Abstract
Plants have evolved a multilayered and sophisticated immune system to establish effective resistance to a variety of pathogens. Phytophthora species are among the most notorious plant pathogens, causing destructive diseases on a variety of agricultural crops. Understanding the plant immune system is crucial for protecting crops from Phytophthora diseases. Here, we summarize the recent work on genes involved in plant resistance against Phytophthora pathogens, including cell surface pattern recognition receptors, cytoplasmic nucleotide-binding leucine-rich repeat receptors, regulator genes, and non-host resistance genes, small RNA, and long non-coding RNA are also discussed in this review. Although the molecular mechanisms of only a small proportion of them have been clarified, emergence of new mechanisms of plant defense will offer exciting opportunities for utilization of these genes in disease resistance breeding as well as generation of disease-resistant crop germplasms.
Similar content being viewed by others
Background
Phytophthora species belong to oomycetes, which have filamentous growth habits and nutritional strategies like fungi, but are evolutionarily distant from fungi and classified in the kingdom stramenopiles. The majority of Phytophthora species are notorious plant pathogens and infect more than 130 known plant species, causing destructive diseases on a variety of agricultural crops including potato, soybean, tomato, pepper, and forests (Tyler et al. 2006). For example, Phytophthora infestans is the causal agent of potato late blight, which was responsible for the Great Irish Famine in the nineteenth century (Kroon et al. 2012). Late blight is also the most economically important disease of potato, resulting in an economic loss more than € 9 billion per year (Haverkort et al. 2016). P. sojae has a narrow host range and causes root and stem rot primarily on soybean, while P. capsici has a broad host range and causes many devastating diseases on a number of vegetables including pepper, tomato, eggplant, and all cucurbits (Tyler 2007; Lamour et al. 2012; Yang et al. 2022). P. capsici is the most important pathogen of solanaceous and cucurbitaceous crops, and it causes total crop loss worldwide (Sanogo et al. 2022). Furthermore, P. ramorum infects multiple species of hardwood trees and ornamentals, leading to a serious threat to the forestry industry (Kamoun et al. 2015). Given the lack of resistant cultivars, Phytophthora diseases are mainly controlled by fungicides, but pathogen isolates that are resistant to commonly used fungicides have been continually reported (Gonzalez-Tobon et al. 2022). On the other hand, breeding resistant varieties is the most economical and effective way to control diseases, but the development of resistant varieties is relatively slow and short-lived due to the rapid and constant evolution of Phytophthora pathogens. Thus, identification and utilization of genes related to plant resistance to facilitate disease resistance breeding is a priority for controlling Phytophthora pathogens.
In the face of the threat from pathogens, plant orchestrate multiple defense strategies ranging from physical barriers to specialized metabolites. Simply, physical barriers of plants include cuticle at the surface, lignin and suberin of cell wall, and papillae or callose at sites of pathogen penetration (Hematy et al. 2009). Plants also produce a wide array of antimicrobial compounds including phytoalexins which are synthesized only upon pathogen attack and glucosinolates that are constitutively accumulated (Burow and Halkier 2017; Li et al. 2023a). Once pathogens penetrate the cell wall, plants could deploy a two-tiered immune system to resist pathogenic microorganisms (Jones and Dangl 2006). The first layer is called pattern-triggered immunity (PTI), which is initiated upon recognition of conserved microbe-/pathogen-associated molecular patterns (M/PAMPs) or damage-associated molecular patterns (DAMPs) by a diverse assortment of cell surface pattern recognition receptors (PRRs). The second layer is effector-triggered immunity (ETI), which is activated through sensing microbial effectors by cytoplasmic nucleotide-binding leucine-rich repeat (NLR) receptors (Bonardi et al. 2012). In such cases, recognized effectors are often referred to as avirulence (Avr) effectors. Activation of PTI or ETI ultimately launch multiple but similar downstream immune responses such as production of reactive oxygen species (ROS), reinforcement of cell wall, biosynthesis of antimicrobial compounds, and production of small silencing RNAs, albeit with distinct amplitudes and dynamics. PTI leads to a basal and broad-spectrum resistance, while ETI is qualitatively much stronger and faster, often results in a localized programmed cell death (PCD) at the infection sites termed hypersensitive response (HR) (Cui et al. 2015; Dong et al. 2022). Recent studies demonstrated that PRR-mediated signaling is important for ETI-associated defense responses, and NLR-mediated signaling leads to up-regulated transcription and/or translation of many PTI components (Yuan et al. 2021). Furthermore, a complex of ETI components, EDS1, PAD4, and ADR1, are also required for PTI triggered by the Arabidopsis cell surface receptor RLP23 (Pruitt et al. 2021). Thus, PTI and ETI can potentiate each other to achieve stronger plant defenses.
In the warfare between hosts and pathogenic microorganisms, Phytophthora pathogens have developed effective strategies to enable the establishment and development of infection. For example, they can evade the plant immune recognition by removal, modification, and mutation of immunogenic microbial molecules including PAMPs and effectors (Wang et al. 2022). Another common strategy is to secrete effector proteins into apoplastic space or host cells for targeting and manipulating different phases of plant immune activation comprising immune signaling transduction and defense execution, facilitating the infection by microbial pathogens (Boevink et al. 2020). Considering Phytophthora pathogens have high evolutionary rates and encode hundreds of effectors, the identification of effective genes related to plant resistance will contribute to disease resistance breeding.
In this review, we mainly focus on discussing the cell surface PRRs and cytoplasmic NLRs which specifically recognize Phytophthora species to activate PTI and ETI (Additional file 1: Table S1). We also summarize an array of genes that participate in plant resistance and defense responses to Phytophthora pathogens, while genes which involve in communal signaling transduction or broad-spectrum immunity through physical and chemical barriers are not included (Additional file 2: Table S2). Furthermore, host small RNA (sRNA) and long non-coding RNA (lncRNA) are also discussed in this review (Additional file 2: Table S2). Based on the known molecular mechanisms underlying interactions between Phytophthora pathogens and their hosts, we highlight recent findings on how plants mobilize the above genes to withstand pathogen infection. Finally, we discuss the utilization of these genes in disease resistance breeding.
Cell surface PRRs
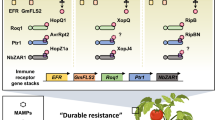
Plants utilize a large repertoire of PRRs at the cell surface to perceive microbial invasion and activate immune responses. For Phytophthora pathogens, certain PAMPs or apoplastic effectors can be recognized by plant PRRs, which are characteristically receptor-like proteins (RLPs) and receptor-like kinases (RLKs) (Fig. 1) (Li et al. 2016). In recent decades, a handful of PAMPs from Phytophthora species have been identified. For example, elicitins are conserved secreted proteins from Phytophthora and Pythium species, and they can elicit HR-like cell death and induce defense responses in several plant species (Kamoun et al. 1993). Correspondingly, a cell surface RLP ELR (elicitin response) isolated from the wild potato Solanum microdontum can mediate specific response to a broad range of elicitins and enhance resistance to P. infestans in potato. The central immune co-receptor BRI1-Associated Kinase 1/Somatic Embryogenesis Receptor Kinase 3 (BAK1/SERK3), associates with ELR to recognize INF1, a well-known elicitin from P. infestans (Du et al. 2015). In addition, Suppressor Of BIR1-1 (SOBIR1) also interacts with ELR and is required for INF1-triggered cell death and basal resistance against P. infestans (Domazakis et al. 2018). More recently, NbEIR1 (Nicotiana benthamiana ELICITIN INSENSITIVE RLK 1) was found to associate with NbBAK1 and NbBSK1 (BRASSINOSTEROID-SIGNALING KINASE 1) to positively regulate the recognition of INF1, INF1-induced defense responses, and resistance to P. capsici (Zhang et al. 2023). In another study, Responsive to ELicitins (REL) was identified in N. benthamiana as an RLP to recognize elicitins through forming complexes with BAK1 and SOBIR1, and REL is required for elicitin-triggered cell death, immune responses, and Phytophthora resistance (Chen et al. 2023). In another case, the plasma membrane lectin RLK, RESISTANT TO DFPM-INHIBITION OF ABSCISIC ACID SIGNALING 2 (RDA2), recognizes the 9-methyl-branched sphingoid bases derived from ceramide of P. infestans and confers Arabidopsis resistance against oomycete pathogens (Kato et al. 2022).
PRRs and NLRs involved in Phytophthora resistance. Cell surface pattern recognition receptors (PRRs) (ELR, EIR1, RE02, RXEG1, REL, ERK1, RLP23, and RDA2) interact with SOBIR1 and BAK1 to form PRR immune complexes upon recognition of pathogen-associated molecular pattern (PAMPs) or apoplastic effectors from Phytophthora pathogens. Activation of PRRs triggers phosphorylation of receptor-like cytoplasmic kinases (RLCKs), RBOHD, PIPs, and Ca2+ channels, leads to production of reactive oxygen species (ROS), influx of calcium, the MAP kinase cascade activation and downstream defense-related gene expression. Other RLKs (LecRK-IX.2, LecRK-I.9, LecRK-IX.1, LecRK-IV.4, LecRK-V.5, and GmLMM1) play positive roles in resistance to Phytophthora pathogens as well. Pathogens also deliver race-specific effector proteins to suppress host immunity in various modes to facilitate infection. In this context, cytoplasmic nucleotide-binding leucine-rich repeat (NLR) receptors (NLRs), including CC-NBD-LRR (CNL) proteins and TIR-NBD-LRR (TNL) proteins, sense a portion of these effectors and activate another layer of plant immunity known as effector-triggered immunity (ETI). In such cases, recognized effectors are referred to as avirulence (Avr) proteins. ETI also leads to a number of overlapping downstream outputs consistent with pattern-triggered immunity (PTI), and usually with stronger amplitudes compared to PTI
XEG1 is an apoplastic effector belonging to a glycoside hydrolase family 12 (GH12) protein produced by Phytophthora spp. and can trigger defense responses, thus it acts as a PAMP in soybean (Glycine max) and solanaceous species (Ma et al. 2015). A leucine-rich repeat (LRR) RLP, Response to XEG1 (RXEG1), specifically recognizes XEG1 and forms a complex with BAK1 and SOBIR1 to regulate XEG1-induced plant immune responses (Wang et al. 2018b). Similarly, RLP23 forms a constitutive complex with SOBIR1 and recruits BAK1 into a tripartite complex upon binding to a conserved 20-amino-acid fragment (nlp20) from necrosis and ethylene-inducing peptide 1-like proteins (NLPs), another class of apoplastic effectors, leading to recognition of nlp20 and enhanced immunity to P. infestans (Albert et al. 2015). SOBIR1 and BAK1 are also involved in the perception of an apoplastic effector VmE02, a novel PAMP that is widely present in oomycetes and fungi, by a receptor-like protein Response to VmE02 (RE02), resulting in VmE02-triggred cell death and enhanced plant resistance to P. capsici (Nie et al. 2021). In another example, N. benthamiana G-type lectin RLK expansin-regulating kinase 1 (ERK1) together with BAK1 and SOBIR1 mediate the perception of a novel apoplastic effector PcEXLX1, an expansin-like protein, to activate multiple immune responses and increase plant resistance to P. capsici (Pi et al. 2022). Furthermore, Arabidopsis lectin receptor kinase LecRK-I.9 localizes at the plasma membrane and plays a positive role in Phytophthora resistance (Bouwmeester et al. 2011). Other LecRKs in Arabidopsis including LecRK-IV.4, LecRK-V.5, LecRK-IX.1, LecRK-IX.2, etc. positively regulate resistance against P. capsici and P. brassicae (Wang et al. 2014, 2015). The soybean malectin-like receptor kinase (RK) GmLMM1 negatively regulates flg22-induced ROS production and XEG1-induced cell death, acting as an important component in PTI regulation and disease resistance to both bacterial and oomycete pathogens (Wang et al. 2020).
Cytoplasmic NLRs
Typical gene-for-gene resistance is activated upon detection of intracellular effectors by cytoplasmic NLR receptors encoded by resistance genes. For Phytophthora pathogens, NLR-mediated resistance is largely based on recognition of RxLR (Arg-any amino acid-Leu-Arg) effectors (Fig. 1) (Anderson et al. 2015). To date, dozens of resistance proteins or NLRs confer Phytophthora resistance have been identified (Fig. 1). Among them, 15 late blight resistance genes which can recognize P. infestans effector have been identified (Paluchowska et al. 2022). For example, the first late blight R gene to be cloned was R1 from S. demissum, which encodes a coiled-coil (CC) NLR that specifically recognizes P. infestans isolates carrying the avirulence gene AVR1 (Ballvora et al. 2002). Besides, the well-characterized Resistance to P. infestans (Rpi) gene R3a originated from S. demissum, encoding a CC NLR protein, recognizes the avirulence gene Avr3a to activate potato resistance to P. infestans (Armstrong et al. 2005; Huang et al. 2005). Another Rpi gene Rpi-blb3 in S. bulbocastanum as well as its orthologs Rpi-abpt, R2, and R2-like in other Solanum spp. can detect the RxLR effector PiAvr2, resulting in HR and enhanced defense responses (Lokossou et al. 2009). Similarly, Rpi-blb1 isolated from S. bulbocastanum as well as its functional homologs Rpi-sto1 and Rpi-pta1 encode a CC NLR receptor, and they can recognize Avrblb1 which is identical to IpiO and resist a broad-spectrum of P. infestans isolates (van der Vossen et al. 2003; Vleeshouwers et al. 2008). Rpi-vnt1.1 and Rpi-vnt1.3 in S. venturii interact with the RxLR effector protein Avrvnt1 to mediate late blight resistance to a broad-spectrum of pathogen isolates (Pel et al. 2009). In addition, a typical CC NLR receptor from S. demissum, R8, recognizes the RxLR effector Avr8 and provides broad-spectrum late blight resistance (Vossen et al. 2016). Recently, Rpi-amr1 was map-based cloned from a wild potato species, S. americanum, and was found to provoke HR and confer late blight resistance in N. benthamiana and cultivated potato by detection of Avramr1 (Lin et al. 2020; Witek et al. 2021). Rpi-amr3 from S. americanum, encoding a typical CC NLR protein, recognizes the broadly conserved avirulence effector Avramr3 in planta and activates a broad-spectrum resistance to multiple Phytophthora pathogens (Witek et al. 2016; Lin et al. 2022).
In addition to P. infestans, several Resistance to P. sojae (Rps) genes have also been mapped in different soybean accessions, but fewer of them have been cloned (Fig. 1). For example, Rps1k or Rps1b encodes a CC NLR protein and recognizes the avirulence gene Avr1k or Avr1b-1, providing stable and broad-spectrum resistance to the oomycete pathogen P. sojae (Shan et al. 2004; Gao and Bhattacharyya 2008). Similarly, two allelic or clustered resistance genes Rps4 and Rps6 are located on chromosome 18 and recognize a single avirulence gene, Avr4/6, to trigger HR cell death and defense responses (Sandhu et al. 2004; Dou et al. 2010). In other cases, Rps10 from the resistant soybean cultivar Wandou 15 was mapped on chromosome 17 of G. max (Zhang et al. 2013). RpsUN1 and RpsUN2 from PI 567139B, a soybean landrace carrying excellent resistance, were mapped on chromosomes 3 and 16, and RpsWY was finely mapped on chromosome 3 of the soybean cultivar Wayao (Lin et al. 2013; Cheng et al. 2017). Rps11 in a soybean landrace, PI 594527, encodes an exceptionally large NLR protein and contributes to broad-spectrum resistance against P. sojae (Wang et al. 2021). Rps8 was reported to interact with the RxLR effector Avr3a, while a specific allele of Avr3a was recognized by Rps3a (Arsenault-Labrecque et al. 2022). More recently, RpsYD29 was map-based cloned from a resistant cultivar “Yudou 29”, and it encodes a C2H2-type zinc finger protein transcription factor which can bind to and activate two SOD1 promoters, leading to enhanced resistance to P. sojae (Li et al. 2023b). Apart from Rpi and Rps genes, few R genes against other species of the genus Phytophthora have been cloned. For oomycete pathogens, the Arabidopsis thaliana Toll/Interleukin-1 receptor (TIR) NLR protein RPP1 recognizes the cognate effector ATR1 from Hyaloperonospora arabidopsidis, leading to HR and enhanced resistance to the downy mildew pathogen (Chou et al. 2011). Similarly, multiple recognition of the CX2CX5G (CCG) effectors from Albugo candida by two Arabidopsis TIR NLR proteins WRR4A and WRR4B triggers HR and confers broad-spectrum resistance to corresponding pathogen races (Redkar et al. 2022).
Regulator genes
Detection of microbial infection by immune receptors activates resistance signaling transduction and ultimately launches effective immune responses against pathogen infection (Wang et al. 2022). To date, a number of genes have been reported to participate in integrated and multilayered plant immune system to antagonize Phytophthora pathogens (Fig. 2). Except for the above-mentioned PRRs and NLRs, two types of genes have also been reported to regulate the recognition process. For example, receptor-like cytoplasmic kinase subfamily VII (RLCK-VII) proteins interact with PRRs to form the PRR complexes and activate a complex array of downstream immune events and defense responses (Liang and Zhou 2018). Among several high-order Arabidopsis rlck-vii mutants, rlck-vii-6 and rlck-vii-8 exhibited greatly enhanced susceptibility to the compatible pathogen P. capsici, and accumulated much higher biomass of the incompatible pathogen P. infestans. Furthermore, Phytophthora culture filtrate (CF)-induced marker gene expression and immune responses were significantly reduced in the rlck-vii-6 mutant, and the RLCK-VII-6 members are specifically required for resistance to Phytophthora pathogens (Liang et al. 2021). Another type of genes encode the “helper” NLRs that translate the defense signal into disease resistance following the recognition of specialized effectors by “sensor” NLRs within the immune network. For instance, the helper NLR NRC4 (NLR required for cell death 4) in N. benthamiana is required for two sensor NLRs Rpi-blb2- and R1-mediated immunity, and it confers disease resistance to P. infestans (Wu et al. 2017). More recently, two helper NLRs NRC2 and NRC4 have been demonstrated to oligomerize into high-molecular-weight resistosomes after the detection of AVRamr3 by Rpi-amr3, and NRC2 also oligomerizes upon AVRamr1-dependent activation by Rpi-amr1 (Ahn et al. 2023). In the signaling transduction and defense execution processes, a series of genes related to Phytophthora pathogens resistance were reported, although a large portion of them confer broad-spectrum immunity to different kinds of pathogens. For example, a collection of Arabidopsis mutants showed that MAPK cascade-involved MPK4, salicylic acid (SA) signaling- and ETI-related EDS1, PAD4, NDR1, jasmonic acid (JA)-related EDS8, ethylene (ET) signaling-involved ETR1, camalexin and indole glucosinolates (iGS) biosynthesis-related PAD3, PAD2, MYB51, CYP81F2, and PEN2 were positive regulators in disease resistance to P. capsici (Wang et al. 2013; Li et al. 2020a).
Regulator genes, NHR genes, sRNAs, and lncRNAs involved in Phytophthora resistance. Plants utilize an assortment of components to regulate Phytophthora resistance. For example, members of receptor-like cytoplasmic kinase subfamily VII (RLCK-VII) act downstream of the PRRs and are targeted by RxLR25. In the signaling transduction process, MAPK cascade-involved MPK4, StMPK7, StMKK1, MAPK3/6, and MKK2, salicylic acid (SA) signaling- and ETI-related EDS1, PAD4, and NDR1, are important regulators in plant immunity. In the defense execution stage, camalexin and indole glucosinolates (iGS) biosynthesis-related PAD3, MYB51, and CYP81F2, pathogenesis-related (PR) proteins P14a, P14b, and P14c, positively regulate immune responses to Phytophthora pathogens. On the contrary, the plasma membrane-associated protein REM1.3 acts as a susceptibility factor that promotes pathogen infection. Besides, different types of genes are also involved in Phytophthora resistance. The transcription factor such as SpWRKY3 promotes PR gene expression, and WRKY1 activates expression of lncRNA33732. GmDIR22, GmACSs, and AtRTP5 involve in the regulation of lignan, ET, and SA/JA biosynthesis, respectively. Endoplasmic reticulum (ER)-localized proteins (GmDAD1, NAC089, RTP1, bZIP28, and bZIP60), Phytophthora effector targets (BPA1, BPLs, StNRL1, PIPs, GmPUB13, and NbRZ-1A), non-host resistance (NHR) genes (PSS1, PEN1, EDR1, EAS, EAH, AtFOLT1, and AtPUB33), sRNAs (miR393, miR159, and miR398b), lncRNAs (lncRNA33732 and nalncFL7), helper NLRs (NRC2 and NRC4), and other regulator genes (Stphot1, StSWAP70, StPM1, StRbohC, FL7, Raf36, StUBA2a/b, RBOHD, EDS8, ETR1, PAD2, and PEN2) are also required for the signaling transduction and defense execution processes
S. tuberosum StMPK7 interacts with StMKK1 (MAPK kinase 1) and is phosphorylated by StMKK1, while StMPK7 phosphorylates and stabilizes an RNA binding protein StUBA2a/b, a positive regulator in plant immunity, to enhance immunity to P. infestans via an SA-dependent signaling pathway (Zhang et al. 2021; Li et al. 2022b). In addition, three members of pathogenesis-related (PR) proteins isolated from tomato, P14a, P14b, and P14c, exhibited antifungal activity against P. infestans both in vitro and in vivo (Niderman et al. 1995). Furthermore, the soybean dirigent gene GmDIR22 from the highly resistant soybean cultivar ‘Suinong 10’ involves in the regulation of lignan biosynthesis, and overexpression of GmDIR22 in a susceptible cultivar enhances its resistance to P. sojae (Li et al. 2017). Expression of S. pimpinellifolium SpWRKY3 was significantly induced upon P. infestans inoculation, while SpWRKY3 reduced ROS accumulation and promoted PR gene expression, leading to enhanced resistance to P. infestans (Cui et al. 2018). GmDAD1 from soybean encodes an endoplasmic reticulum (ER)-localized protein, and expression of GmDAD1 in soybean hairy roots and N. benthamiana enhance their resistance to P. sojae and P. parasitica, respectively (Yan et al. 2019). In another example, the ER stress regulator NAC089 in Arabidopsis translocates from the ER to the nucleus in response to culture filtrate (CF) of P. capsici and positively regulate immune activation and PCD, contributing to host resistance against the oomycete pathogen P. capsici (Ai et al. 2021b).
Alternatively, genes that negatively regulate plant resistance against Phytophthora pathogens, termed susceptibility (S) factors or S genes, have also been reported in recent years (Fig. 2). For instance, the plasma membrane-associated protein REM1.3 in S. tuberosum accumulates around noncallosic haustoria for the development of extrahaustorial membrane (EHM) during host colonization by P. infestans, thus it acts as a susceptibility factor that promotes infection (Bozkurt et al. 2014). T-DNA insertion mutation of the Resistance to Phytophthora parasitica 1 (RTP1) gene encoding a susceptibility factor in Arabidopsis results in restricted cell death, increased ROS accumulation, and accelerated PR1 expression during P. parasitica infection, and RTP1 negatively modulates activation of unfolded protein response (UPR) and ER stress through interaction with bZIP60 and bZIP28, leading to susceptibility to P. parasitica (Pan et al. 2016; Qiang et al. 2021). Similarly, the Arabidopsis thaliana Resistant to Phytophthora 5 (AtRTP5) gene encodes a WD40 repeat domain-containing protein, T-DNA insertion mutation of AtRTP5 activates SA biosynthesis and SA-dependent responses (Li et al. 2020b). Raf-like kinase Raf36 interacts with MAPK kinase 2 (MKK2), a positive immune regulator, and mediates host susceptibility to P. parasitica in Arabidopsis and N. benthamiana upstream of MKK2 (Li et al. 2022a). Interestingly, S. tuberosum Stphot1 (blue light phototropin 1) interacts with potato NPH3/RPT2-like 1 (StNRL1), a susceptibility factor in plant immunity, to suppress INF1-triggered cell death and promote degradation of a guanine nucleotide exchange factor StSWAP70, a positive immune regulator, leading to enhanced infection by P. infestans (He et al. 2018; Naqvi et al. 2022). More recently, S. tuberosum PLASMA MEMBRANE PROTEIN 1 (StPM1) was reported to associate with the NADPH oxidase StRbohC to promote its degradation, and knockout of StPM1 leads to elevated expression of defense-related genes and reduced disease symptoms, suggesting that it acts as a novel susceptibility factor in potato immunity and resistance to Phytophthora pathogens (Bi et al. 2023).
Owing to the markedly accelerated functional genomic research on Phytophthora effectors, increasing numbers of effectors-targeted novel regulatory components of plant immune system have been identified (Fig. 2). For example, Type2 soybean 1-aminocyclopropane-1-carboxylate synthase (GmACS) isoforms are host targets of the P. sojae RxLR effector PsAvh238, GmACSs significantly promote ET biosynthesis and contribute to soybean resistance against P. sojae infection (Yang et al. 2019). A. thaliana BPA1 (binding partner of ACD11) and four BPA1-Like proteins (BPLs) are targeted by the P. capsici effector RxLR207, BPA1 and BPLs function redundantly and negatively regulate ROS production, ROS-mediated defense response, and pathogen resistance (Li et al. 2019). In another case, the PIP2-family aquaporin proteins including N. benthamiana NbPIP2;2 and G. max GmPIP2-13 are phosphorylated and degraded by the P. sojae crinkling- and necrosis-inducing (CRN) effector CRN78, and NbPIP2;2 is conserved in higher plants and positively regulates H2O2 production, transportation, and plant immunity (Ai et al. 2021a). The soybean E3 ubiquitin ligase GmPUB13 is a host target for the P. sojae RxLR effector Avr1d, which inhibits the enzyme activity of GmPUB13 and stabilizes GmPUB13 to facilitate pathogen infection, suggesting that GmPUB13 acts as a susceptibility factor (Lin et al. 2021). More recently, NbRZ-1A is targeted by the P. sojae effector PsFYVE1 to regulate plant immunity-related genes (NbNSL1, NbHCT, NbEIN2, and NbSUS4) at both pre-mRNA alternative splicing and transcription levels to promote infection, indicating that NbRZ-1A positively regulates plant resistance (Lu et al. 2023).
Non-host resistance genes
Non-host resistance (NHR) protects all members of a particular plant species from all isolates of a given pathogen species that cause diseases in other plant species. As the most common form of plant immunity, NHR provides the most durable, robust, and broad-spectrum resistance to almost all non-adapted or non-host plant pathogens (Uma et al. 2011). In recent decades, several genes have been identified to involve in NHR against Phytophthora species (Fig. 2). For example, Phytophthora sojae susceptible 1 (PSS1) encoding a glycine-rich protein in Arabidopsis was reported to confer a novel NHR against the hemibiotrophic oomycete pathogen P. sojae at both pre- and post-haustorial levels, while penetration deficient 1 (PEN1) provided NHR only at the pre-haustorial level against this soybean pathogen (Sumit et al. 2012; Wang et al. 2018a). ENHANCED DISEASE RESISTANCE 1 (EDR1) in A. thaliana encodes a putative MAPK kinase kinase, and loss of EDR1 leads to increased SA signaling and callose deposition upon P. infestans inoculation, suggesting that EDR1 acts as a negative regulator in post-invasive NHR (Geissler et al. 2015). Moreover, a subset of pepper 5-epi-aristolochene synthase (EAS) and 5-epi-aristolochene-1,3-dihydroxylase (EAH) gene family members contribute to phytoalexin capsidiol accumulation and NHR of pepper (Capsicum spp.) against potato late blight pathogen P. infestans (Lee et al. 2017). A. thaliana PSS30-encoded AtFOLT1 is responsible for transport of folate from the cytosol to plastids, and T-DNA insertion mutation of AtFOLT1 leads to reduced folate levels and loss of non-host immunity against P. sojae (Kambakam et al. 2021). More recently, the A. thaliana E3 ubiquitin ligase AtPUB33 was proposed to complement loss of activity of the predicted ortholog potato U-box-kinase protein (StUBK), which was targeted by the P. infestans RxLR effector Pi06087, and AtPUB33 contributed to NHR against P. infestans (He et al. 2019; McLellan et al. 2022).
sRNAs and lncRNAs
Plant sRNAs are important small non-coding RNAs and have been found to regulate gene expression in diverse biological processes including host-pathogen interactions, and the most famous category is microRNA (miRNA) (Hou and Ma 2020). In the last decade, sRNAs have been reported to regulate host resistance to Phytophthora pathogens (Fig. 2). For instance, expression of miR393 was induced by cell-wall component(s) or PAMPs of P. sojae, and knockdown of miR393 in soybean led to enhanced P. sojae infection and reduced expression of isoflavonoid biosynthetic genes, suggesting that miR393 was required for soybean defense against P. sojae (Wong et al. 2014). Repression of GAMYB expression by miR159 is highly conserved, and loss-of-function of miR159 in N. tabacum strongly activates expression of defense genes including a suite of PR genes and results in enhanced resistance to P. parasitica infection (Zheng et al. 2020). Moreover, A. thaliana miR398b targets and suppresses expression of Cu/Zn-Superoxidase Dismutase 1 (CSD1) and CSD2, mediating plant susceptibility to the oomycete pathogen P. parasitica, while A. thaliana core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase (AtC2GnT) transcripts inhibits the miR398b-CSDs module to elevate plant resistance against Phytophthora pathogens (Gou et al. 2022). In addition to sRNAs, lncRNAs whose lengths are more than 200 nucleotides have also been proven to play an important role in response to Phytophthora species (Fig. 2). Tomato transcription factor WRKY1 activates lncRNA33732, and lncRNA33732 induces the expression of respiratory burst oxidase (RBOH) and accumulation of H2O2 to enhance tomato resistance to P. infestans, comprising the WRKY1-lncRNA33732-RBOH module involved in Phytophthora resistance (Cui et al. 2019). Recent research has shown that A. thaliana RNA-binding protein BPL3 directly binds to and stabilizes the cis-natural antisense lncRNA of FORKED-LIKE7 (FL7) (nalncFL7) to suppress transcript accumulation of FL7, a positive immune regulator that can increase the phosphorylation levels of MPK3/6, demonstrating nalncFL7 negatively regulates plant immunity to P. capsici (Ai et al. 2023).
Utilization of resistance-related genes
Research advances on plant immune system and molecular plant–microbe interactions have provided novel opportunities to generate Phytophthora-resistant crop germplasms. For example, to confer broad-spectrum resistance, an anticipated strategy is to transform plants with known PRRs which recognize conserved M/PAMPs. In the case of ELR which mediates extracellular recognition of elicitin, transfer of ELR from the wild potato into cultivated potato led to enhanced resistance to P. infestans (Du et al. 2015). RLP23 bound to the conserved 20-amino-acid fragment nlp20, and stable expression of RLP23 in transgenic potato lines enhanced their resistance to the destructive oomycete pathogen P. infestans (Albert et al. 2015). In addition, stacking NLRs that are able to sense Avr effectors can provide durable disease resistance and minimize the evasion of existing resistance by mutation or variation of pathogens (Li et al. 2021). For example, several NLR genes e.g. Rpi-sto1, Rpi-vnt1.1, and Rpi-blb3 were jointly introduced into a susceptible potato cultivar, and the resulting triple gene transformants exhibited broad-spectrum Phytophthora resistance (Zhu et al. 2012). Similarly, introduction of Rps11 into the susceptible soybean variety 93Y21 led to full resistance to several P. sojae isolates, and stable transformation of RpsYD29 into soybean Williams 82 substantially enhanced resistance to the P. sojae strain PsMC1 (Wang et al. 2021; Li et al. 2023b). Both stable transgenic plants carrying Rpi-amr3 in a potato line (Line 26, Solynta B.V.) and transgenic potato cv. Maris Piper plants carrying Rpi-amr1 showed full resistance against diverse P. infestans isolates (Witek et al. 2016, 2021). More importantly, transformation of Rpi-amr3 into two lines of the potato cv. Maris Piper also resulted in effective protection against potato late blight in field conditions (Lin et al. 2022).
In other cases, silencing or inactivation of an S gene can weaken the compatible interactions between pathogens and hosts, suggesting an effective strategy to generate broad-spectrum resistance. For example, virus-induced gene silencing (VIGS) of the S gene REM1.3 enhanced plant resistance to P. infestans in N. benthamiana, and expression of an antisense REM1.3 construct in transgenic potato showed reduced P. infestans infection (Bozkurt et al. 2014). Silencing of two S genes GmPUB13 and its homolog GmPUB13L in soybean by RNA interference (RNAi) resulted in decreased infection by P. sojae (Lin et al. 2021). Besides, transfer of NHR genes from non-host plants to host plants could be a suitable strategy for development of broad-spectrum disease resistance. For example, transformation of Arabidopsis PSS1 into the soybean cultivar Williams 82 resulted in enhanced resistance to Fusarium virguliforme, a fungal pathogen which causes sudden death syndrome in soybean (Wang et al. 2018a). Stable overexpression of AtPUB33 in N. benthamiana and potato led to a significant decrease in P. infestans colonization (McLellan et al. 2022). Moreover, inhibition or silencing of sRNAs and lncRNAs can also be an efficient strategy for generating pathogen-resistant crops. For example, miR159 loss-of-function plants were obtained through expressing an miR159 decoy MIM159, and MIM159 transgenic tobacco plants were highly resistant to P. parasitica (Zheng et al. 2020). Tomato miR482b and miR482c were simultaneously silenced through the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system, resulted in increased expression of target genes and reduced P. infestans infection (Hong et al. 2021).
Conclusions
Phytophthora pathogens are destructive threats to global agricultural production and ecosystem, causing devastating diseases on a variety of agricultural crops and forests. Identification and utilization of genes related to plant resistance is crucial for protecting crops from Phytophthora diseases. In this review, we summarize the current knowledge of roughly-defined different types of genes related to plant Phytophthora resistance, including PRRs, NLRs, regulator genes, and NHR genes, plus the non-coding RNAs. Overall, based on the increasing knowledge of mechanisms underlying plant immune system and molecular plant-microbe interactions, a variety of Phytophthora resistance-related genes have been identified and explored in detail. Utilization of these genes for disease resistance breeding to generate disease-resistant varieties are facilitated by rapidly developing biotechnologies including RNA silencing and genome editing. Novel discoveries in the research on plant immunity will offer exciting opportunities to engineer disease resistance in crops in the coming decades.
Availability of data and materials
Not applicable.
Abbreviations
- Avr:
-
Avirulence
- CC:
-
Coiled-coil
- CF:
-
Culture filtrate
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- CRN:
-
Crinkling- and necrosis-inducing
- DAMPs:
-
Damage-associated molecular patterns
- EHM:
-
Extrahaustorial membrane
- ER:
-
Endoplasmic reticulum
- ET:
-
Ethylene
- ETI:
-
Effector-triggered immunity
- HR:
-
Hypersensitive response
- iGS:
-
Indole glucosinolates
- JA:
-
Jasmonic acid
- lncRNA:
-
Long non-coding RNA
- LRR:
-
Leucine-rich repeat
- miRNA:
-
MicroRNA
- M/PAMPs:
-
Microbe-/pathogen-associated molecular patterns
- NHR:
-
Non-host resistance
- NLPs:
-
Ethylene-inducing peptide 1-like proteins
- NLRs:
-
Nucleotide-binding leucine-rich repeat receptors
- PCD:
-
Programmed cell death
- PR:
-
Pathogenesis-related
- PRRs:
-
Pattern recognition receptors
- PTI:
-
Pattern-triggered immunity
- RK:
-
Receptor kinase
- RLKs:
-
Receptor-like kinases
- RLPs:
-
Receptor-like proteins
- RLCK:
-
Receptor-like cytoplasmic kinase
- RNAi:
-
RNA interference
- ROS:
-
Reactive oxygen species
- RxLR:
-
Arg-any amino acid-Leu-Arg
- SA:
-
Salicylic acid
- sRNA:
-
Small RNA
- TIR:
-
Toll/Interleukin-1 receptor
- UPR:
-
Unfolded protein response
- VIGS:
-
Virus-induced gene silencing
References
Ahn HK, Lin X, Olave-Achury AC, Derevnina L, Contreras MP, Kourelis J, et al. Effector-dependent activation and oligomerization of plant NRC class helper NLRs by sensor NLR immune receptors Rpi-amr3 and Rpi-amr1. EMBO J. 2023;42: e111484.
Ai G, Xia Q, Song T, Li T, Zhu H, Peng H, et al. A Phytophthora sojae CRN effector mediates phosphorylation and degradation of plant aquaporin proteins to suppress host immune signaling. PLoS Pathog. 2021a;17: e1009388.
Ai G, Zhu H, Fu X, Liu J, Li T, Cheng Y, et al. Phytophthora infection signals-induced translocation of NAC089 is required for endoplasmic reticulum stress response-mediated plant immunity. Plant J Cell Mol Biol. 2021b;108:67–80.
Ai G, Li T, Zhu H, Dong X, Fu X, Xia C, et al. BPL3 binds the long non-coding RNA nalncFL7 to suppress FORKED-LIKE7 and modulate HAI1-mediated MPK3/6 dephosphorylation in plant immunity. Plant Cell. 2023;35:598–616.
Albert I, Bohm H, Albert M, Feiler CE, Imkampe J, Wallmeroth N, et al. An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat Plants. 2015;1:15140.
Anderson RG, Deb D, Fedkenheuer K, McDowell JM. Recent progress in RXLR effector research. Mol Plant-Microbe Interact. 2015;28:1063–72.
Armstrong MR, Whisson SC, Pritchard L, Bos JI, Venter E, Avrova AO, et al. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci USA. 2005;102:7766–71.
Arsenault-Labrecque G, Santhanam P, Asselin Y, Cinget B, Lebreton A, Labbe C, et al. RXLR effector gene Avr3a from Phytophthora sojae is recognized by Rps8 in soybean. Mol Plant Pathol. 2022;23:693–706.
Ballvora A, Ercolano MR, Weiss J, Meksem K, Bormann CA, Oberhagemann P, et al. The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J. 2002;30:361–71.
Bi W, Liu J, Li Y, He Z, Chen Y, Zhao T, et al. CRISPR/Cas9-guided editing of a novel susceptibility gene in potato improves Phytophthora resistance without growth penalty. Plant Biotechnol J. 2023;6:66.
Boevink PC, Birch PRJ, Turnbull D, Whisson SC. Devastating intimacy: the cell biology of plant–Phytophthora interactions. New Phytol. 2020;228:445–58.
Bonardi V, Cherkis K, Nishimura MT, Dangl JL. A new eye on NLR proteins: focused on clarity or diffused by complexity? Curr Opin Immunol. 2012;24:41–50.
Bouwmeester K, de Sain M, Weide R, Gouget A, Klamer S, Canut H, et al. The lectin receptor kinase LecRK-I.9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog. 2011;7:e1001327.
Bozkurt TO, Richardson A, Dagdas YF, Mongrand S, Kamoun S, Raffaele S. The plant membrane-associated REMORIN1.3 accumulates in discrete perihaustorial domains and enhances susceptibility to Phytophthora infestans. Plant Physiol. 2014;165:1005–18.
Burow M, Halkier BA. How does a plant orchestrate defense in time and space? Using glucosinolates in Arabidopsis as case study. Curr Opin Plant Biol. 2017;38:142–7.
Chen Z, Liu F, Zeng M, Wang L, Liu H, Sun Y, et al. Convergent evolution of immune receptors underpins distinct elicitin recognition in closely related Solanaceous plants. Plant Cell. 2023;35:1186–201.
Cheng Y, Ma Q, Ren H, Xia Q, Song E, Tan Z, et al. Fine mapping of a Phytophthora-resistance gene RpsWY in soybean (Glycine max L.) by high-throughput genome-wide sequencing. Theor Appl Genet. Theoretische und angewandte Genetik. 2017;130:1041–51.
Chou S, Krasileva KV, Holton JM, Steinbrenner AD, Alber T, Staskawicz BJ. Hyaloperonospora arabidopsidis ATR1 effector is a repeat protein with distributed recognition surfaces. Proc Natl Acad Sci. 2011;108:13323–8.
Cui H, Tsuda K, Parker JE. Effector-triggered immunity: from pathogen perception to robust defense. Annu Rev Plant Biol. 2015;66:487–511.
Cui J, Xu P, Meng J, Li J, Jiang N, Luan Y. Transcriptome signatures of tomato leaf induced by Phytophthora infestans and functional identification of transcription factor SpWRKY3. Theor Appl Genet. Theoretische und angewandte Genetik. 2018; 131:787–800.
Cui J, Jiang N, Meng J, Yang G, Liu W, Zhou X, et al. LncRNA33732-respiratory burst oxidase module associated with WRKY1 in tomato- Phytophthora infestans interactions. Plant J Cell Mol Biol. 2019;97:933–46.
Domazakis E, Wouters D, Visser RGF, Kamoun S, Joosten M, Vleeshouwers V. The ELR-SOBIR1 complex functions as a two-component receptor-like kinase to mount defense against Phytophthora infestans. Mol Plant-Microbe Interact. 2018;31:795–802.
Dong X, Ai G, Xia C, Pan W, Yin Z, Dou D. Different requirement of immunity pathway components by oomycete effectors-induced cell death. Phytopathol Res. 2022;4:4.
Dou D, Kale SD, Liu T, Tang Q, Wang X, Arredondo FD, et al. Different domains of Phytophthora sojae effector Avr4/6 are recognized by soybean resistance genes Rps4 and Rps6. Mol Plant-Microbe Interact. 2010;23:425–35.
Du J, Verzaux E, Chaparro-Garcia A, Bijsterbosch G, Keizer LC, Zhou J, et al. Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat Plants. 2015;1:15034.
Gao H, Bhattacharyya MK. The soybean-Phytophthora resistance locus Rps1-k encompasses coiled coil-nucleotide binding-leucine rich repeat-like genes and repetitive sequences. BMC Plant Biol. 2008;8:29.
Geissler K, Eschen-Lippold L, Naumann K, Schneeberger K, Weigel D, Scheel D, et al. Mutations in the EDR1 gene alter the response of Arabidopsis thaliana to Phytophthora infestans and the bacterial PAMPs flg22 and elf18. Mol Plant-Microbe Interact. 2015;28:122–33.
Gonzalez-Tobon J, Childers RR, Rodriguez A, Fry W, Myers KL, Thompson JR, et al. Searching for the mechanism that mediates mefenoxam-acquired resistance in Phytophthora infestans and how it is regulated. Phytopathology. 2022;112:1118–33.
Gou X, Zhong C, Zhang P, Mi L, Li Y, Lu W, et al. miR398b and AtC2GnT form a negative feedback loop to regulate Arabidopsis thaliana resistance against Phytophthora parasitica. Plant J Cell Mol Biol. 2022;111:360–73.
Haverkort AJ, Boonekamp PM, Hutten R, Jacobsen E, Lotz LAP, Kessel GJT, et al. Durable late blight resistance in potato through dynamic varieties obtained by cisgenesis: scientific and societal advances in the DuRPh project. Potato Res. 2016;59:35–66.
He Q, Naqvi S, McLellan H, Boevink PC, Champouret N, Hein I, et al. Plant pathogen effector utilizes host susceptibility factor NRL1 to degrade the immune regulator SWAP70. Proc Natl Acad Sci USA. 2018;115:E7834–43.
He Q, McLellan H, Hughes RK, Boevink PC, Armstrong M, Lu Y, et al. Phytophthora infestans effector SFI3 targets potato UBK to suppress early immune transcriptional responses. New Phytol. 2019;222:438–54.
Hematy K, Cherk C, Somerville S. Host-pathogen warfare at the plant cell wall. Curr Opin Plant Biol. 2009;12:406–13.
Hong Y, Meng J, He X, Zhang Y, Liu Y, Zhang C, et al. Editing miR482b and miR482c simultaneously by CRISPR/Cas9 enhanced tomato resistance to Phytophthora infestans. Phytopathology. 2021;111:1008–16.
Hou Y, Ma W. Natural host-induced gene silencing offers new opportunities to engineer disease resistance. Trends Microbiol. 2020;28:109–17.
Huang S, van der Vossen EA, Kuang H, Vleeshouwers VG, Zhang N, Borm TJ, et al. Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J Cell Mol Biol. 2005;42:251–61.
Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–9.
Kambakam S, Ngaki MN, Sahu BB, Kandel DR, Singh P, Sumit R, et al. Arabidopsis non-host resistance PSS30 gene enhances broad-spectrum disease resistance in the soybean cultivar Williams 82. Plant J Cell Mol Biol. 2021;107:1432–46.
Kamoun S, Klucher KM, Coffey MD, Tyler BM. A gene encoding a host-specific elicitor protein of Phytophthora parasitica. Mol Plant-Microbe Interact. 1993;6:573–81.
Kamoun S, Furzer O, Jones JD, Judelson HS, Ali GS, Dalio RJ, et al. The Top 10 oomycete pathogens in molecular plant pathology. Mol Plant Pathol. 2015;16:413–34.
Kato H, Nemoto K, Shimizu M, Abe A, Asai S, Ishihama N, et al. Recognition of pathogen-derived sphingolipids in Arabidopsis. Science. 2022;376:857–60.
Kroon LP, Brouwer H, de Cock AW, Govers F. The genus Phytophthora anno 2012. Phytopathology. 2012;102:348–64.
Lamour KH, Stam R, Jupe J, Huitema E. The oomycete broad-host-range pathogen Phytophthora capsici. Mol Plant Pathol. 2012;13:329–37.
Lee HA, Kim S, Kim S, Choi D. Expansion of sesquiterpene biosynthetic gene clusters in pepper confers nonhost resistance to the Irish potato famine pathogen. New Phytol. 2017;215:1132–43.
Li L, Yu Y, Zhou Z, Zhou JM. Plant pattern-recognition receptors controlling innate immunity. Sci China Life Sci. 2016;59:878–88.
Li N, Zhao M, Liu T, Dong L, Cheng Q, Wu J, et al. A novel soybean dirigent gene GmDIR22 contributes to promotion of lignan biosynthesis and enhances resistance to Phytophthora sojae. Front Plant Sci. 2017;8:1185.
Li Q, Ai G, Shen D, Zou F, Wang J, Bai T, et al. A Phytophthora capsici effector targets ACD11 binding partners that regulate ROS-mediated defense response in Arabidopsis. Mol Plant. 2019;12:565–81.
Li Q, Wang J, Bai T, Zhang M, Jia Y, Shen D, et al. A Phytophthora capsici effector suppresses plant immunity via interaction with EDS1. Mol Plant Pathol. 2020a;21:502–11.
Li W, Zhao D, Dong J, Kong X, Zhang Q, Li T, et al. AtRTP5 negatively regulates plant resistance to Phytophthora pathogens by modulating the biosynthesis of endogenous jasmonic acid and salicylic acid. Mol Plant Pathol. 2020b;21:95–108.
Li Q, Wang B, Yu J, Dou D. Pathogen-informed breeding for crop disease resistance. J Integr Plant Biol. 2021;63:305–11.
Li J, Deng F, Wang H, Qiang X, Meng Y, Shan W. The Raf-like kinase Raf36 negatively regulates plant resistance against the oomycete pathogen Phytophthora parasitica by targeting MKK2. Mol Plant Pathol. 2022a;23:530–42.
Li T, Zhang H, Xu L, Chen X, Feng J, Wu W, et al. StMPK7 phosphorylates and stabilizes a potato RNA-binding protein StUBA2a/b to enhance plant defence responses. Hortic Res. 2022b;9:uhac177.
Li J, Wang C, Yang L, Qiu F, Li Y, Zheng Y, et al. Enhancing tomato resistance by exploring early defense events against Fusarium wilt disease. Phytopathol Res. 2023a;5:24.
Li W, Zheng X, Cheng R, Zhong C, Zhao J, Liu TH, et al. Soybean ZINC FINGER PROTEIN03 targets two SUPEROXIDE DISMUTASE1s and confers resistance to Phytophthora sojae. Plant Physiol. 2023b;192:633–47.
Liang X, Zhou J-M. Receptor-like cytoplasmic kinases: central players in plant receptor kinase–mediated signaling. Annu Rev Plant Biol. 2018;69:267–99.
Liang X, Bao Y, Zhang M, Du D, Rao S, Li Y, et al. A Phytophthora capsici RXLR effector targets and inhibits the central immune kinases to suppress plant immunity. New Phytol. 2021;232:264–78.
Lin F, Zhao M, Ping J, Johnson A, Zhang B, Abney TS, et al. Molecular mapping of two genes conferring resistance to Phytophthora sojae in a soybean landrace PI 567139B. Theor Appl Genet. Theoretische und angewandte Genetik. 2013; 126:2177–85.
Lin X, Song T, Fairhead S, Witek K, Jouet A, Jupe F, et al. Identification of Avramr1 from Phytophthora infestans using long read and cDNA pathogen-enrichment sequencing (PenSeq). Mol Plant Pathol. 2020;21:1502–12.
Lin Y, Hu Q, Zhou J, Yin W, Yao D, Shao Y, et al. Phytophthora sojae effector Avr1d functions as an E2 competitor and inhibits ubiquitination activity of GmPUB13 to facilitate infection. Proc Natl Acad Sci USA. 2021;118:66.
Lin X, Olave-Achury A, Heal R, Pais M, Witek K, Ahn HK, et al. A potato late blight resistance gene protects against multiple Phytophthora species by recognizing a broadly conserved RXLR-WY effector. Mol Plant. 2022;15:1457–69.
Lokossou AA, Park TH, van Arkel G, Arens M, Ruyter-Spira C, Morales J, et al. Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Mol Plant-Microbe InteractI. 2009;22:630–41.
Lu X, Yang Z, Song W, Miao J, Zhao H, Ji P, et al. The Phytophthora sojae effector PsFYVE1 modulates immunity-related gene expression by targeting host RZ-1A protein. Plant Physiol. 2023;191:925–45.
Ma Z, Song T, Zhu L, Ye W, Wang Y, Shao Y, et al. A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell. 2015;27:2057–72.
McLellan H, Harvey SE, Steinbrenner J, Armstrong MR, He Q, Clewes R, et al. Exploiting breakdown in nonhost effector-target interactions to boost host disease resistance. Proc Natl Acad Sci USA. 2022;119: e2114064119.
Naqvi S, He Q, Trusch F, Qiu H, Pham J, Sun Q, et al. Blue-light receptor phototropin 1 suppresses immunity to promote Phytophthora infestans infection. New Phytol. 2022;233:2282–93.
Niderman T, Genetet I, Bruyere T, Gees R, Stintzi A, Legrand M, et al. Pathogenesis-related PR-1 proteins are antifungal. Isolation and characterization of three 14-kilodalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against Phytophthora infestans. Plant Physiol. 1995;108:17–27.
Nie J, Zhou W, Liu J, Tan N, Zhou JM, Huang L. A receptor-like protein from Nicotiana benthamiana mediates VmE02 PAMP-triggered immunity. New Phytol. 2021;229:2260–72.
Paluchowska P, Sliwka J, Yin Z. Late blight resistance genes in potato breeding. Planta. 2022;255:127.
Pan Q, Cui B, Deng F, Quan J, Loake GJ, Shan W. RTP1 encodes a novel endoplasmic reticulum (ER)-localized protein in Arabidopsis and negatively regulates resistance against biotrophic pathogens. New Phytol. 2016;209:1641–54.
Pel MA, Foster SJ, Park TH, Rietman H, van Arkel G, Jones JD, et al. Mapping and cloning of late blight resistance genes from Solanum venturii using an interspecific candidate gene approach. Mol Plant-Microbe Interact. 2009;22:601–15.
Pi L, Yin Z, Duan W, Wang N, Zhang Y, Wang J, et al. A G-type lectin receptor-like kinase regulates the perception of oomycete apoplastic expansin-like proteins. J Integr Plant Biol. 2022;64:183–201.
Pruitt RN, Locci F, Wanke F, Zhang L, Saile SC, Joe A, et al. The EDS1-PAD4-ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature. 2021;598:495–9.
Qiang X, Liu X, Wang X, Zheng Q, Kang L, Gao X, et al. Susceptibility factor RTP1 negatively regulates Phytophthora parasitica resistance via modulating UPR regulators bZIP60 and bZIP28. Plant Physiol. 2021;186:1269–87.
Redkar A, Cevik V, Bailey K, Zhao H, Kim DS, Zou Z, et al. The Arabidopsis WRR4A and WRR4B paralogous NLR proteins both confer recognition of multiple Albugo candida effectors. New Phytol. 2022;237:532–47.
Sandhu D, Gao H, Cianzio S, Bhattacharyya MK. Deletion of a disease resistance nucleotide-binding-site leucine-rich-repeat-like sequence is associated with the loss of the Phytophthora resistance gene Rps4 in soybean. Genetics. 2004;168:2157–67.
Sanogo S, Lamour K, Kousik S, Lozada DN, Parada Rojas CH, Quesada-Ocampo L, et al. Phytophthora capsici, 100 years later: research mile markers from 1922 to 2022. Phytopathology. 2022;6:66.
Shan W, Cao M, Leung D, Tyler BM. The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol Plant-Microbe Interact. 2004;17:394–403.
Sumit R, Sahu BB, Xu M, Sandhu D, Bhattacharyya MK. Arabidopsis nonhost resistance gene PSS1 confers immunity against an oomycete and a fungal pathogen but not a bacterial pathogen that cause diseases in soybean. BMC Plant Biol. 2012;12:87.
Tyler BM. Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol Plant Pathol. 2007;8:1–8.
Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, Aerts A, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–6.
Uma B, Rani TS, Podile AR. Warriors at the gate that never sleep: non-host resistance in plants. J Plant Physiol. 2011;168:2141–52.
van der Vossen E, Sikkema A, Hekkert B, Gros J, Stevens P, Muskens M, et al. An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J Cell Mol Biol. 2003;36:867–82.
Vleeshouwers VG, Rietman H, Krenek P, Champouret N, Young C, Oh SK, et al. Effector genomics accelerates discovery and functional profiling of potato disease resistance and phytophthora infestans avirulence genes. PLoS ONE. 2008;3: e2875.
Vossen JH, van Arkel G, Bergervoet M, Jo K-R, Jacobsen E, Visser RGF. The Solanum demissum R8 late blight resistance gene is an Sw-5 homologue that has been deployed worldwide in late blight resistant varieties. Theor Appl Genet. 2016;129:1785–96.
Wang Y, Bouwmeester K, van de Mortel JE, Shan W, Govers F. A novel Arabidopsis-oomycete pathosystem: differential interactions with Phytophthora capsici reveal a role for camalexin, indole glucosinolates and salicylic acid in defence. Plant Cell Environ. 2013;36:1192–203.
Wang Y, Bouwmeester K, Beseh P, Shan W, Govers F. Phenotypic analyses of Arabidopsis T-DNA insertion lines and expression profiling reveal that multiple L-type lectin receptor kinases are involved in plant immunity. Mol Plant-Microbe Interact. 2014;27:1390–402.
Wang Y, Cordewener JH, America AH, Shan W, Bouwmeester K, Govers F. Arabidopsis lectin receptor kinases LecRK-IX.1 and LecRK-IX.2 are functional analogs in regulating Phytophthora resistance and plant cell death. Mol Plant-Microbe Interact. 2015;28:1032–48.
Wang B, Sumit R, Sahu BB, Ngaki MN, Srivastava SK, Yang Y, et al. Arabidopsis novel glycine-rich plasma membrane PSS1 protein enhances disease resistance in transgenic soybean plants. Plant Physiol. 2018a;176:865–78.
Wang Y, Xu Y, Sun Y, Wang H, Qi J, Wan B, et al. Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat Commun. 2018b;9:594.
Wang D, Liang X, Bao Y, Yang S, Zhang X, Yu H, et al. A malectin-like receptor kinase regulates cell death and pattern-triggered immunity in soybean. EMBO Rep. 2020;21: e50442.
Wang W, Chen L, Fengler K, Bolar J, Llaca V, Wang X, et al. A giant NLR gene confers broad-spectrum resistance to Phytophthora sojae in soybean. Nat Commun. 2021;12:6263.
Wang Y, Pruitt RN, Nurnberger T, Wang Y. Evasion of plant immunity by microbial pathogens. Nat Rev Microbiol. 2022;20:449–64.
Witek K, Jupe F, Witek AI, Baker D, Clark MD, Jones JD. Accelerated cloning of a potato late blight-resistance gene using RenSeq and SMRT sequencing. Nat Biotechnol. 2016;34:656–60.
Witek K, Lin X, Karki HS, Jupe F, Witek AI, Steuernagel B, et al. A complex resistance locus in Solanum americanum recognizes a conserved Phytophthora effector. Nat Plants. 2021;7:198–208.
Wong J, Gao L, Yang Y, Zhai J, Arikit S, Yu Y, et al. Roles of small RNAs in soybean defense against Phytophthora sojae infection. Plant J Cell Mol Biol. 2014;79:928–40.
Wu CH, Abd-El-Haliem A, Bozkurt TO, Belhaj K, Terauchi R, Vossen JH, et al. NLR network mediates immunity to diverse plant pathogens. Proc Natl Acad Sci USA. 2017;114:8113–8.
Yan Q, Si J, Cui X, Peng H, Jing M, Chen X, et al. GmDAD1, a conserved Defender Against Cell Death 1 (DAD1) from soybean, positively regulates plant resistance against Phytophthora pathogens. Front Plant Sci. 2019;10:107.
Yang B, Wang Y, Guo B, Jing M, Zhou H, Li Y, et al. The Phytophthora sojae RXLR effector Avh238 destabilizes soybean Type2 GmACSs to suppress ethylene biosynthesis and promote infection. New Phytol. 2019;222:425–37.
Yang K, Yan Q, Wang Y, Peng H, Jing M, Dou D. GmPAO-mediated polyamine catabolism enhances soybean Phytophthora resistance without growth penalty. Phytopathol Res. 2022;4:35.
Yuan M, Jiang Z, Bi G, Nomura K, Liu M, Wang Y, et al. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature. 2021;592:105–9.
Zhang J, Xia C, Duan C, Sun S, Wang X, Wu X, et al. Identification and candidate gene analysis of a novel phytophthora resistance gene Rps10 in a Chinese soybean cultivar. PLoS ONE. 2013;8: e69799.
Zhang H, Li F, Li Z, Cheng J, Chen X, Wang Q, et al. Potato StMPK7 is a downstream component of StMKK1 and promotes resistance to the oomycete pathogen Phytophthora infestans. Mol Plant Pathol. 2021;22:644–57.
Zhang Y, Yin Z, Pi L, Wang N, Wang J, Peng H, et al. A Nicotiana benthamiana receptor-like kinase regulates Phytophthora resistance by coupling with BAK1 to enhance elicitin-triggered immunity. J Integr Plant Biol. 2023;6:66.
Zheng Z, Wang N, Jalajakumari M, Blackman L, Shen E, Verma S, et al. miR159 represses a constitutive pathogen defense response in tobacco. Plant Physiol. 2020;182:2182–98.
Zhu S, Li Y, Vossen JH, Visser RG, Jacobsen E. Functional stacking of three resistance genes against Phytophthora infestans in potato. Transgenic Res. 2012;21:89–99.
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the National Natural Science Foundation of China (32001959), the Natural Science Foundation of Jiangsu Province (BK20200286), and the Special Fund on Technology Innovation of Carbon Dioxide Peaking and Carbon Neutrality of Jiangsu Province ( BE2022306).
Author information
Authors and Affiliations
Contributions
QL, DD, and JY contributed to the study conception and design. HZ and GA drew the diagrams and tables. QL and HZ performed the data collection. QL and DD analyzed the data and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Supplementary Information
Additional file 1: Table S1
. Representative PRR and NLR genes related to Phytophthora resistance.
Additional file 2: Table S2
. Representative regulator genes, NHR genes, sRNAs, and lncRNAs related to Phytophthora resistance.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, Q., Zhu, H., Ai, G. et al. Plant genes related to Phytophthora pathogens resistance. Phytopathol Res 6, 15 (2024). https://doi.org/10.1186/s42483-024-00229-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42483-024-00229-w