, Phillips C, Vanstone JR. Identification of Staphylococci by polymerase chain reaction directly from a positive blood culture and effect on patient care. Can J Hosp Pharm. 2021;74:43–9.
Zboromyrska Y, De la Calle C, Soto M, Sampietro-Colom L, Soriano A, Alvarez-Martínez MJ, Almela M, Marco F, Arjona R, Cobos-Trigueros N, Morata L, Mensa J, Martínez JA, Mira A, Vila J. Rapid diagnosis of staphylococcal catheter-related bacteraemia in direct blood samples by real-time PCR. PLoS ONE. 2016;11:e0161684.
Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–61.
Patel M, Weinheimer JD, Waites KB, Baddley JW. Active surveillance to determine the impact of methicillin-resistant Staphylococcus aureus colonization on patients in intensive care units of a Veterans Affairs Medical Center. Infect Control Hosp Epidemiol. 2008;29:503–9.
Hu F-P, Guo Y, Zhu D-M, Wang F, Jiang X-F, Xu Y-C, et al. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005–2014. Clin Microbiol Infect. 2016;22:S9–14.
Sutcliffe CG, Grant LR, Reid A, Douglass G, Brown LB, Kellywood K, et al. High burden of Staphylococcus aureus among Native American individuals on the White Mountain Apache Tribal lands. Open Forum Infect Dis. 2020;7: ofaa061.
Ippolito G, Leone S, Lauria FN, Nicastri E, Wenzel RP. Methicillin-resistant Staphylococcus aureus: the superbug. Int J Infect Dis. 2010;14:S7–11.
Vaudaux P, Huggler E, Arhin FF, Moeck G, Renzoni A, Lew DP. Comparative activity of oritavancin against meticillin-resistant Staphylococcus aureus (MRSA) bloodstream isolates from Geneva University Hospital. Int J Antimicrob Agents. 2009;34:540–3.
Saravolatz LD, Stein GE. Delafloxacin: a new anti–methicillin-resistant Staphylococcus aureus Fluoroquinolone. Clin Infect Dis. 2019;68:1058–62.
Zhang S, Sun X, Chang W, Dai Y, Ma X. Systematic review and meta-analysis of the epidemiology of vancomycin-intermediate and heterogeneous vancomycin-intermediate Staphylococcus aureus isolates. PLoS ONE. 2015;10:e0136082.
Gould IM, Cauda R, Esposito S, Gudiol F, Mazzei T, Garau J. Management of serious meticillin-resistant Staphylococcus aureus infections: what are the limits? Int J Antimicrob Agents. 2011;37:202–9.
Chimento GF, Finger S, Barrack RL. Gram stain detection of infection during revision arthroplasty. J Bone Joint Surg Brit. 1996;78:838–9.
O’Horo JC, Thompson D, Safdar N. Is the gram stain useful in the microbiologic diagnosis of VAP? A meta-analysis. Clin Infect Dis. 2012;55:551–61.
Prabhoo R, Chaddha R, Iyer R, Mehra A, Ahdal J, Jain R. Overview of methicillin resistant Staphylococcus aureus mediated bone and joint infections in India. Orthop Rev. 2019;11:8070.
Chalmers JD, Taylor JK, Singanayagam A, Fleming GB, Akram AR, Mandal P, et al. Epidemiology, antibiotic therapy, and clinical outcomes in health care–associated pneumonia: a UK cohort study. Clin Infect Dis. 2011;53:107–13.
Souweine B, Veber B, Bedos JP, Gachot B, Dombret MC, Regnier B, Wolff M. Diagnostic accuracy of protected specimen brush and bronchoalveolar lavage in nosocomial pneumonia: impact of previous antimicrobial treatments. Crit Care Med. 1998;26:236–44.
Cepheid. Xpert™ MRSA/SA blood culture: Package insert, 300–7188, revision A. Cepheid: Sunnyvale; 2008.
Chen K, Malik AA, Sheng Y-J, Ahmed S, Sun C, Deng C-L, Ojha SC. Clinical utility of molecular tests for guiding therapeutic decisions in bloodstream staphylococcal infections: a meta-analysis. Front Pediatr. 2021;2021:796.
Brown J, Paladino JA. Impact of rapid methicillin-resistant Staphylococcus aureus polymerase chain reaction testing on mortality and cost effectiveness in hospitalized patients with bacteraemia. Pharmacoeconomics. 2010;28:567–75.
Chen K, Ahmed S, Sun C, Sheng Y-J, Wu G, Deng C-L, Ojha SC. Accuracy of molecular amplification assays for diagnosis of staphylococcal pneumonia: a systematic review and meta-analysis. J Clin Microbiol. 2021;59:e03003-e3020.
Noto MJ, Kreiswirth BN, Monk AB, Archer GL. Gene acquisition at the insertion site for SCC mec, the genomic island conferring methicillin resistance in Staphylococcus aureus. J Bacteriol. 2008;190:1276–83.
Bauer KA, West JE, Balada-Llasat J-M, Pancholi P, Stevenson KB, Goff DA. An antimicrobial stewardship program’s impact with rapid polymerase chain reaction methicillin-resistant Staphylococcus aureus/S. aureus blood culture test in patients with S. aureus bacteremia. Clin Infect Dis. 2010;51:1074–80.
McInnes MDF, Moher D, Thombs BD, McGrath TA, Bossuyt PM, The PRISMA-DTA Group, Clifford T, Cohen JF, Deeks JJ, Gatsonis C, Hooft L, Hunt HA, Hyde CJ, Korevaar DA, Leeflang MMG, Macaskill P, Reitsma JB, Rodin R, Rutjes AWS, Salameh JP, Stevens A, Takwoingi Y, Tonelli M, Weeks L, Whiting P, Willis BH. Preferred reporting items for a systematic review and meta-analysis of diagnostic test accuracy studies: the PRISMA-DTA statement. JAMA. 2018;319:388–96.
Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–36.
Cochrane T. Review manager (RevMan) 5.4. Copenhagen: The Nordic Cochrane Centre; 2020.
Zamora J, Abraira V, Muriel A, Khan K, Coomarasamy A. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol. 2006;6:31.
Higgins JP, Thomas J, Chandler J, Cumpston M, Li T, Page MJ, Welch VA, editors. Cochrane handbook for systematic reviews of interventions. Chichester: John Wiley & Sons; 2011.
Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58:882–93.
Lodise TP Jr, McKinnon PS, Levine DP, Rubak MJ. Impact of empirical-therapy selection on outcomes of intravenous drug users with infective endocarditis caused by methicillin-susceptible Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51:3731–3.
Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis. 2003;2003(36):53–9.
Huang H, Weintraub A, Fang H, Nord CE. Comparison of a commercial multiplex real-time PCR to the cell cytotoxicity neutralization assay for diagnosis of Clostridium difficile infections. J Clin Microbiol. 2009;47:3729–31.
El Helali N, Nguyen J-C, Ly A, Giovangrandi Y, Trinquart L. Diagnostic accuracy of a rapid real-time polymerase chain reaction assay for universal intrapartum group B streptococcus screening. Clin Infect Dis. 2009;49:417–23.
Ulrich MP, Christensen DR, Coyne SR, Craw PD, Henchal EA, Sakai SH, Swenson D, Tholath J, Tsai J, Weir AF, Norwood DA. Evaluation of the Cepheid GeneXpert® system for detecting Bacillus anthracis. J Appl Microbiol. 2006;100:1011–6.
Parente DM, Cunha CB, Mylonakis E, Timbrook TT. The clinical utility of methicillin-resistant Staphylococcus aureus (MRSA) nasal screening to rule out MRSA pneumonia: a diagnostic meta-analysis with antimicrobial stewardship implications. Clin Infect Dis. 2018;67:1–7.
Vandercam B, Jeumont S, Cornu O, Yombi JC, Lecouvet F, Lefèvre P, Irenge LM, Gala JL. Amplification-based DNA analysis in the diagnosis of prosthetic joint infection. J Mol Diagn. 2008;10:537–43.
Prina E, Ranzani OT, Torres A. Community-acquired pneumonia. Lancet. 2015;386:1097–108.
Parta M, Goebel M, Thomas J, Matloobi M, Stager C, Musher DM. Impact of an assay that enables rapid determination of Staphylococcus species and their drug susceptibility on the treatment of patients with positive blood culture results. Infect Control Hosp Epidemiol. 2010;31:1043–8.
Acknowledgements
Funding
This research and the journal’s Rapid Service Fee was funded in part by the National Science Foundation of China (grant no. 82150410452) and the Doctoral Research Fund to S.C.O.
Author Contributions
Suvash Chandra Ojha and Ke Chen conceptualized and drafted the study. Suvash Chandra Ojha, Ke Chen, and Changfeng Sun conducted the literature search, analyzed data, and edited the manuscript. Yun-Jian Sheng, Sarfraz Ahmad, and Cun-Liang Deng reviewed the manuscript. All authors read and approved the final manuscript.
Disclosures
Suvash Chandra Ojha, Ke Chen, Changfeng Sun, Sarfraz Ahmad, Yun-Jian Sheng and Cun-Liang Deng confirm that they have no conflicts of interest to disclose.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any new studies with human participants or animals performed by any of the authors.
Data Availability
All data generated or analyzed during this study are included in the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Ojha, S.C., Chen, K., Sun, C. et al. Clinical Relevance of Xpert MRSA/SA in Guiding Therapeutic Decisions for Staphylococcal Infections: A Diagnostic Test Accuracy Analysis. Infect Dis Ther 11, 1205–1227 (2022). https://doi.org/10.1007/s40121-022-00632-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40121-022-00632-w