Abstract
Every year, despite the use of chemicals, significant crop loss is caused by pathogenic microbes. Plant innate resistance to pathogens depends on two sets of genetically encoded immune receptors that sense invaders and trigger signaling cascades leading to reinforcement of physical barriers and production of various antimicrobial compounds. In the past 30 years, the molecular cloning and characterization of plant immune receptors have deepened our understanding of the plant immune system and more importantly, have provided means to improve crop protection against devastating pathogens. Here, we review the molecular characterization of selected immune receptors that can detect multiple species of bacterial pathogens through an expanded recognition range, or through the detection of conserved pathogen activities or host targets. These recent structural and molecular insights about the activation of immune receptors provide the necessary framework to design their concomitant deployment in crops, in order to lower selective pressure on pathogen populations and prevent evasion from recognition. Hence, these few immune receptors emerge as high potential genetic resources to provide durable and environmentally safe protection against important bacterial diseases of solanaceous crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Crop yield is significantly reduced each year, worldwide, because of wounds and diseases caused by pests and pathogens (Popp and Hantos 2011; Bailey-Serres et al. 2019). Disease management relies heavily on chemical warfare, i.e., the use of antibiotics, fungicides, and pesticides to control populations of pathogenic microbes and invertebrate herbivores. To achieve sustainable food security while preserving the environment, both crop losses to disease and usage of harmful chemicals should be reduced. Plants possess an innate immune system, based on the perception of non-self or damaged-self molecules, that activates the reinforcement of physical barriers and the production of antimicrobials, hence efficiently warding off pathogens (Zhang et al. 2020; Zhou and Zhang 2020). Plant inherent resistance to most microbial pathogens has been the focus of research for the past 50 years, as it provides a simple and environmentally safe protection to minimize yield losses (Dangl et al. 2013; Kourelis and van der Hoorn 2018).
In plants, two broad classes of receptors frame the immune system: surface-localized pattern-recognition receptors (PRRs) monitor microbe-derived molecules at the interface between the plant cell and its immediate environment, and intracellular nucleotide-binding leucine-rich repeat receptors (NLRs) monitor microbe-derived molecules once/if the cell wall barrier has been breached by the pathogen (Boutrot and Zipfel 2017; Monteiro and Nishimura 2018). PRRs and NLRs can act in concert to drive signaling cascades leading to a massive reprogramming of gene expression, and the consequent production of defense-related hormones and secondary metabolites that render the plant immune to pathogen infection (Lu and Tsuda 2020; Zhou and Zhang 2020). PRR and NLR repertoires vary among plant species depending on their co-evolution with the pathogen populations in their environment (Boutrot and Zipfel 2017; Monteiro and Nishimura 2018). However, pioneer studies have demonstrated that both types of receptors could be transferred across species, genera or even families of plants to confer robust disease resistance to plants lacking these receptors (Tai et al. 1999; Lacombe et al. 2010; Horvath et al. 2012). Thereby these studies also supported the concept that the defense responses are broadly conserved in the plant kingdom, downstream of specific, receptor-dependent recognition events. A widely accepted idea ensues that crop disease resistance could be significantly improved by addition of immune receptors to the innate repertoire of a given species (Dangl et al. 2013).
Adapted pathogens have acquired diverse systems to inhibit or manipulate the host immune system. Both eukaryotic and prokaryotic pathogens can deliver proteins, termed effectors, into the host cell. Collectively, the effector activities dampen the host defense responses and contribute to establish a favorable environment to promote pathogen growth. Effector repertoires vary among pathogen species and even isolates (Baltrus et al. 2017; Peeters et al. 2013; Schwartz et al. 2015). However, certain effector activities such as proteases or acetyltransferase are broadly conserved, highlighting common requirements necessary for pathogens to achieve their life cycle on a plant (Toruño et al. 2016; Xin et al. 2018). Effectors are rapidly evolving in pathogen populations, partly because of their numbers and redundancy (White et al. 2009; Peeters et al. 2013; Dillon et al. 2019). The selection pressure exerted by the presence of matching host immune receptors that monitor their presence also contributes to rapid effector evolution, enabling a pathogen to evade recognition and re-gain virulence in a given plant genotype (Gassmann et al. 2020; McDonald and Linde 2002; Horvath et al. 2015). The evolutionary arms race between plants and pathogens drives the diversity of both effector and immune receptor repertoires (Boller and He 2009). Hence, the durability of genetic disease resistance in crops would benefit from the concomitant deployment of multiple immune receptors that recognize conserved effectors (Dangl et al. 2013; Jayaraman et al. 2016). Although recent advances in molecular cloning techniques can now allow the rapid construction and transfer of multigenic cassettes into crop genomes, the minimal number of additional immune receptors required to confer robust and long-lasting resistance to one or multiple pathogens in crops remains to be experimentally determined in controlled and field conditions (Dangl et al. 2013).
Recent acceleration in the number of immune receptors cloned and in our understanding of their activation mechanisms revealed that some receptors can provide resistance to multiple pathogens. Three concepts underlie this cross-resistance to multiple pathogens conferred by individual immune receptors: (1) the immune receptor detects molecules highly conserved in a whole class of microbes; (2) the immune receptor monitors the presence of effectors conserved in different pathogen species or (3) the immune receptor monitors modifications of key components of the immune system targeted by pathogen effectors. Here we review these three types of cross-protection conferred by individual immune receptors. We focus on bacterial diseases that affect solanaceous crop production, namely the bacterial speck disease caused by Pseudomonas syringae, the bacterial wilt disease caused by several species of Ralstonia (R. solanacearum, R. pseudosolanacearum and R. syzygii; hereafter denoted R. solanacearum for simplicity) and the bacterial spot disease caused by several species of Xanthomonas (X. euvesicatoria, X. gardneri, and X. perforans) (Mansfield et al. 2012; Prior et al. 2016; Timilsina et al. 2020). We further suggest that stacking these immune receptors in elite crop cultivars would contribute to the development of crops immune to a wide range of bacterial diseases.
2 Plant immune receptors: features and roles in disease resistance
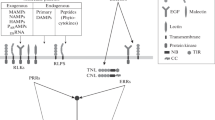
Two large families of modular proteins form the core of the plant immune system. The first group encompasses receptor-like kinases (RLKs) and receptor-like proteins (RLPs) that possess diverse types of extracellular domains able to bind peptides, carbohydrates or other molecules present in the apoplast. Multiple RLKs and RLPs detect various pathogen- or damaged-derived molecules indicative of danger and are collectively termed pattern-recognition receptors (Gust et al. 2017). The prototypical PRR FLAGELLIN SENSING 2 (FLS2), identified in the model plant Arabidopsis thaliana (hereafter Arabidopsis), harbors a leucine-rich repeat (LRR) ectodomain that binds to a conserved fragment of the bacterial flagellin (Gómez-Gómez and Boller 2000; Chinchilla et al. 2006). Similar mechanisms of perception mediated through binding to ectodomains of RLKs or RLPs are well characterized for fragments of the fungal cell wall polymer chitin, the bacterial cold shock protein or lipidic metabolites (Boutrot and Zipfel 2017; Kutschera et al. 2019). Ligand binding triggers conformational changes transmitted through the transmembrane domain leading to the formation of receptor complexes in specialized nanodomains of the plasma membrane and activation of kinase activities in the cytoplasm (Wan et al. 2019b). These rapid events are accompanied by transient influx of Ca2+ and production of reactive oxygen species (ROS). The phosphorylation cascade initiated at the receptor complex is amplified through networks of receptor-like cytoplasmic kinases (RLCKs), calcium-dependent protein kinases and mitogen activated protein kinases (MAPKs) and culminates, through the activation of transcription factors, in massive changes in gene expression (Boutrot and Zipfel 2017; Bjornson et al. 2021). Thus, within hours, the detection of danger signals by PRRs leads to reinforcement of the cell wall through enhanced activity of callose synthases, the production of antimicrobial compounds and a systemically heightened level of defense (Zhou and Zhang 2020).
Adapted pathogens secrete effector proteins in the host cell in part to breach the defense signaling triggered by PRR activation (Macho 2016). Hence the second set of plant immune receptors regroups intracellular NLRs that monitor the pathogenic effector presence or activities in the host cell (Kourelis and van der Hoorn 2018; Monteiro and Nishimura 2018). Each plant species possesses hundreds of NLR genes encoding modular proteins that harbor a conserved nucleotide-binding domain and a carboxy-terminal LRR domain which are both required for intramolecular regulation of the receptor activity. NLRs are further classified depending on the N-terminal domain as Toll-interleukin-1 receptors (TIR)-NLRs or TNLs and coiled-coil (CC)-NLRs or CNLs (Monteiro and Nishimura 2018). Both TNLs and CNLs act as molecular switches that undergo conformational changes upon direct or indirect sensing of effectors. These changes lead to oligomerization of the NLRs into structures termed resistosomes, that are reminiscent of the mammalian apoptosome formed during inflammation or programed cell death (Wang et al. 2019a, b; Ma et al. 2020; Martin et al. 2020; Duxbury et al. 2021). The TNL resistosomes activate a NAD+-hydrolase activity of the TIR domains, while the CNL resistosomes form pores at the plasma membrane allowing ion fluxes (Wan et al. 2019a; Wang et al. 2019a, b; Bi et al. 2021). Further signaling downstream of NLR activation by effectors remains to be fully characterized but depends on a network of helper NLRs and key signaling components such as the lipase-like ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) or the chaperone SUPPRESSOR OF THE G2 ALLELE OF SKP1 (SGT1) (Shirasu 2009; Wu et al. 2018; Lapin et al. 2020). Importantly, TNL- and CNL-dependent responses include Ca2+ influx, production of ROS, and hormonal and transcriptional changes similar to PRR-dependent signaling (Lu and Tsuda 2020). The overlaps in responses triggered by both types of immune receptors can be integrated in a general surveillance system model, where PRRs and NLRs act in concert and feedback into the signaling network to fine-tune the defense responses in time and space (Lu and Tsuda 2020). The detailed understanding of this intricate surveillance system, based on the function of PRRs and NLRs, now provides relevant tools and methods to improve crop resistance to diseases.
3 Immune receptors recognize conserved features of pathogens
Plant PRRs detect microbe-associated molecular patterns (MAMPs) that include exposed or intracellular components of bacteria, such as the building block of the flagellum (flagellin) or of the cell wall (peptidoglycan), and the elongation factor-Tu (EF-Tu) or the cold shock proteins (Boller and Felix 2009). The latter two proteins can be shed or released from dying bacterial cells in the plant apoplast, betraying the bacterial presence. Pioneer studies based on bacterial fractionation in the early 2000s demonstrated that plant cell suspensions can differentially recognize specific prokaryotic epitopes and respond to this elicitation by rapid alkalinization of the apoplastic space and the production of defense-associated molecules such as ROS or the hormone ethylene (Felix et al. 1999; Felix and Boller 2003; Kunze et al. 2004). The purified or synthesized epitopes were consequently used to screen the plant genetic diversity and identify the matching receptors.
3.1 Arabidopsis EFR confers resistance to multiple bacterial pathogens across plant families
Short EF-Tu peptides from Erwinia amylovora, Agrobacterium tumefaciens, Xylella fastidiosa or P. syringae elicit medium alkalinization in Arabidopsis cell suspensions, but not in tobacco, tomato, Citrus or Medicago tissues, revealing the restricted presence of the EF-Tu RECEPTOR (EFR) in the Brassicaceae family (Table 1) (Kunze et al. 2004). EFR was identified from a forward genetic screen for loss of EF-Tu active epitope elf18-induced seedling growth inhibition in Arabidopsis (Zipfel et al. 2006). EFR belongs to group XII LRR-RLK and bares strong homology with the rice PRR XANTHOMONAS RESISTANCE 21 (XA21) that plays a key role in rice resistance against Xanthomonas oryzae oryzae (Song et al. 1995; Zipfel et al. 2006; Holton et al. 2015). The EF-Tu active epitope elf18 binding to the EFR LRR ectodomain has been demonstrated through elegant chimeric receptor studies, revealing the modular functionality and stepwise activation of PRRs through ligand-binding, changes in the conformation of the trans- and juxta-membrane domains and activation of the intracellular kinase domain (Albert et al. 2010a; Albert and Felix 2010).
More importantly, the molecular cloning of EFR from Arabidopsis revealed the important role of PRRs for effective restriction of bacterial infection. Transgenic tobacco expressing EFR are less susceptible to P. syringae pathovars. Similarly, transgenic EFR expression in tomato confers resistance to R. solanacearum and X. perforans (Lacombe et al. 2010). Hence, the disease resistance conferred by EFR could be transferred to susceptible species outside of the Brassicaceae family, supporting the broad conservation of the plant defense responses downstream of the receptor activation. This first breakthrough was followed by numerous studies based on the transfer of PRRs between plant species. Thus, EFR can confer resistance to P. syringae, R. solanacearum and/or Xanthomonas species in tomato, potato, rice, wheat, Medicago, and sweet orange and could even protect apple rootstock from the fire blight caused by E. amylovora (Lacombe et al. 2010; Lu et al. 2015; Schoonbeek et al. 2015; Schwessinger et al. 2015; Boschi et al. 2017; Pfeilmeier et al. 2019; Mitre et al. 2021; Piazza et al. 2021). Of note, the constitutive expression of EFR in the transgenic plants did not alter growth nor the associated microbiome and did not cause yield penalties (Pfeilmeier et al. 2019; Piazza et al. 2021). Moreover, EFR has also been successfully deployed together with the pepper NLR BACTERIAL SPOT RESISTANCE 2 (Bs2) in transgenic tomato for the concomitant field management of bacterial wilt and bacterial spot, highlighting the advantages of stacking different types of immune receptors to achieve durable resistance in crops (Kunwar et al. 2018).
3.2 Soybean GmFLS2 expands the range of flagellin perception across plant families
The restricted presence of EFR in species from the Brassicaceae family indicates its relatively recent evolution (Zipfel et al. 2006). Conversely, most species across the angiosperms and some gymnosperms, possess at least one flagellin receptor (Albert et al. 2010b). Flagellin is the building block of the bacterial flagellum, an essential structure that allows bacteria to move toward nutrients and evade stress (Panopoulos and Schroth 1974; Ramos et al. 2004). Flagellin can leak in the outer medium during flagellum assembly; plants also secrete glycosidases and proteases to release active epitopes of flagellin in the apoplast (Buscaill et al. 2019). Two conserved regions of the bacterial flagellin have been shown to differentially elicit defense responses in plants. The first region at the N-terminal of flagellin contains the immunogenic epitopes flg15 and flg22 that are recognized in tomato and Arabidopsis cell suspensions (Felix et al. 1999). Later, a second region, inferred from the natural variation of the flagellin-encoding FLAGELLIN C (fliC) gene in the P. syringae pv. tomato T1 strain, was identified and its active epitope termed flgII-28 (Cai et al. 2011). Interestingly, flgII-28 detection is restricted to some Solanaceae species including tomato, pepper, and potato, but not tobacco (Clarke et al. 2013). Although highly conserved, the flg22 or flgII-28 sequences of some phytopathogenic bacteria harbor amino acid changes that prevent detection by the plant receptors. For instance, flg22, flgII-28 or even the whole fliC protein of R. solanacearum do not trigger defense responses in Arabidopsis, tobacco, tomato, pepper, or eggplant (Felix et al. 1999; Wei et al. 2017). Hence, R. solanacearum could evade recognition, even from species like tomato that possesses both the receptors for flg15/flg22 and for flgII-28 (Robatzek et al. 2007; Hind et al. 2016). On another hand, to the best of our knowledge, whether the specific X. euvesicatoria flagellin epitopes are detected in a FLS2-dependent manner has not been reported. However, the flg22 peptides from the related species X. campestris and X. axonopodis pv. citri are immunogenic in Arabidopsis and tomato (Sun et al. 2006; Bhattarai et al. 2016).
Interestingly, in the Fabaceae family, soybean (Glycine max) but not common bean, peanut, pea nor Medicago, can perceive R. solanacearum purified flagellin and the corresponding flg22 peptide (Wei et al. 2020). In accordance, soybean appears to be a nonhost for R. solanacearum, as no pathogenic strain could be isolated from soybean so far (Wei et al. 2020). This indicates that the natural resistance of soybean to bacterial wilt may be due, at least in part, to the detection of the R. solanacearum flagellin. There are two FLS2 orthologs in G. max and both could confer R. solanacearum flg22 perception in Nicotiana benthamiana, with the full response requiring co-expression of GmFLS2 with the co-receptor BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED KINASE 1 (GmBAK1) (Table 1). GmFLS2b was further characterized to pinpoint the residues of the ectodomain required for the binding of R. solanacearum flg22. Tomato hairy root transiently expressing GmFLS2b and GmBAK1 from a multigenic cassette displayed responsiveness to R. solanacearum flg22 and the resulting plants were more resistant to R. solanacearum infection. This study, along with another from other researchers, demonstrates further that GmFLS2 has a broader range of flg22 detection, as the changes that enable R. solanacearum flg22 detection do not affect the perception of the typical flg22 peptide from P. syringae (Tian et al. 2020; Wei et al. 2020). Whether GmFLS2 could detect X. euvesicatoria or X. perforans flg22 remains to be determined.
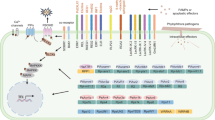
As demonstrated with Arabidopsis EFR and soybean GmFLS2, the transfer of lacking PRRs and/or PRRs with extended recognition could both widen the range of pathogenic molecules being perceived by a given species and enhance durable crop resistance to bacterial pathogens (Fig. 1).
Possible minimum stack of immune receptors to deploy for tomato resistance to bacterial speck, bacterial wilt and bacterial spot diseases. Transgenic tomato expressing immune receptors (PRRs and NLRs) from a multigenic cassette could gain immunity to multiple bacterial diseases through the direct recognition of conserved pathogenic features (MAMPs or effectors) and the monitoring of conserved host factors targeted by the pathogens. Arabidopsis EFR (yellow) and soybean GmFLS2 (orange) extend the recognition range of EF-Tu and flagellin. Nicotiana benthamiana Roq1 (green) directly recognizes the homologous effectors HopQ1, XopQ and RipB of P. syringae, Xanthomonas spp., and R. solanacearum, respectively. Solanum lycopersicoides Ptr1 (blue) and NbZAR1 (dark grey) confer resistance by monitoring the modification of host factors targeted by AvrRpt2 and RipBN, and HopZ1 and XopJ4, respectively. Dotted lines and question marks indicate possible interactions between immune receptors and MAMPs/effectors
4 Immune receptors recognize conserved effectors or modifications of conserved targets
The comparison of the type III effector repertoires in P. syringae, X. euvesicatoria and R. solanacearum highlights several broad families of effectors with conserved enzymatic activities, suggesting that activities such as acetyltransferases, ribosyltransferases, ubiquitin ligases, cysteine proteases, and hydrolases are required across genera for bacteria to colonize plant tissues (Wroblewski et al. 2009; Peeters et al. 2013; Schwartz et al. 2015; Dillon et al. 2019). In the last few years, NLRs that sense such conserved effectors have been cloned and shown to confer resistance to multiple species of pathogens.
4.1 Nicotiana benthamiana Roq1 confers resistance to P. syringae, R. solanacearum and X. euvesicatoria
Several species of Xanthomonas, and different strains of P. syringae and R. solanacearum harbor homologous effectors with a predicted nucleoside hydrolase (NH) domain, termed Xanthomonas outer protein XopQ, hypersensitive response and pathogenicity-dependent outer protein HopQ1 and Ralstonia-injected protein RipB, respectively (Roden et al. 2004; Li et al. 2013; Peeters et al. 2013). Although the ability of these effectors to cleave nucleosides has not been clearly established, structural and recombinant protein studies indicate that XopQ can hydrolyze substrates containing a ribosyl group, while HopQ1 can hydrolyze a cytokinin precursor in vitro (Li et al. 2013; Hann et al. 2014; Yu et al. 2014; Gupta et al. 2015). Strikingly, the transient expression of XopQ, HopQ1 or RipB triggers rapid defense responses in Nicotiana spp. (Wei et al. 2007; Schwartz et al. 2015; Nakano and Mukaihara 2019). These responses depend on the key TNL regulators SGT1 and EDS1, indicating that these effectors are likely recognized by the Nicotiana immune system (Wei et al. 2007; Adlung et al. 2016; Nakano and Mukaihara 2019). Indeed, the major contribution of XopQ, HopQ1 and RipB to the avirulence of X. euvesicatoria, P. syringae pv. tomato DC3000 and R. solanacearum RS1002 was further demonstrated using multiple mutant strains lacking the NH effectors (Wei et al. 2007; Adlung et al. 2016; Nakano and Mukaihara 2019).
Taking advantage of the limited number of TNLs predicted in the N. benthamiana genome, Schultink and colleagues designed a rapid reverse genetic screen based on virus-induced silencing of multiple genes to identify the XopQ matching immune receptor, RECOGNITION OF XOPQ1 (Roq1) (Table 1) (Schultink et al. 2017). Roq1 encodes a typical TNL protein that directly interacts with XopQ and HopQ1 through its LRR and post-LRR domains (Schultink et al. 2017; Qi et al. 2018; Martin et al. 2020). In the presence of XopQ, Roq1 oligomerizes, forming a tetrameric resistosome that results in activation of the TIR domain and downstream immune signaling (Qi et al. 2018; Martin et al. 2020). Interestingly, rather than depending on the effector NH activity, the recognition event is controlled by the structural features of the effector’s active site, where Roq1 makes multiple contacts. Considering the high degree of amino acid identity in the active site region of XopQ, HopQ1 and RipB (Yu et al. 2014), it is likely that HopQ1 and RipB activate Roq1 through similar interactions.
The high potential of Roq1 as a source of resistance to multiple bacterial pathogens has recently been tested in transgenic tomato lines (Thomas et al. 2020). Homozygous tomato plants expressing Roq1 from a constitutive promoter are more resistant to X. perforans, X. euvesicatoria and P. syringae, as demonstrated by up to 100 times lower bacterial titer in leaf tissues and clearly reduced disease symptoms after infection. Moreover, X. perforans disease severity was strongly reduced in the Roq1-expressing tomato plants in consecutive field trials, while no impact on the plant growth, morphology or fruit yield could be observed. Furthermore, in a controlled environment, Roq1-expressing tomato showed only weak disease symptoms after R. solanacearum infection, while the wild-type plants completely wilted (Thomas et al. 2020). Hence, Roq1 stands out among the NLRs as a single gene conferring resistance to a broad range of bacterial pathogens (Fig. 1). This is reminiscent of the broad resistance conferred by EFR, highlighting the gain that could be harnessed for crop protection from immune receptors evolved to recognize conserved features of pathogens.
4.2 Solanum lycopersicoides Ptr1 confers resistance to P. syringae and R. solanacearum
Besides the direct binding of effectors to matching NLRs, effector presence in the plant cell can also be monitored through the results of the effector’s enzymatic activity (Kourelis and van der Hoorn 2018). For example, the P. syringae effector AvrRpt2 belongs to a broad clade of cysteine proteases that contains homologous effectors in other phytopathogenic bacteria such as Acidovorax citrulli, E. amylovora or R. solanacearum (Zhao et al. 2006; Eschen-Lippold et al. 2016). The conserved catalytic triad cysteine, histidine and aspartate residues are essential for the protease activity and the recognition of AvrRpt2 in plants (Axtell et al. 2003). When naturally delivered by the bacterial type III secretion system or heterologously expressed in Arabidopsis, AvrRpt2 cleaves RESISTANCE TO PSEUDOMONAS SYRINGAE PV. MACULICOLA 1-INTERACTING PROTEIN 4 (RIN4) and thereby activates a CNL termed RESISTANCE TO PSEUDOMONAS SYRINGAE 2 (RPS2) (Axtell et al. 2003; Mackey et al. 2003). Solanum lycopersicum possesses three homologs of Arabidopsis RIN4 but fails to recognize P. syringae AvrRpt2 (Mazo-Molina et al. 2019). However, the careful observation of disease symptoms during a natural outbreak of bacterial speck disease in introgression lines derived from S. lycopersicum and S. lycopersicoides recently led to the discovery of individual plants resistant to P. syringae race 1 strain carrying AvrRpt2 (Mazo-Molina et al. 2019). Using molecular markers and transcriptomic datasets, the researchers cloned the gene responsible for the resistance from S. lycopersicoides, PSEUDOMONAS TOMATO RACE 1 (Ptr1) (Table 1) (Mazo-Molina et al. 2020). Like Arabidopsis RPS2, Ptr1 requires the cleavage of SlRIN4 for its activation by AvrRpt2. Interestingly, Ptr1 can also recognize an AvrRpt2 homolog from R. solanacearum, RipBN. Indeed, transient expression of RipBN in S. lycopersicoides induced a typical hypersensitive response and the growth of RipBN-carrying R. solanacearum strain was inhibited in transgenic tomato expressing Ptr1 (Mazo-Molina et al. 2019).
Ptr1 has homologs in diverse solanaceous species, including functional homologs in potato, and N. benthamiana (Mazo-Molina et al. 2020). However, in many S. lycopersicum and S. pennellii accessions Ptr1 has been pseudogenized due to a small deletion near the start codon of the gene. On the other hand, although AvrRpt2 is present in a range of P. syringae strains, RipBN presence is restricted to R. solanacearum strains mainly found in Africa (Eschen-Lippold et al. 2016; Mazo-Molina et al. 2020). Nonetheless, it is not excluded that Ptr1 could detect other effectors from the large cysteine protease family that includes AvrRpt2 and RipBN, as well as HopN1 and HopX1 in P. syringae and related effectors from X. euvesicatoria and R. solanacearum (Nimchuk et al. 2007; Shindo and van der Hoorn 2008; Mazo-Molina et al. 2019; Sang et al. 2020). Additionally, RIN4 is an important hub of the plant immune signaling pathway and as such has been shown to interact with or be modified by several effectors (Toruño et al. 2019). Ptr1 could therefore be used as an important genetic source of resistance to develop tomato cultivars with heightened resistance to both bacterial speck and bacterial wilt diseases (Fig. 1).
4.3 Nicotiana benthamiana NbZAR1 confers resistance to P. syringae and X. perforans
Many pathogen effectors function as suppressors or modulators of the plant immune system (Toruño et al. 2016; Macho 2016; Xin et al. 2018). For example, the P. syringae effectors AvrPto and AvrPtoB directly impair the function of the plant PRR complexes at the plasma membrane, through the inhibition of their intracellular kinase domain or by promoting their proteasome-dependent degradation, respectively (Xiang et al. 2008; Göhre et al. 2008; Gimenez-Ibanez et al. 2009). The P. syringae effector AvrPphB and the X. campestris pv. campestris effector XopAC can cleave or uridylylate, respectively, members of the group VII RLCKs that contribute to immune signaling downstream of PRRs (Swiderski and Innes 2001; Shao et al. 2003; Feng et al. 2012; Rao et al. 2018). On another hand, the group XII RLCK HOPZ-EFFECTOR-TRIGGERED IMMUNITY DEFICIENT 1 (ZED1) does not participate in PRR signaling but is acetylated by the P. syringae effector HopZ1a (Lewis et al. 2013, 2014). Rather than affecting the plant defense response, ZED1 modification by HopZ1a leads to the activation of the CNL HOPZ-ACTIVATED RESISTANCE 1 (ZAR1) and plant resistance. (Lewis et al. 2010, 2013). HopZ1a also acetylates several group VII RLCKs, promoting their interaction with a ZED1-ZAR1 complex (Bastedo et al. 2019). Another ternary complex containing the group VII RLCK AVRPPHB SUSCEPTIBLE 1-LIKE 2 (PBL2), the group XII RLCK RESISTANCE-RELATED KINASE 1 (RKS1) and ZAR1 is formed when X. campestris XopAC uridylylates PBL2 (Wang et al. 2015). The structure of this ternary complex was determined using gel filtration and cryo-electron microscopy and revealed how activated ZAR1 oligomerizes in a wheel-shape resistosome (Wang et al. 2019a, b). Through its association with different RLCKs, ZAR1 also contributes to the recognition of several other P. syringae effectors from 5 distinct families: HopX1, HopF1/HopF2, HopO1 and HopBA1 (Seto et al. 2017; Laflamme et al. 2020; Martel et al. 2020). Together these recent findings highlight the crucial function of ZAR1 as a versatile sensor of different effector activities targeting the RLCKs and thus its potential to provide durable disease resistance in crops (Fig. 1).
Phylogenetic analyses indicate the conservation of ZAR1 sequence in several dicot species and more broadly across angiosperms (Baudin et al. 2017; Schultink et al. 2019; Adachi et al. 2020). However, the function of the ZAR1 orthologs has not yet been investigated, except for N. benthamiana NbZAR1 (Table 1) (Schultink et al. 2019). Nicotiana benthamiana is resistant to the tomato bacterial spot pathogen X. perforans, due to the recognition of several effectors including AvrBsT, XopQ and XopJ4 (Roden et al. 2004; Schwartz et al. 2015). XopJ4 belongs to the same acetyltransferase family as HopZ1a, and its expression triggers a robust cell death in N. benthamiana leaves. By screening an ethylmethanesulfonate (EMS)-mutagenized population of N. benthamiana for loss of XopJ4-induced cell death and subsequent genetic analysis, Schultink and colleagues identified two mutations in NbZAR1 that caused the loss of XopJ4 recognition (Schultink et al. 2019). In the zar1 mutant plants, a X. perforans strain lacking AvrBsT and XopQ could grow 100 times more than in the wild-type N. benthamiana, revealing the contribution of NbZAR1 to X. perforans resistance. Unexpectedly, neither Arabidopsis ZAR1 nor tomato SlZAR1 could restore the XopJ4-induced cell death in N. benthamiana zar1 plants. Conversely, NbZAR1 expressed in combination with Arabidopsis ZED1 could restore HopZ1a recognition in zar1 plants, suggesting that NbZAR1 could also contribute to P. syringae resistance. However, for XopJ4 recognition NbZAR1 interacts with and functions with one of the 4 group XII RLCKs present in N. benthamiana, XOPJ4 IMMUNITY 2 (JIM2) (Schultink et al. 2019). Considering the ability of NbZAR1 to function with different RLCK partners together with the fact that RLCKs are the targets of several effector families, we suggest that NbZAR1, like Roq1 and Ptr1, is a promising source of resistance to multiple bacterial pathogens. However, the transfer of NbZAR1 to solanaceous crops such as tomato and the gain of disease resistance it confers remains to be tested, both in controlled and field conditions.
5 Conclusion
The resistance conferred by the transfer of single immune receptor could and has been broken down by simple loss or mutation of the rapidly evolving pathogen effectors (Gassmann et al. 2020; McDonald and Linde 2002; Horvath et al. 2015). Hence, durable crop disease resistance can only be achieved through the deployment of stacks of immune receptors that recognize different components of a given pathogen to minimize the selection pressure on pathogen populations in the field (Dangl et al. 2013). The question remains about the features and, importantly, the number of immune receptors to transfer into a given crop for robust and durable disease resistance. The recent works highlighted in this review collectively revealed that individual immune receptors can sense multiple pathogens, through the detection of either essential bacterial features, conserved virulence activities or by monitoring key components of the defense signaling pathways. This knowledge opens the possibility to minimize the number of immune receptors to include in “resistance stacks”, by selecting these multitasking immune receptors able to launch the defense program in response to different species or genera of pathogens (Fig. 1). With current advances in synthetic biology, molecular breeding, and crop transformation technologies, deploying these immune receptors from multigenic cassettes in elite crop cultivars is now within reach to develop crops resistant to a wide range of diseases, thus contributing to sustainable food security.
References
Adachi H, Sakai T, Kourelis J, Hernandez JLG, Maqbool A, Kamoun S (2020) Jurassic NLR: conserved and dynamic evolutionary features of the atypically ancient immune receptor ZAR1. bioRxiv. https://doi.org/10.1101/2020.10.12.333484
Adlung N, Prochaska H, Thieme S, Banik A, Blüher D, John P, Nagel O, Schulze S, Gantner J, Delker C, Stuttmann J, Bonas U (2016) Non-host resistance induced by the Xanthomonas effector XopQ is widespread within the genus Nicotiana and functionally depends on EDS1. Front Plant Sci 7:1796. https://doi.org/10.3389/fpls.2016.01796
Albert M, Felix G (2010) Chimeric receptors of the Arabidopsis thaliana pattern recognition receptors EFR and FLS2. Plant Signal Behav 5:1430–1432. https://doi.org/10.4161/psb.5.11.13312
Albert M, Jehle AK, Lipschis M, Mueller K, Zeng Y, Felix G (2010a) Regulation of cell behaviour by plant receptor kinases: pattern recognition receptors as prototypical models. Eur J Cell Biol 89:200–207. https://doi.org/10.1016/j.ejcb.2009.11.015
Albert M, Jehle AK, Mueller K, Eisele C, Lipschis M, Felix G (2010b) Arabidopsis thaliana pattern recognition receptors for bacterial elongation factor Tu and flagellin can be combined to form functional chimeric receptors. J Biol Chem 285:19035–19042. https://doi.org/10.1074/jbc.M110.124800
Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ (2003) Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol Microbiol 49:1537–1546. https://doi.org/10.1046/j.1365-2958.2003.03666.x
Bailey-Serres J, Parker JE, Ainsworth EA, Oldroyd GED, Schroeder JI (2019) Genetic strategies for improving crop yields. Nature 575:109–118. https://doi.org/10.1038/s41586-019-1679-0
Baltrus DA, McCann HC, Guttman DS (2017) Evolution, genomics and epidemiology of Pseudomonas syringae: challenges in bacterial molecular plant pathology. Mol Plant Pathol 18:152–168. https://doi.org/10.1111/mpp.12506
Bastedo DP, Khan M, Martel A, Seto D, Kireeva I, Zhang J, Masud W, Millar D, Lee JY, Lee AH-Y, Gong Y, Santos-Severino A, Guttman DS, Desveaux D (2019) Perturbations of the ZED1 pseudokinase activate plant immunity. PLoS Pathog 15:e1007900. https://doi.org/10.1371/journal.ppat.1007900
Baudin M, Hassan JA, Schreiber KJ, Lewis JD (2017) Analysis of the ZAR1 immune complex reveals determinants for immunity and molecular interactions. Plant Physiol 174:2083–2053. https://doi.org/10.1104/pp.17.00441
Bhattarai K, Louws FJ, Williamson JD, Panthee DR (2016) Differential response of tomato genotypes to Xanthomonas-specific pathogen-associated molecular patterns and correlation with bacterial spot (Xanthomonas perforans) resistance. Hortic Res 3:16035. https://doi.org/10.1038/hortres.2016.35
Bi G, Su M, Li N, Liang Y, Dang S, Xu J, Hu M, Wang J, Zou M, Deng Y, Li Q, Huang S, Li J, Chai J, He K, Chen Y, Zhou J-M (2021) The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184:3528–3541. https://doi.org/10.1016/j.cell.2021.05.003
Bjornson M, Pimprikar P, Nürnberger T, Zipfel C (2021) The transcriptional landscape of Arabidopsis thaliana pattern-triggered immunity. Nat Plants 7:579–586. https://doi.org/10.1038/s41477-021-00874-5
Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60:379–406. https://doi.org/10.1146/annurev.arplant.57.032905.105346
Boller T, He SY (2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324:742–744. https://doi.org/10.1126/science.1171647
Boschi F, Schvartzman C, Murchio S, Ferreira V, Siri MI, Galván GA, Smoker M, Stransfeld L, Zipfel C, Vilaró FL, Dalla-Rizza M (2017) Enhanced bacterial wilt resistance in potato through expression of Arabidopsis EFR and introgression of quantitative resistance from Solanum commersonii. Front Plant Sci 8:1642. https://doi.org/10.3389/fpls.2017.01642
Boutrot F, Zipfel C (2017) Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu Rev Phytopathol 55:257–286. https://doi.org/10.1146/annurev-phyto-080614-120106
Buscaill P, Chandrasekar B, Sanguankiattichai N, Kourelis J, Kaschani F, Thomas EL, Morimoto K, Kaiser M, Preston GM, Ichinose Y, van der Hoorn RAL (2019) Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides. Science 364:eaav0748. https://doi.org/10.1126/science.aav0748
Cai R, Lewis J, Yan S, Liu H, Clarke CR, Campanile F, Almeida NF, Studholme DJ, Lindeberg M, Schneider D, Zaccardelli M, Setubal JC, Morales-Lizcano NP, Bernal A, Coaker G, Baker C, Bender CL, Leman S, Vinatzer BA (2011) The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathog 7:e1002130. https://doi.org/10.1371/journal.ppat.1002130
Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G (2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18:465–476. https://doi.org/10.1105/tpc.105.036574
Clarke CR, Chinchilla D, Hind SR, Taguchi F, Miki R, Ichinose Y, Martin GB, Leman S, Felix G, Vinatzer BA (2013) Allelic variation in two distinct Pseudomonas syringae flagellin epitopes modulates the strength of plant immune responses but not bacterial motility. New Phytol 200:847–860. https://doi.org/10.1111/nph.12408
Dangl JL, Horvath DM, Staskawicz BJ (2013) Pivoting the plant immune system from dissection to deployment. Science 345:746–751. https://doi.org/10.1126/science.1236011
Dillon MM, Almeida RND, Laflamme B, Martel A, Weir BS, Desveaux D, Guttman DS (2019) Molecular evolution of Pseudomonas syringae type III secreted effector proteins. Front Plant Sci 10:418. https://doi.org/10.3389/fpls.2019.00418
Duxbury Z, Wu C, Ding P (2021) A comparative overview of the intracellular guardians of plants and animals: NLRs in innate immunity and beyond. Annu Rev Plant Biol 72:155–184. https://doi.org/10.1146/annurev-arplant-080620-104948
Eschen-Lippold L, Jiang X, Elmore JM, Mackey D, Shan L, Coaker G, Scheel D, Lee J (2016) Bacterial AvrRpt2-like cysteine proteases block activation of the Arabidopsis mitogen-activated protein kinases, MPK4, and MPK11. Plant Physiol 171:2223–2238. https://doi.org/10.1104/pp.16.00336
Felix G, Boller T (2003) Molecular sensing of bacteria in plants: the highly conserved RNA-binding motif RNP-1 of bacterial cold shock proteins is recognized as an elicitor signal in tobacco. J Biol Chem 278:6201–6208. https://doi.org/10.1074/jbc.M209880200
Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18:265–276. https://doi.org/10.1046/j.1365-313x.1999.00265.x
Feng F, Yang F, Rong W, Wu X, Zhang J, Chen S, He C, Zhou J-M (2012) A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485:114–118. https://doi.org/10.1038/nature10962
Gassmann W, Dahlbeck D, Chesnokova O, Minsavage GV, Jones JB, Staskawicz BJ (2020) Molecular evolution of virulence in natural field strains of Xanthomonas campestris pv. vesicatoria. J Bacteriol 182:7053–7059. https://doi.org/10.1128/JB.182.24.7053-7059.2000
Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP (2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol 19:423–429. https://doi.org/10.1016/j.cub.2009.01.054
Göhre V, Spallek T, Häweker H, Mersmann S, Mentzel T, Boller T, de Torres M, Mansfield JW, Robatzek S (2008) Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol 18:1824–1832. https://doi.org/10.1016/j.cub.2008.10.063
Gómez-Gómez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5:1003–1011. https://doi.org/10.1016/S1097-2765(00)80265-8
Gupta MK, Nathawat R, Sinha D, Haque AS, Sankaranarayanan R, Sonti RV (2015) Mutations in the predicted active site of Xanthomonas oryzae pv. oryzae XopQ differentially affect virulence, suppression of host innate immunity, and induction of the HR in a nonhost plant. Mol Plant Microbe Interact 28:195–206. https://doi.org/10.1094/MPMI-09-14-0288-R
Gust AA, Pruitt R, Nürnberger T (2017) Sensing danger: key to activating plant immunity. Trends Plant Sci 22:779–797. https://doi.org/10.1016/j.tplants.2017.07.005
Hann DR, Rathjen JP (2007) Early events in the pathogenicity of Pseudomonas syringae on Nicotiana benthamiana. Plant J 49:607–618. https://doi.org/10.1111/j.1365-313X.2006.02981.x
Hann DR, Domínguez-Ferreras A, Motyka V, Dobrev PI, Schornack S, Jehle A, Felix G, Chinchilla D, Rathjen JP, Boller T (2014) The Pseudomonas type III effector HopQ1 activates cytokinin signaling and interferes with plant innate immunity. New Phytol 201:585–598. https://doi.org/10.1111/nph.12544
Hind SR, Strickler SR, Boyle PC, Dunham DM, Bao Z, O’Doherty IM, Baccile JA, Hoki JS, Viox EG, Clarke CR, Vinatzer BA, Schroeder FC, Martin GB (2016) Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat Plants 2:16128. https://doi.org/10.1038/nplants.2016.128
Holton N, Nekrasov V, Ronald PC, Zipfel C (2015) The phylogenetically-related pattern recognition receptors EFR and XA21 recruit similar immune signaling components in monocots and dicots. PLoS Pathog 11:e1004602. https://doi.org/10.1371/journal.ppat.1004602
Horvath DM, Stall RE, Jones JB, Pauly MH, Vallad GE, Dahlbeck D, Staskawicz BJ, Scott JW (2012) Transgenic resistance confers effective field level control of bacterial spot disease in tomato. PLoS ONE 7:e42036. https://doi.org/10.1371/journal.pone.0042036
Horvath DM, Pauly MH, Hutton SF, Vallad GE, Scott JW, Jones JB, Stall RE, Dahlbeck D, Staskawicz BJ, Tricoli D, Deynze AV (2015) The pepper Bs2 gene confers effective field resistance to bacterial leaf spot and yield enhancement in Florida tomatoes. Acta Hortic 1069:47–51. https://doi.org/10.17660/ActaHortic.2015.1069.5
Jayaraman J, Segonzac C, Cho H, Jung G, Sohn KH (2016) Effector-assisted breeding for bacterial wilt resistance in horticultural crops. Hortic Environ Biotechnol 57:415–423. https://doi.org/10.1007/s13580-016-0191-9
Kourelis J, van der Hoorn RAL (2018) Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30:285–299. https://doi.org/10.1105/tpc.17.00579
Kunwar S, Iriarte F, Fan Q, da Silva EE, Ritchie L, Nguyen NS, Freeman JH, Stall RE, Jones JB, Minsavage GV, Colee J, Scott JW, Vallad GE, Zipfel C, Horvath D, Westwood J, Hutton SF, Paret ML (2018) Transgenic expression of EFR and Bs2 genes for field management of bacterial wilt and bacterial spot of tomato. Phytopathol 108:1402–1411. https://doi.org/10.1094/PHYTO-12-17-0424-R
Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16:3496–3507. https://doi.org/10.1105/tpc.104.026765
Kutschera A, Dawid C, Gisch N, Schmid C, Raasch L, Gerster T, Schäffer M, Smakowska-Luzan E, Belkhadir Y, Vlot AC, Chandler CE, Schellenberger R, Schwudke D, Ernst RK, Dorey S, Hückelhoven R, Hofmann T, Ranf S (2019) Bacterial medium-chain 3-hydroxy fatty acid metabolites trigger immunity in Arabidopsis plants. Science 364:178–181. https://doi.org/10.1126/science.aau1279
Lacombe S, Rougon-Cardoso A, Sherwood E, Peeters N, Dahlbeck D, van Esse HP, Smoker M, Rallapalli G, Thomma BPHJ, Staskawicz B, Jones JDG, Zipfel C (2010) Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat Biotechnol 28:365–369. https://doi.org/10.1038/nbt.1613
Laflamme B, Dillon MM, Martel A, Almeida RND, Desveaux D, Guttman DS (2020) The pan-genome effector-triggered immunity landscape of a host-pathogen interaction. Science 367:763–768. https://doi.org/10.1126/science.aax4079
Lapin D, Bhandari DD, Parker JE (2020) Origins and immunity networking functions of EDS1 family proteins. Annu Rev Phytopathol 58:253–276. https://doi.org/10.1146/annurev-phyto-010820-012840
Lewis JD, Wu R, Guttman DS, Desveaux D (2010) Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet 6:e1000894. https://doi.org/10.1371/journal.pgen.1000894
Lewis JD, Lee AH-Y, Hassan JA, Wan J, Hurley B, Jhingree JR, Wang PW, Lo T, Youn J-Y, Guttman DS, Desveaux D (2013) The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proc Natl Acad Sci USA 110:18722–18727. https://doi.org/10.1073/pnas.1315520110
Lewis JD, Lo T, Bastedo P, Guttman DS, Desveaux D (2014) The rise of the undead: pseudokinases as mediators of effector-triggered immunity. Plant Signal Behav 9:e27563. https://doi.org/10.4161/psb.27563
Li W, Chiang Y-H, Coaker G (2013) The HopQ1 effector’s nucleoside hydrolase-like domain is required for bacterial virulence in Arabidopsis and tomato, but not host recognition in tobacco. PLoS ONE 8:e59684. https://doi.org/10.1371/journal.pone.0059684
Lu Y, Tsuda K (2020) Intimate association of PRR- and NLR-mediated signaling in plant immunity. Mol Plant Microbe Interact 34:3–14. https://doi.org/10.1094/MPMI-08-20-0239-IA
Lu F, Wang H, Wang S, Jiang W, Shan C, Li B, Yang J, Zhang S, Sun W (2015) Enhancement of innate immune system in monocot rice by transferring the dicotyledonous elongation factor Tu receptor EFR. J Integr Plant Biol 57:641–652. https://doi.org/10.1111/jipb.12306
Ma S, Lapin D, Liu L, Sun Y, Song W, Zhang X, Logemann E, Yu D, Wang J, Jirschitzka J, Han Z, Schulze-Lefert P, Parker JE, Chai J (2020) Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science 370:eabe3069. https://doi.org/10.1126/science.abe3069
Macho AP (2016) Subversion of plant cellular functions by bacterial type-III effectors: beyond suppression of immunity. New Phytol 210:51–57. https://doi.org/10.1111/nph.13605
Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112:379–389. https://doi.org/10.1016/S0092-8674(03)00040-0
Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, Toth I, Salmond G, Foster GD (2012) Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13:614–629. https://doi.org/10.1111/j.1364-3703.2012.00804.x
Martel A, Laflamme B, Seto D, Bastedo DP, Dillon MM, Almeida RND, Guttman DS, Desveaux D (2020) Immunodiversity of the Arabidopsis ZAR1 NLR is conveyed by receptor-like cytoplasmic kinase sensors. Front Plant Sci 11:1290. https://doi.org/10.3389/fpls.2020.01290
Martin R, Qi T, Zhang H, Liu F, King M, Toth C, Nogales E, Staskawicz BJ (2020) Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Science 370:eabd9993. https://doi.org/10.1126/science.abd9993
Mazo-Molina C, Mainiero S, Hind SR, Kraus CM, Vachev M, Maviane-Macia F, Lindeberg M, Saha S, Strickler SR, Feder A, Giovannoni JJ, Smart CD, Peeters N, Martin GB (2019) The Ptr1 locus of Solanum lycopersicoides confers resistance to race 1 strains of Pseudomonas syringae pv. tomato and to Ralstonia pseudosolanacearum by recognizing the type III effectors AvrRpt2 and RipBN. Mol Plant Microbe Interact 32:949–960. https://doi.org/10.1094/MPMI-01-19-0018-R
Mazo-Molina C, Mainiero S, Haefner BJ, Bednarek R, Zhang J, Feder A, Shi K, Strickler SR, Martin GB (2020) Ptr1 evolved convergently with RPS2 and Mr5 to mediate recognition of AvrRpt2 in diverse solanaceous species. Plant J 103:1433–1445. https://doi.org/10.1111/tpj.14810
McDonald BA, Linde C (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Ann Rev Phytopathol 40:349–379. https://doi.org/10.1146/annurev.phyto.40.120501.101443
Mitre LK, Teixeira-Silva NS, Rybak K, Magalhães DM, de Souza-Neto RR, Robatzek S, Zipfel C, de Souza AA (2021) The Arabidopsis immune receptor EFR increases resistance to the bacterial pathogens Xanthomonas and Xylella in transgenic sweet orange. Plant Biotechnol J 19:1294–1296. https://doi.org/10.1111/pbi.13629
Monteiro F, Nishimura MT (2018) Structural, functional, and genomic diversity of plant NLR proteins: an evolved resource for rational engineering of plant immunity. Annu Rev Phytopathol 56:243–267. https://doi.org/10.1146/annurev-phyto-080417-045817
Nakano M, Mukaihara T (2019) The type III effector RipB from Ralstonia solanacearum RS1000 acts as a major avirulence factor in Nicotiana benthamiana and other Nicotiana species. Mol Plant Pathol 20:1237–1251. https://doi.org/10.1111/mpp.12824
Nimchuk ZL, Fisher EJ, Desveaux D, Chang JH, Dangl JL (2007) The HopX (AvrPphE) family of Pseudomonas syringae type III effectors require a catalytic triad and a novel N-terminal domain for function. Mol Plant Microbe Interact 20:346–357. https://doi.org/10.1094/MPMI-20-4-0346
Panopoulos NJ, Schroth MN (1974) Role of flagellar motility in the invasion of bean leaves by Pseudomonas phaseolicola. Phytopathology 64:1389–1397. https://doi.org/10.1094/Phyto-64-1389
Peeters N, Carrère S, Anisimova M, Plener L, Cazalé A-C, Genin S (2013) Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genom 14:859. https://doi.org/10.1186/1471-2164-14-859
Pfeilmeier S, George J, Morel A, Roy S, Smoker M, Stransfeld L, Downie JA, Peeters N, Malone JG, Zipfel C (2019) Expression of the Arabidopsis thaliana immune receptor EFR in Medicago truncatula reduces infection by a root pathogenic bacterium, but not nitrogen-fixing rhizobial symbiosis. Plant Biotechnol J 17:569–579. https://doi.org/10.1111/pbi.12999
Piazza S, Campa M, Pompili V, Costa LD, Salvagnin U, Nekrasov V, Zipfel C, Malnoy M (2021) The Arabidopsis pattern recognition receptor EFR enhances fire blight resistance in apple. Hortic Res 8:204. https://doi.org/10.1038/s41438-021-00639-3
Popp J, Hantos K (2011) The impact of crop protection on agricultural production. Stud Agric Econ 113:47–66. https://doi.org/10.22004/ag.econ.102401
Prior P, Ailloud F, Dalsing BL, Remenant B, Sanchez B, Allen C (2016) Genomic and proteomic evidence supporting the division of the plant pathogen Ralstonia Solanacearum into three species. BMC Genom 17:90. https://doi.org/10.1186/s12864-016-2413-z
Qi T, Seong K, Thomazella DPT, Kim JR, Pham J, Seo E, Cho M-J, Schultink A, Staskawicz BJ (2018) NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc Natl Acad Sci USA 115:E10979–E10987. https://doi.org/10.1073/pnas.1814856115
Ramos HC, Rumbo M, Sirard J-C (2004) Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol 12:509–517. https://doi.org/10.1016/j.tim.2004.09.002
Rao S, Zhou Z, Miao P, Bi G, Hu M, Wu Y, Feng F, Zhang X, Zhou J-M (2018) Roles of receptor-like cytoplasmic kinase VII members in pattern-triggered immune signaling. Plant Physiol 177:1679–1690. https://doi.org/10.1104/pp.18.00486
Robatzek S, Bittel P, Chinchilla D, Köchner P, Felix G, Shiu S-H, Boller T (2007) Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol Biol 64:539–547. https://doi.org/10.1007/s11103-007-9173-8
Roden JA, Belt B, Ross JB, Tachibana T, Vargas J, Mudgett MB (2004) A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection. Proc Natl Acad Sci USA 101:16624–16629. https://doi.org/10.1073/pnas.0407383101
Sang Y, Yu W, Zhuang H, Wei Y, Derevnina L, Yu G, Luo J, Macho AP (2020) Intra-strain elicitation and suppression of plant immunity by Ralstonia solanacearum type-III effectors in Nicotiana benthamiana. Plant Commun 1:100025. https://doi.org/10.1016/j.xplc.2020.100025
Schoonbeek H-j, Wang H-H, Stefanato FL, Craze M, Bowden S, Wallington E, Zipfel C, Ridout CJ (2015) Arabidopsis EF-Tu receptor enhances bacterial disease resistance in transgenic wheat. New Phytol 206:606–613. https://doi.org/10.1111/nph.13356
Schultink A, Qi T, Lee A, Steinbrenner AD, Staskawicz B (2017) Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J 92:787–795. https://doi.org/10.1111/tpj.13715
Schultink A, Qi T, Bally J, Staskawicz B (2019) Using forward genetics in Nicotiana benthamiana to uncover the immune signaling pathway mediating recognition of the Xanthomonas perforans effector XopJ4. New Phytol 221:1001–1009. https://doi.org/10.1111/nph.15411
Schwartz AR, Potnis N, Timilsina S, Wilson M, Patané J, Martins JJ, Minsavage GV, Dahlbeck D, Akhunova A, Almeida N, Vallad GE, Barak JD, White FF, Miller SA, Ritchie D, Goss E, Bart RS, Setubal JC, Jones JB, Staskawicz BJ (2015) Phylogenomics of Xanthomonas field strains infecting pepper and tomato reveals diversity in effector repertoires and identifies determinants of host specificity. Front Microbiol 6:535. https://doi.org/10.3389/fmicb.2015.00535
Schwessinger B, Bahar O, Thomas N, Holton N, Nekrasov V, Ruan D, Canlas PE, Daudi A, Petzold CJ, Singan VR, Kuo R, Chovatia M, Daum C, Heazlewood JL, Zipfel C, Ronald PC (2015) Transgenic expression of the dicotyledonous pattern recognition receptor EFR in rice leads to ligand-dependent activation of defense responses. PLoS Pathog 11:e1004872. https://doi.org/10.1371/journal.ppat.1004809
Seto D, Koulena N, Lo T, Menna A, Guttman DS, Desveaux D (2017) Expanded type III effector recognition by the ZAR1 NLR protein using ZED1-related kinases. Nat Plants 3:17027. https://doi.org/10.1038/nplants.2017.27
Shao F, Golstein C, Ade J, Stoutemeyer M, Dixon JE, Innes RW (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301:1230–1233. https://doi.org/10.1126/science.1085671
Shindo T, van der Hoorn RAL (2008) Papain-like cysteine proteases: key players at molecular battlefields employed by both plants and their invaders. Mol Plant Pathol 9:119–125. https://doi.org/10.1111/j.1364-3703.2007.00439.x
Shirasu K (2009) The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol 60:139–164. https://doi.org/10.1146/annurev.arplant.59.032607.092906
Song W-Y, Wang G-L, Chen L-L, Kim H-S, Pi L-Y, Holsten T, Gardner J, Wang B, Zhai W-X, Zhu L-H, Fauquet C, Ronald P (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270:1804–1806. https://doi.org/10.1126/science.270.5243.1804
Sun W, Dunning FM, Pfund C, Weingarten R, Bent AF (2006) Within-species flagellin polymorphism in Xanthomonas campestris pv campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2–dependent defenses. Plant Cell 18:764–779. https://doi.org/10.1105/tpc.105.037648
Swiderski MR, Innes RW (2001) The Arabidopsis PBS1 resistance gene encodes a member of a novel protein kinase subfamily. Plant J 26:101–112. https://doi.org/10.1046/j.1365-313x.2001.01014.x
Tai TH, Dahlbeck D, Clark ET, Gajiwala P, Pasion R, Whalen MC, Stall RE, Staskawicz BJ (1999) Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc Natl Acad Sci USA 96:14153–14158. https://doi.org/10.1073/pnas.96.24.14153
Thomas NC, Hendrich CG, Gill US, Allen C, Hutton SF, Schultink A (2020) The immune receptor Roq1 confers resistance to the bacterial pathogens Xanthomonas, Pseudomonas syringae, and Ralstonia in tomato. Front Plant Sci 11:463. https://doi.org/10.3389/fpls.2020.00463
Tian S-N, Liu D-D, Zhong C-L, Xu H-Y, Yang S, Fang Y, Ran J, Liu J-Z (2020) Silencing GmFLS2 enhances the susceptibility of soybean to bacterial pathogen through attenuating the activation of GmMAPK signaling pathway. Plant Sci 292:110386. https://doi.org/10.1016/j.plantsci.2019.110386
Timilsina S, Potnis N, Newberry EA, Liyanapathiranage P, Iruegas-Bocardo F, White FF, Goss EM, Jones JB (2020) Xanthomonas diversity, virulence and plant-pathogen interactions. Nat Rev Microbiol 18:415–427. https://doi.org/10.1038/s41579-020-0361-8
Toruño TY, Stergiopoulos I, Coaker G (2016) Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu Rev Phytopathol 54:419–441. https://doi.org/10.1146/annurev-phyto-080615-100204
Toruño TY, Shen M, Coaker G, Mackey D (2019) Regulated disorder: posttranslational modifications control the RIN4 plant immune signaling hub. Mol Plant Microbe Interact 32:56–64. https://doi.org/10.1094/MPMI-07-18-0212-FI
Wan L, Essuman K, Anderson RG, Sasaki Y, Monteiro F, Chung E-H, Nishimura EO, DiAntonio A, Milbrandt J, Dangl JL, Nishimura MT (2019a) TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science 365:799–803. https://doi.org/10.1126/science.aax1771
Wan W-L, Fröhlich K, Pruitt RN, Nürnberger T, Zhang L (2019b) Plant cell surface immune receptor complex signaling. Curr Opin Plant Biol 50:18–28. https://doi.org/10.1016/j.pbi.2019.02.001
Wang G, Roux B, Feng F, Guy E, Li L, Li N, Zhang X, Lautier M, Jardinaud M-F, Chabannes M, Arlat M, Chen S, He C, Noël LD, Zhou J-M (2015) The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe 18:285–295. https://doi.org/10.1016/j.chom.2015.08.004
Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, Qi Y, Wang H-W, Zhou J-M, Chai J (2019a) Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364:eaav5870. https://doi.org/10.1126/science.aav5870
Wang J, Wang J, Hu M, Wu S, Qi J, Wang G, Han Z, Qi Y, Gao N, Wang H-W, Zhou J-M, Chai J (2019b) Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364:eaav5868. https://doi.org/10.1126/science.aav5868
Wei C-F, Kvitko BH, Shimizu R, Crabill E, Alfano JR, Lin N-C, Martin GB, Huang H-C, Collmer A (2007) A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J 51:32–46. https://doi.org/10.1111/j.1365-313X.2007.03126.x
Wei Y, Caceres-Moreno C, Jimenez-Gongora T, Wang K, Sang Y, Lozano-Duran R, Macho AP (2017) The Ralstonia solanacearum csp22 peptide, but not flagellin-derived peptides, is perceived by plants from the Solanaceae family. Plant Biotechnol J 16:1349–1362. https://doi.org/10.1111/pbi.12874
Wei Y, Balaceanu A, Rufian JS, Segonzac C, Zhao A, Morcillo RJL, Macho AP (2020) An immune receptor complex evolved in soybean to perceive a polymorphic bacterial flagellin. Nat Commun 11:3763. https://doi.org/10.1038/s41467-020-17573-y
White FF, Potnis N, Jones JB, Koebnik R (2009) The type III effectors of Xanthomonas. Mol Plant Pathol 10:749–766. https://doi.org/10.1111/j.1364-3703.2009.00590.x
Wroblewski T, Caldwell KS, Piskurewicz U, Cavanaugh KA, Xu H, Kozik A, Ochoa O, McHale LK, Lahre K, Jelenska J, Castillo JA, Blumenthal D, Vinatzer BA, Greenberg JT, Michelmore RW (2009) Comparative large-scale analysis of interactions between several crop species and the effector repertoires from multiple pathovars of Pseudomonas and Ralstonia. Plant Physiol 150:1733–1749. https://doi.org/10.1104/pp.109.140251
Wu C-H, Derevnina L, Kamoun S (2018) Receptor networks underpin plant immunity. Science 360:1300–1301. https://doi.org/10.1126/science.aat2623
Xiang T, Zong N, Zou Y, Wu Y, Zhang J, Xing W, Li Y, Tang X, Zhu L, Chai J, Zhou J-M (2008) Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol 18:74–80. https://doi.org/10.1016/j.cub.2007.12.020
Xin X-F, Kvitko B, He SY (2018) Pseudomonas syringae: what it takes to be a pathogen. Nat Rev Microbiol 16:316–328. https://doi.org/10.1038/nrmicro.2018.17
Yu S, Hwang I, Rhee S (2014) The crystal structure of type III effector protein XopQ from Xanthomonas oryzae complexed with adenosine diphosphate ribose. Proteins 82:2910–2914. https://doi.org/10.1002/prot.24656
Zhang J, Coaker G, Zhou J-M, Dong X (2020) Plant immune mechanisms: from reductionistic to holistic points of view. Mol Plant 13:1358–1378. https://doi.org/10.1016/j.molp.2020.09.007
Zhao Y, He S-Y, Sundin GW (2006) The Erwinia amylovora avrRpt2EA gene contributes to virulence on pear and AvrRpt2EA is recognized by Arabidopsis RPS2 when expressed in Pseudomonas syringae. Mol Plant Microbe Interact 19:644–654. https://doi.org/10.1094/MPMI-19-0644
Zhou J-M, Zhang Y (2020) Plant immunity: danger perception and signaling. Cell 181:978–989. https://doi.org/10.1016/j.cell.2020.04.028
Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T, Felix G (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125:749–460. https://doi.org/10.1016/j.cell.2006.03.037
Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Sciences and ICT (Projects No. 2018R1A5A1023599 and No. 2020R1A2C1101419).
Author information
Authors and Affiliations
Contributions
CS conceived the manuscript; BK, JC and CS compiled the literature and wrote the manuscript; BK and JC prepared the illustrations.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Additional information
Communicated by Seon-In Yeom.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, B., Choi, J. & Segonzac, C. Tackling multiple bacterial diseases of Solanaceae with a handful of immune receptors. Hortic. Environ. Biotechnol. 63, 149–160 (2022). https://doi.org/10.1007/s13580-021-00415-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13580-021-00415-1