Abstract
A potassium-containing hexaazamacrocyclic dication, [M•H•K]2+, is able to add in the gas phase mono- and dicarboxylate anions as well as inorganic anions by forming the corresponding monocharged adducts, the structure of which markedly depends on the basicity of the anion. With anions, such as acetate or fluoride, the neutral hexaazamacrocycle M acts as an acceptor of monosolvated K+ ion. With less basic anions, such as trifluoroacetate or chloride, the protonated hexaazamacrocycle [M•H]+ performs the unusual functions of an acceptor of contact K+/anion pairs.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ion-selective interactions of inorganic species with organic host molecules (e.g., enzymes, antibodies, or membrane transporters) play an important role in many chemical and biological processes occurring in aqueous solutions. In particular, this is true for the interaction of cations with anionic carboxylate groups as it influences protein association and enzymatic activity [1–12].
So far, model studies have centered mainly on the recognition of cations using synthetic host compounds, such as macrocyclic polyether [13–16] and polyamines [17]. Polyazamacrocycles, such as M (Figure 1), have been successfully employed as chiral solvating agent for monocarboxylic acids in CDCl3 [18], but not in aqueous solutions because the strong solvation capability of water inhibits the formation of the corresponding noncovalent adducts. However, in water, macrocycle M can efficiently form noncovalent adducts with polycarboxylic acids, although the process is very sensitive to the state of charge of both components and, therefore, to the pH of the solution [19, 20].
Indeed, the hexaazamacrocycle M is a very basic acceptor which, in water at neutral pH, can accommodate more than two protons onto their NH groups [20]. Under these conditions, multiprotonated M is able to form 1:1 complexes only with dicarboxylates having suitable geometry and electronic properties [20–25]. Its selectivity has been interpreted in terms of geometric and electronic requirements for multiple hydrogen bondings with the dicarboxylate anion, thereby the intuition that multiprotonated M can mimic the active center of polycarboxylate carrier proteins.
Since Na+ and K+ are the most abundant monovalent ions in biological systems, it would be of interest to investigate whether alkali-containing polyammonium ions can interact with anionic species. This may be an arduous task in aqueous solution because of the unfavorable alkali cation desolvation energy. In this paper, we will show that an alkali-containing polyammonium ion can be conveniently generated in the gas phase and its interaction with inorganic and organic anions investigated.
Experimental
Materials
All the chemicals employed in the present study were purchased from commercial sources. The racemate of the hexaazamacrocycle M was synthesized and purified according to our modifications of the procedures reported in the literature [20, 26, 27] (see Experimental Details and Scheme S1 in the Supplementary Material (SM) section).
Electrospray Ionization Mass Spectrometry Experiments
The electrospray ionization mass spectrometry (ESI-MS) experiments were carried out in a Bruker Esquire 6000 quadrupole ion trap. High resolving power spectra and ion-molecule reactions (mass range: 50–1000 m/z) were carried out by using an APEX 47e FT-ICR mass spectrometer equipped with an ESI source (Bruker, Billerica, MA, USA) and a resonance cell (“infinity cell”) situated between the poles of a superconducting magnet (4.7 T). Operating conditions of the ESI source were as follows: ion spray voltage = +4.0 kV; nebulizer gas (N2) = 15 psi; drying gas (N2) = 5 psi; end plate = +4.0 kV; capillary exit 100 V; dry temperature = 300 °C. Methanolic solutions containing M (10–5 M) together with the appropriate acid or its derivative (10–5 M) were infused via a syringe pump at a flow rate of 3 μL min–1. The relative abundance of peaks results from their area acquired in profile mode. In each acquisition, the final spectra are the average of about 10 scans. The [M•K•YCOOH]+ and [M•K•HX]+ ions were submitted to collision induced dissociation (CID) (collision energies employed up to 40 eV, lab frame). During the CID experiments, the nominal pressure of He in the collision chamber was 1.4 × 10–5 mbar.
Computational Details
A preliminary investigation on the potential energy surfaces (PES’s) of the [M•H]+, [M•Na]+, [M•K]+, and [M•H2]2+ ions were carried out using classic molecular dynamics (MD) simulations based on Amber [28] force field using partial charge from RESP [29] fit. According to the procedure exploited by Valden et al. [30], a simulated annealing approach was performed in order to investigate the conformational space. The minimum energy conformers found by MD were optimized at quantum mechanics level. For optimized geometries and frequencies calculations, we have used Density Functional Theory approach using B3LYP [31] exchange-correlation functional and the 6-31G* basis set. We employed the ORCA [32] package with SCF convergence criteria set as TightSCF (energy change 1e-08; max density change 1e-07; rms-density change 1e-09) and a high precision for the integration grids. Geometry optimizations were carried out without any constraint.
The ab-initio molecular dynamic simulations (AIMD) were carried out with Terachem package [33] using BLYP/6-31G* level of theory and including DFT-D3 dispersion corrections [34]. The trajectory was evolved according to the Born-Oppeneimer [35] scheme. Dynamic simulations were performed for 10 ps, using a time-step of 1 ft, at 400 K temperature controlled via Nose-Hoover thermostat [36, 37]. The trajectory was save each step.
Results and Discussion
The [M•H•K]2+ ion
In a recent study [38], protonated M, formed in a mass spectrometer by ESI, was found to be capable of selectively trapping ppb amounts of the K+ ions ubiquitously present in ultrapure methanol (≥99.9%). The same species was exclusively observed even when 10–5 M sodium chloride was added to the methanolic solution containing 10–5 M potassium chloride (see, for instance, Figure S1 of the SM section). Such an unexpected result has been attributed to the favorable free energy balance between the K+ ion desolvation and its coordination to [M•H]+, which allows formation of the [M•H•K]2+ adduct by the time of the Coulombic explosion of the ESI-MS nanodroplets. Under the same conditions, formation of the [M•H•Na]2+ analogue is kinetically prevented by the over 17 kcal mol–1 higher Na+ desolvation free energy.
The [M•H•K]2+ (m/z 237) dication was accompanied by comparable yields of the [M•H2]2+ (m/z 218) one. More intense signals were also observed corresponding to the monocharged [M•H]+ (m/z 435), [M•Na]+ (m/z 457), and [M•K]+ (m/z 473) ions (Figure S1 of the SM section).
The [M•K•YCOOH]+ Complexes
When acetic acid (10–5 M) is added to the methanolic solution, another signal corresponding to [M•K•CH3COOH]+ (m/z 533) shows up at the expenses of the K+-containing ions (Figure S2 of the SM section). Similar adducts are efficiently formed by replacing acetic acid with an equimolar amount of other mono- and dicarboxylic acids (YCOOH in Scheme 1) (see, for instance, Figure S3 of the SM section). Signals corresponding to their [M•Na•YCOOH]+ analogues (m/z 517 (Y = CH3)) are never detected despite the high abundance of the [M•Na]+ ion and the B3LYP/6-31G*-calculated exoergicity for its addition to YCOOH [e.g., ΔG300 (kcal mol-1) = –2.7 (Y = CH3); –6.0 (Y = CF3)].
These pieces of evidence confirm the view that the [M•K•YCOOH]+ ion is predominantly, if not exclusively, generated by the strongly exoergic addition of the YCOO- anion to [M•H•K]2+ (Table 1) [38]. This conclusion is further corroborated by the fact that a similar behavior was observed when replacing the carboxylic acid with the same concentration of its sodium or ammonium salt (see, for instance, Figures S4 and S5 of the SM section). In contrast, no similar adducts were observed when the acid was replaced by its ester or amide, which cannot generate the conjugate base of the acid in methanol.
Structure and Dynamics of the [M•K•YCOOH]+ Complexes
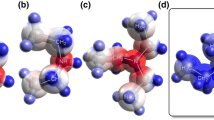
Collision induced dissociation (CID) experiments were carried out to gather some information on the structure of the [M•K•YCOOH]+ complexes. As a matter of fact, most of them did not show any tendency to fragment even at high collision energies (up to 40 eV, lab frame) (path a in Scheme 1). This behavior is rather unusual in view of the ability of similar metal bound adducts to readily release neutral fragments when collisionally excited [39–41]. However, it should be considered that the excited ions have a definite time to decompose in the collision cell. For large complexes, as those involved here, the lifetime for dissociation may exceed their residence time in the cell so that efficient dissociation of the ions may not be observed at the thermodynamic threshold. In contrast, when at least one of the methyl hydrogens of acetic acid is replaced by F or Cl, an extensive fragmentation is observed with formation of the [M•K•HF]+ (path b in Scheme 1; see, for instance, Figure S6 of the SM section) or [M•K•HCl]+ ion (path c in Scheme 1), respectively. In the same fashion, the [M•K•H2O]+ fragment is readily formed by CID of the adducts where the acetic acid is replaced by lactic acid or by a dicarboxylic acid (path d in Scheme 1). In contrast, no fragmentation is observed under the same conditions when one methyl hydrogen of acetic acid is replaced by the OH or CN group (path a in Scheme 1).
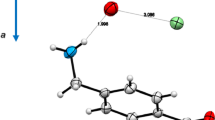
Some insights into the behavior of the two simplest members of the [M•K•YCOOH]+ family (i.e., [M•K•CH3COOH]+ and [M•K•CF3COOH]+ ) are obtained from ab initio molecular dynamic (AIMD) simulations starting from the corresponding global minima arising from classical MD simulation and optimized at the B3LYP/6-31G* level of theory. The trajectory of AIMD simulation for [M•K•CH3COOH]+ indicates that the K–O1 distance (O1 and O2 denote the carbonyl and the hydroxyl oxygens of the acid, respectively) oscillates between 2.5 and 3.0 Å during the entire lapse of time (10 ps) (Figure 2a), whereas the proton tends to reside on the O2 oxygen after some oscillations toward the proximate amino group of M (Figure 2b).
In contrast, the trajectory of AIMD simulation for [M•K•CF3COOH]+ points to a fast proton transfer from the O2 oxygen to the proximate amino group of M (Figure 3a). At the same time, the O2 atom gets closer to K+ (Figure 3b) while the entire CF3COO– moiety of the complex revolves around the C–O2 bond by allowing the establishment of weak K+–F interactions ( Figure 3c).
Accordingly, the most stable B3LYP/6-31G*-optimized [M•K•CH3COOH]+ and [M•K•CF3COOH]+ structures are depicted in Figure 4a and b, respectively. It should be noted that the structure of Figure 4b, wherein a F atom interacts with the K+ center and the proton of the acid with the amino group of M, clearly exhibits the premise for the effective CID-induced formation of [M•K•HF]+ through the formal loss of the CF2COO fragment.
As pointed out before, multiprotonated M, formed in water at neutral pH, does not exhibit any tendency to add monocarboxylate anions, although it is able to efficiently trap dicarboxylates of suitable geometry and electronic properties [20–25]. A reason for such a different behavior can be found in the large dehydration penalty to be paid by the charged species involved, which can be offset only by the establishment of multiple electrostatic interactions within the adduct, provided that it does not demand any significant structural distortion of the diprotonated acceptor. This requirement cannot be easily fulfilled by the monocarboxylates because of the large difference between the O–O distance [2.28 Å (CH3COO–); 2.29 Å (CF3COO–)] and the proton–proton distance in the most stable [M•H2]2+ isomer (4.12 Å; Figure S7 of MS), whereas it is more easily satisfied by the dicarboxylates (e.g., the O-–O- distance in the B3LYP/6-31G*-optimized structure of the oxalate dianion amounts to 3.17 Å). The geometrical constraints preventing the establishment of multiple electrostatic interactions between the monocarboxylates and [M•H2]2+ are much less cogent for the [M•H•K]2+ acceptor since here the K+ is located slightly above the center of the macrocycle cavity at a distance of 3.19 Å from the closest NH2 + proton (Figure S7 of MS). This distance is not far from the distance between the oxygens of monocarboxylates (ca. 2.3 Å) so that an intense enough chelation of the monocarboxylate by the [M•H•K]2+ dication can take place without involving a significant structural distortion of the multicharged receptor.
Structure of the [M•K•HX]+ Complexes
That the addition of anions to the [M•H•K]2+ dication is a general process is demonstrated by the ESI-MS spectra of methanolic solution of M containing 10–5 M of inorganic ammonium salts NH4X, with X = F, Cl, Br, I, NO3, and HSO4. In all cases, intense signals attributed to the corresponding [M•K•HX]+ adducts were observed.
As for YCOO–, the multidentate NO3 –, and SO4H– anions can efficiently interact with both the K+ and the proton centers of [M•H•K]2+. The monodentate halide anions can be accommodated between the same centers to yield two limiting structures depending upon the capability of the anion to accept or not the proton from the receptor. If it does accept, the [M•K•HX]+ complex can be regarded as a HX molecule on the potassiated M. If it does not, the [M•K•HX]+ structure can be seen as a contact K+X– ion pair on the protonated M. In any case, similarly to some [M•K•YCOOH]+ complexes (path a in Scheme 1), the [M•K•HX]+ adducts were found to be highly reluctant toward CID fragmentation even at high collision energies (up to 40 eV, lab frame).
The most stable B3LYP/6-31G*-optimized structures of [M•K•HX]+ (X = F, Cl) are illustrated in Figure 5a and b, respectively. In both, the halogen atom resides between the K+ and the proton centers of [M•H•K]2+. In the [M•K•HF]+ structure, the proton is essentially bound to the fluoride anion while interacting with the pyridine nitrogen of M. However, a close inspection of Figure 5a reveals that the H–F distance (1.00 Å) is significantly longer and that of H–N (1.56 Å) much shorter than the corresponding distances in most common F-H–N interactions (i.e., 0.94 Å and 1.77 Å) respectively, [42]. This would mean that in [M•K•HF]+ the HF molecule is strongly polarized by the [M•K]+ environment.
In contrast, in [M•K•HCl]+ (Figure 5b), the proton is placed on the NH group of the receptor (HN-H distance =1.09 Å) while interacting with the chloride anion (Cl–H distance =1.91 Å). This arrangement points to the existence of the contact K+Cl– ion pair over protonated M [43]. This view is consistent with the calculated K–Cl distance (3.07 Å), which is essentially equal to the sum of the K+ and Cl– ionic radii (3.14 Å).
An indirect piece of evidence for the different structures in Figure 5a and b can be found in the different reactivity of the ESI-formed [M•K•HX]+ (X = F, Cl, Br, I) ions towards CD3COOH, present in the reaction cell of the FT-ICR spectrometer at pressure ranging from 3 × 10–8 to 5 × 10–8 mbar. As shown in Figure S8, a very efficient CD3COOH-to-HF displacement is observed with [M•K•HF]+ without any sign of H/D exchange. The same process is not observed with [M•K•HBr]+ and [M•K•HI]+, while extensive H/D exchange exclusively takes place with incorporation of up to three deuteriums. Both reaction channels are opened to [M•K•HCl]+ with the H/D exchange path kinetically prevailing over the substitution one (Figure S9). The corresponding kinetic parameters are listed in Table 2.
The kinetic data of Figures S8 and S9 and of Table 2 are consistent with the structural features of [M•K•HF]+ (Figure 5a), on one side, and of [MH+•K+•X-]+ (X=Cl, Br, I) (Figure 5b), on the other. The presence of the polarized HF molecule in [M•K•HF]+ (Figure 5a) favors its fast displacement induced by the interaction between the K+ center and the most basic site of CD3COOH (i.e., O1). Instead, when approaching the zwitterionic [MH+•K+•X-]+ (X = Cl, Br, I) ions (Figure 5b), the CD3COOH acid can establish a double X––H-O2 and the HN+-H–O1 interaction wherein a prototropic transfer may take place, depending upon the ability of the X– center to abstract the proton from the acetic acid (300 K gas phase basicity (GB300) of acetic acid = 341.8 kcal mol–1) [44]. This, in turn, is related to the 300 K gas phase basicity of the X– ion, which increases in the order: I– (309.3 kcal mol–1) < Br- (318.4 kcal mol–1) < Cl- (323.7 kcal mol–1) [44]. Therefore, when the prototropic rearrangement is energetically more demanding (X– = I-; Br-), the CD3COOH moiety can acquire some positive charge from the HN+-H–O1 interaction, which increases the acidity of its CD3 deuteriums [45]. The consequence is an enhanced probability of consecutive H/D scrambling between the CD3 group and the unprotonated NH functionalities of the receptor. When, instead, the prototropic transfer is energetically more favored (X– = Cl–), the D/H exchange pathway undergoes competition by the loss of the so-formed neutral HCl moiety (Table 2).
Conclusions
The present study demonstrates that the [M•H•K]2+ dication is able to add mono- and dicarboxylate anions as well as inorganic anions, such as halides, nitrate, etc., by forming the corresponding monocharged adducts in the gas phase. Several stable structures can be formed depending on the relative basicities of the amino groups of M and of the anion coordinated to the K+ center. With anions, such as trifluoroacetate (GB300 = 317.4 kcal mol–1) or chloride (GB300 = 323.7 kcal mol–1) [45], the protonated hexaazamacrocycle [M•H]+ performs the functions of a host for contact ion pairs (e.g., K+Cl–). With more basic anions, such as acetate (GB300 = 341.7 kcal mol–1) or fluoride (GB300 = 365.7 kcal mol–1) [45], the neutral hexaazamacrocycle M acts as a host for monosolvated K+ ion (e.g., K+•••FH). Easy interconversion between the two structural families is expected to take place by proton transfer between the amino groups of M and the anion coordinated to the K+ center when they are endowed with comparable basicities.
Such a prototropic rearrangement may provide an additional explanation of the unexpected reluctance of some [M•K•YCOOH]+ (path a in Scheme 1) and [M•K•HX]+ adducts to undergo CID fragmentation. It may be conceivable that when warmed up by collisions with an inert gas, the loss of the neutral acid molecule is prevented by the fast proton transfer from the acid to the proximate amino group of the macrocycle. In this case, no fragmentation takes place since the loss of the so-formed ion pair is too energy-demanding (e.g., ΔG300 = +22.4 kcal mol–1 for the loss of K+Cl– from the [MH+•K+•Cl–]+ adduct). However, even when the contact ion pair is already present, e.g., K+CF3COO–in the [MH+•K+•CF3COO–]+ adduct (Figure 4b), nontheless an extensive CID fragmentation is observed that is favored by the structural preorganization of a potential leaving moiety (i.e., CF2COO).
References
Perez-Jimenez, R., Godoy-Ruiz, R., Ibarra-Molero, B., Sanchez-Ruiz, J.M.: The efficiency of different salts to screen charge interactions in proteins: a Hofmeister effect? Biophys. J. 86, 2414–2429 (2004)
Zhang, Y.J., Furyk, S., Bergbreiter, D.E., Cremer, P.S.: Specific ion effects on the water solubility of macromolecules: PNIPAM and the Hofmeister series. J. Am. Chem. Soc. 127, 14505–14510 (2005)
Bostrom, M., Williams, D.R.M., Ninham, B.W.: Surface tension of electrolytes: specific ion effects explained by dispersion forces. Langmuir 17, 4475–4478 (2001)
Kunz, W., Lo Nostro, P., Ninham, B.: The present state of affairs with Hofmeister effects. Curr. Opin. Colloid Interface Sci. 9, 1–18 (2004)
Collins, K.D.: Ions from the Hofmeister series and osmolytes: effects on proteins in solution and in the crystallization process. Methods 34, 300–311 (2004)
Lo Nostro, P., Ninham, B.W.: Hofmeister phenomena: an update on ion specificity in biology. Chem. Rev. 112, 2286–2322 (2012)
Vrbka, L., Jungwirth, P., Bauduin, P., Touraud, D., Kunz, W.: Specific ion effects at protein surfaces: a molecular dynamics study of bovine pancreatic trypsin inhibitor and horseradish peroxidase in selected salt solutions. J. Phys. Chem. B 110, 7036–7043 (2006)
Lund, M., Vrbka, L., Jungwirth, P.: Specific ion binding to nonpolar surface patches of proteins. J. Am. Chem. Soc. 130, 11582–11583 (2008)
Jungwirth, P., Tobias, D.J.: Specific ion effects at the air/water interface. Chem. Rev. 106, 1259–1281 (2006)
Hess, B., van der Vegt, N.F.A.: Cation specific binding with protein surface charges. Proc. Natl. Acad. Sci. U. S. A. 106, 13296–13300 (2009)
Vrbka, L., Vondrášek, J., Jagoda-Cwiklik, B., Vácha, R., Jungwirth, P.: Quantification and rationalization of the higher affinity of sodium over potassium to protein surfaces. Proc. Natl. Acad. Sci. U. S. A. 103, 15440–15444 (2006)
Annapureddy, H.V.R., Dang, L.X.: Molecular mechanism of specific ion interactions between alkali cations and acetate anion in aqueous solution: a molecular dynamics study. J. Phys. Chem. B 116, 7492–7498 (2012)
Christensen, J.J., Eatough, D.J., Izatt, R.M.: Thermodynamics of cation-macrocyclic compound interaction. Struct. Bond. Berlin, Germany 16, 161–189 (1973)
Izatt, R.M., Bradshaw, J.S., Nielsen, J.D., Lamb, J.J., Christensen, J.: Thermodynamic and kinetic data for cation-macrocycle interaction. J. Chem. Rev. 85, 271–339 (1985)
Izatt, R.M., Pawlak, K., Bradshaw, J.S., Bruening, R.L.: Thermodynamic and kinetic data for macrocycle interactions with cations and anions. Chem. Rev. 91, 1721–1785 (1991)
Gokel, G.W., Leevy, W.M., Weber, M.E.: Crown ethers: sensors for ions and molecular scaffolds for materials and biological models. Chem. Rev. 104, 2723–2750 (2004)
Korendovych, I.V., Roesner, R.A., Rybak-Akimova, E.V.: Molecular recognition of neutral and charged guests using metallomacrocyclic hosts. Adv. Inorg. Chem. 59, 109–173 (2007)
Gonzalez-Alvarez, A., Alfonso, I., Gotor, V.: An azamacrocyclic receptor as efficient polytopic chiral solvating agent for carboxylic acids. Tetrahedron Lett. 47, 6397–6400 (2006)
Mateus, P., Bernier, N., Delgado, R.: Recognition of anions by polyammonium macrocyclic and cryptand receptors: Influence of the dimensionality on the binding behavior. Coord. Chem. Rev. 254, 1726–1747 (2010)
Gonzalez-Alvarez, A., Alfonso, I., Diaz, P., Garcia-Espana, E., Gotor-Fernandez, V., Gotor, V.: A simple helical macrocyclic polyazapyridinophane as a stereoselective receptor of biologically important dicarboxylates under physiological conditions. J. Org. Chem. 73, 374–382 (2008)
González-Álvarez, A., Alfonso, I., Díaz, P., García-España, E., Gotor, V.: A highly enantioselective abiotic receptor for malate dianion in aqueous solution. Chem. Commun. 11, 1227–1229 (2006)
Busto, E., Gonzalez-Alvarez, A., Gotor-Fernandez, V., Alfonso, I., Gotor, V.: Optically active macrocyclic hexaazapyridinophanes decorated at the periphery: synthesis and applications in the NMR enantiodiscrimination of carboxylic acids. Tetrahedron 66, 6070–6077 (2010)
Gospodarowicz, K., Holynska, M., Paluch, M., Lisowski, J.: Novel chiral hexaazamacrocycles for the enantiodiscrimination of carboxylic acids. Tetrahedron 68, 9930–9935 (2012)
Kimura, E., Sakonara, A., Yatsunami, T., Kodama, M.: Macromonocyclic polyamines as specific receptors for tricarboxylate-cycle anions. J. Am. Chem. Soc. 103, 3041–3045 (1981)
Kimura, E., Kodama, M., Yatsunami, T.: Macromonocyclic polyamines as biological polyanion complexons. 2. Ion-pair association with phosphate and nucleotides. J. J. Am. Chem. Soc. 104, 3182–3187 (1982)
Fitzsimmons, P.M., Jackels, S.C.: Helical hexaazamacrocyclic ligands containing pyridyl and (+ or –)-trans-diaminocyclohexyl groups: effect of ligand constraints upon metal ion binding. Inorg. Chim. Acta 246, 301–310 (1996)
Gonzalez-Alvarez, A., Alfonso, I., Lopez-Ortiz, F., Aguirre, A., Garcia-Granda, S., Gotor, V.: Selective host amplification from a dynamic combinatorial library of oligoimines for the syntheses of different optically active polyazamacrocycles. Eur. J. Org. Chem. 5, 1117–1127 (2004)
Case, D.A., Darden, T.A., Cheatham, T.E. III,, Simmerling, C.L., Wang, J., Duke, R.E., Luo, R., Walker, R.C., Zhang, W., Merz, K.M., Roberts, B., Hayik, S., Roitberg, A., Seabra, G., Swails, J., Götz, A.W., Kolossváry, I., Wong, K.F., Paesani, F., Vanicek, J., Wolf, R.M., Liu, J., Wu, X.; Brozell, S.R., Steinbrecher, T., Gohlke, H., Cai, Q.; Ye, X., Wang, J., Hsieh, M.J., Cui, G., Roe, D.R., Mathews, D.H., Seetin, M.G., Salomon-Ferrer, R., Sagui, C., Babin, V., Luchko, T., Gusarov, S., Kovalenko, A., Kollman P.A.: AMBER 12, University of California, San Francisco (2012)
Cornell, W.D., Cieplak, P., Bayly, C.I., Kollman, P.A.: Application of RESP charges to calculate conformational energies, hydrogen bond energies, and free energies of solvation. J. Am. Chem. Soc. 115, 9620–9631 (1993)
Vaden, T.D., de Boer, T.S.J.A., Simons, J.P., Snoek, L.C., Suhai, S., Paizs, B.: Vibrational spectroscopy and conformational structure of protonated polyalanine peptides isolated in the gas phase. J. Phys. Chem. A 112, 4608–4616 (2008)
Stephens, P.J., Devlin, F.J., Chabalowski, C.F., Frisch, M.J.: Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. J. Phys. Chem. 98, 11623–11627 (1994)
Neese, F.: The ORCA program system. WIREs Comput Mol. Sci. 2, 73–78 (2012)
Ufimtsev, I.S., Martinez, T.J.: Quantum chemistry on graphical processing units. 3. Analytical energy gradients, geometry optimization, and first principles molecular dynamics. J. Chem. Theory Comput. 5, 261–2628 (2009)
Grimme, S., Antony, J., Ehrlich, S., Krieg, H.: A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 132, 154104 (2010)
Marx, D., Hutter, J.: Ab initio molecular dynamics: basic theory and advanced methods. Cambridge University Press, Cambridge (2009)
Hoover, W.: Constant-pressure equations of motion. Phys. Rev. A 34, 2499–2500 (1986)
Nose, S.: An extension of the canonical ensemble molecular dynamics method. Mol. Phys. 52, 255–256 (1986)
Fraschetti, C., Filippi, A., Crestoni, M.E., Marcantoni, E., Glucini, M., Guarcini, L., Montagna, M., Guidoni, L., Speranza, M.: Protonated hexaazamacrocycles as selective K+ receptors. J. Am. Soc. Mass Spectrom. 26, 1186–1190 (2015)
Filippi, A., Speranza, M., Paladini, A., De Carolis, R., Giardini Guidoni, A., Laganà, A., Satta, M.: Diastereoselective fragmentation of chiral α-aminophosphonic acids/metal ion aggregates. J. Mass Spectrom. 41, 98–102 (2006)
Wu, L., Tao, W.A., Cooks, R.G.: Ligand and metal-ion effects in metal-ion clusters used for chiral analysis of α-hydroxy acids by the kinetic method. Anal. Bioanal. Chem. 373, 618–627 (2002)
Wu, L., Meurer, E.C., Young, B., Yang, P., Eberlin, M.N., Cooks, R.G.: Isomeric differentiation and quantification of α, β-amino acid-containing tripeptides by the kinetic method: alkali metal-bound dimeric cluster ions. Int. J. Mass Spectrom. 231, 103–111 (2004)
Liu, G., Zhang, X.: The argon matrix-induced frequency shift of νs(F-H) of the F-H–A complexes: Comparison between computation and experiment. J. Molec. Struct.: THEOCHEM 817, 147–152 (2007)
Cametti, M., Nissinen, M., Dalla Cort, A., Mandolini, L., Rissanen, K.: Recognition of alkali metal halide contact ion pairs by uranyl-salophen receptors bearing aromatic sidearms. J. Am. Chem. Soc. 127, 3831–3837 (2005)
NIST Chemistry Webbook. NIST Standard Reference Database. Available at: http://webbook.nist.gov/chemistry. Accessed 5 Feb 2015
Grabowski, J.J., Cheng, X.: Gas-phase formation of the enolate monoanion of acetic acid by proton abstraction. J. Am. Chem. Soc. 111, 3106–3108 (1989)
Acknowledgments
The authors acknowledge support for this work by the Ministero dell’Istruzione dell’Università e della Ricerca of Italy (PRIN 2010–2011: CUP B81J1200283001, prot. 2010ERFKXL_006), the University of Rome, La Sapienza (FAR 2013-2014), and the University of Camerino (FAR 2011-2012).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 920 kb)
Rights and permissions
About this article
Cite this article
Fraschetti, C., Filippi, A., Crestoni, M.E. et al. Contact Ion Pairs on a Protonated Azamacrocycle: the Role of the Anion Basicity. J. Am. Soc. Mass Spectrom. 27, 615–621 (2016). https://doi.org/10.1007/s13361-015-1327-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-015-1327-3