Abstract
In the present work, the kinetic intermediates of holo- and apo-myoglobin were studied by correlating the ion-neutral collision cross section and time resolved H/D back exchange rate simultaneously in a trapped ion mobility spectrometer coupled to a mass spectrometer (HDX-TIMS-MS). The high mobility resolution of the TIMS cell permitted the observation of multiple IMS bands and complementary molecular dynamics simulations resulted in the assignment of candidate structures for each experimental condition studied (e.g., holo [M + 8H]+8–[M + 9H]+9 and apo [M + 9H]+9–[M + 19H]+19). Inspection of the kinetic intermediates suggests that the tertiary structure of apomyoglobin unfolds quickly upon the loss of the Fe protoporphyrin IX that stabilizes the interactions between the A, G, and H helices. In the absence of the porphyrin heme, the apomyoglobin unfolds to Xn kinetic intermediates that vary in the extent of unfolding as a result of the observed charge state.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since the determination of the three-dimensional structure in 1957 [1], myoglobin (Mb) has been extensively studied and is commonly used as a protein model to better understand the mechanisms involved in protein folding [2–5]. In particular, Mb functionality (and hence the tertiary structure) results from the presence of a prosthetic group, Fe protophorphyrin IX, which reversibly binds molecular oxygen for transportation and delivery to muscle tissue [6]. The heme-cavity interactions play an important role during Mb folding via transitions from native (N) to molten globule (MG) to acid-unfolded (UA) forms [7, 8]. Although the Mb tertiary structure in the N state is generally well accepted, during the unfolding process there is no defined tertiary structure in the MG state [9] before it further unfolds to the rod-like, completely denatured state [7].
The multiple kinetic intermediate states of Mb (and other globular proteins) have triggered multiple experimental approaches to better understand the folding process, including optical spectroscopy (e.g., circular dichroism and time-resolved fluorescence and infra-red spectroscopy), hydrogen-deuterium exchange combined with nuclear magnetic resonance and ion mobility spectrometry (IMS), electron capture dissociation and hydrogen-deuterium exchange (HDX) combined with mass spectrometry [10–17]. In particular, ESI-MS experiments have shown non-cooperative unfolding for Mb, which involves multiple protein conformations [14, 18, 19]. Moreover, changes in the H/D exchange rate for holo and apo charge states have also been identified as a function of time, providing general characteristics pertaining to structural features [20]. IMS has been widely used to study the conformational properties of globular proteins, and when complemented with molecular dynamics, it provides a powerful tool for structural biology [21–35]. Recently, HDX experiments in a traveling wave ion guide at short labeling times (0.1–10 ms) enabled the interrogation of apo Mb conformers [36].
With the recent development of trapped ion mobility spectrometry (TIMS) [37, 38], higher mobility separation [39, 40] and the possibility to interrogate and simultaneously measure the ion-neutral collision cross section (CCS) as a function of the time after the molecular ion formation (gas-phase time after desolvation or trapped time in the TIMS) has permitted kinetic studies of molecular ions in the millisecond to second time scale as well as the influence of the collision partner on the conformational space accessible to the molecular ions in the gas phase [41, 42]. In the current study, the unique potential of TIMS to hold ions while interacting with bath gas molecules (“TIMS” thermostat) is utilized to study the folding motifs of holo- and apoMb by measuring the CCS and HDX under the same folding conditions (e.g., solvent conditions and time after molecular ion formation). A novel candidate structure generation algorithm was applied for a more efficient sampling of the conformational space of proteins and the generation of candidate structures for each IMS bands with candidate filtering using the HDX data [43]. A folding pathway is proposed for the native (N) to molten globule (MG) to acid-unfolded (UA) forms including (Xn) kinetic intermediates.
Experimental
Materials and Reagents
Equine holoMb was purchased from Sigma-Aldrich (St. Louis, MO, USA) and used as received. All solvents and ammonium acetate salts used in these studies were analytical grade or better and purchased from Fisher Scientific (Pittsburg, PA, USA). Low concentration tune mix (G2421A; Agilent Technologies, Santa Clara, CA, USA) was used as a mobility calibration standard and used as received. HoloMb and apoMb samples were prepared by diluting an equine holomyoglobin stock, 100 μM, in 10 mM ammonium acetate (pH 6 and pH 4.5) to a final concentration of 1 μM. The pH was adjusted using acetic acid. Deuterated solutions of Mb were prepared using analytical grade D2O (Sigma-Aldrich) in 10 mM ammonium acetate (pH 6 and pH 4.5) to a final concentration of 1 μM for HDX back exchange experiments.
HDX-TIMS-MS Analysis
Details regarding the TIMS operation and specifics compared with traditional IMS can be found elsewhere [37–39, 41, 42]. Briefly, TIMS mobility separation is based on holding the ions stationary using an electric field against a moving gas. The separation in a TIMS device can be described by the center of the mass frame using the same principles as in a conventional IMS drift tube [44]. Since mobility separation is related to the number of ion-neutral collisions (or drift time in traditional drift tube cells), the mobility separation in a TIMS device depends on the bath gas drift velocity, ion confinement, and ion elution parameters. Using the same principle, HDX-TIMS-MS provides the possibility to measure the rate of back exchange by varying the time the molecular ions spend in the TIMS cell. The mobility, K, of an ion in a TIMS cell is described by:
where vg, E, Velution, and Vbase are the velocity of the gas, applied electric field, elution, and base voltages, respectively. The constant A can be determined using calibration standards of known reduced mobilities (e.g., from low concentration tune mix: m/z = 322 Ko = 1.376 cm2V−1 s−1, m/z = 622 Ko = 1.013 cm2V−1 s−1, and m/z = 922 Ko = 0.835 cm2V−1 s−1). In TIMS operation, multiple geometric isomers/conformers are trapped simultaneously at different E values resulting from a voltage gradient applied across the TIMS tunnel. After thermalization, geometric isomers/conformers are eluted by decreasing the electric field in stepwise decrements (referred to as the “ramp”). Each isomer/conformer eluting from the TIMS cell can be described by a characteristic voltage (i.e., Velution – Vbase). Eluted ions are then mass-analyzed and detected by a maXis impact Q-ToF mass spectrometer (Bruker Daltonics Inc., Billerica, MA, USA).
In a TIMS device, the total analysis time can be described as:
where, Ttrap is the thermalization/trapping time, ToF is the time after the mobility separation, and Vramp and Tramp are the voltage range and time required to vary the electric field, respectively. The elution voltage can be experimentally determined by varying the ramp time for a constant ramp voltage. This procedure also determines the time ions spend outside the separation region To (e.g., ion trapping and time-of-flight). The same rf (880 kHz and 200 Vp-p) was applied to all electrodes including the entrance funnel, the mobility separating section, and the exit funnel. An electrospray ionization source (ESI Apollo II design, Bruker Daltonics, Inc., MA, USA) was used for all the analyses. TIMS separation was performed using nitrogen as a bath gas at ca. 300 K, and the gas flow velocity was controlled by the pressure difference between entrance funnel P1 = 2.6 mbar and the exit funnel P2 = 1.0 mbar. P1 and P2 values were held constant for all experiments. The TIMS cell was operated using a fill/trap/ramp/wait sequence of 1–10/1–10/5–1950/50 ms. Average mobility resolution at 10/10/500/50 was 160–190.
Reduced mobility values (K0) were correlated with CCS (Ω) using the equation:
where z is the charge of the ion, kB is the Boltzmann constant, N* is the number density and mI and mb refer to the masses of the ion and bath gas, respectively [44].
Theoretical
A novel candidate structure generation algorithm was used for a more efficient sampling of the conformational space of proteins and the generation of candidate structures for each IMS band with candidate filtering using the HDX data (more details in [43]). The 1YMB protein data bank entry for Mb was utilized as the starting structure with the presence or absence of the heme, for the holo and apo theoretical calculations, respectively [45]. Briefly, molecular dynamics simulations were used to reproduce the experimental conditions (e.g., “TIMS” thermostat) and to generate the identity vectors that define Mb conformational space. This approach is similar to that previously described in reference [46], with the main characteristic that the initial search targets the generation of the identity vectors, followed by charged assignment (based on the accessible surface area), and energy minimization. Once the candidate structures are identified for each IMS band (a minimum of four CM was considered per IMS band [43]), charge assignment is performed by scoring the accessible surface area based on the basicity/acidity of the chemical groups. That is, charge assignment is based on the score of the amino acid residues from each tri-dimensional structure [47, 48]. For example, solvent accessibility and the pka of the acidic (aspartic acid and glutamic acid) and basic (arginine, histidine, and lysine) amino acid residues are primarily utilized to assign the protonation and deprotonation sites. It is known that charge localization can influence electrostatic interactions and, therefore, the conformational dynamics of molecular ions [49, 50]. To account for the charge state influence on the CCS, energy optimization steps are performed after the charge site assignments for all candidate structures. All simulations were performed using an NVT thermostat using AMBER03 force field in YASARA software (http://www.yasara.org). The molecular dynamic simulations yielded identity vectors that cover the transition from the N to the denatured UA states.
Theoretical CCSHe were calculated using the MOBCAL software for helium [51]. To account for the reported 15% underestimation of CCS that occurs when using the PA algorithm [17, 52], all CCS calculations were scaled accordingly. Correlation between CCSHe and CCSN2 was performed using previously reported CCS values for denatured and native proteins [24]. That is, CCS(N2) = 0.9916*CCS(He) + 524.66 (native) and CCS(N2) = 1.174 CCS(He) + 466.34 (denatured) equations were used for holoMb and apoMb conversion, respectively.
Results and Discussion
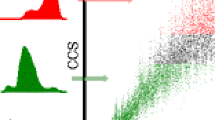
Confirmation of the presence of the holo and apo forms of Mb in the starting solution was performed using mass spectrometry (see Supplemental Figure 1). An acid-induced transition from holoMb to apoMb was used to generate both forms, which was easily confirmed by the mass shift related to the loss of the prosthetic group in apoMb. The acid-induced transition is also accompanied by protein unfolding and yields a characteristic charge state distributions (in good agreement with previous experiments [14, 18]). In particular, the loss of the heme, which influences overall protein flexibility, accompanied by the deviation from physiological conditions results in a large distribution of charge states for apoMb (e.g., [M + 9H]+9–[M + 19H]+19). Changes in the protein flexibility as a function of the charge state for the holo and apoMb were easily detected by TIMS-MS (see Figure 1 for IMS projections of the holo [M + 8H]+8–[M + 9H]+9 and apo [M + 9H]+9–[M + 19H]+19 forms of Mb). Previous IMS studies of Mb reported IMS bands with a similar trend [19, 53, 54]; however, a larger number of mobility bands was observed as a result of the higher mobility resolution of the TIMS analyzer. The presence of the heme, in particular the mobility band corresponding to the [M + 8H]+8 of the holoMb, illustrates the structural rigidity that can inhibit the adoption of multiple conformers. To simplify the discussion, the peak width of the holoMb [M + 8H]+8 was used as the criterion to propose the minimum width of an IMS band corresponding to a single resolved conformation, or a population of conformations indistinguishable by IMS. The experimental data also illustrates that larger CCS values were observed for ions of higher charge state (see Table 1). In addition, inspection of Figure 1 shows that the charge state has a large influence on the protein stability. For example, as the charge state increases, the holoMb changes from a single to a bimodal IMS distribution (in good agreement with [53]). In the case of the apoMb, the loss of the heme destabilizes the interaction between the A, G, and H helices and makes the charge state effect more pronounced.
IMS data reflect changes in the protein folding (average of size) and can provide an indirect measurement of the solution states; however, when complemented with theoretical calculations, a more detailed description can be achieved (e.g., discrimination between a protein expansion preserving a folded state or an extended state is possible). That is, changes in the CCS values can be interpreted in a similar fashion to the radius of gyration in order to describe a change in a protein conformation, typically an increase in size [55]. Deviations in the radius of gyration for N and MG states of the holoMb (19 ± 1 and 23 ± 2, respectively) are in good correspondence with the small CCS variation for the [M + 8H]+8 and [M + 9H]+9 holoMb conformers; this small change in CCS is in good agreement with a previous observation of similar folding motifs for the N and MG states [56]. In the case of apoMb, larger differences in CCS from the [M + 9H]+9 suggest the presence of several intermediate conformations of varying degrees of unfolding that occur in the N → MG → UA transition pathway. Here, the increase in CCS observed across the apoMb charge state distributions, particularly [M + 10H]+10–[M + 14H]+14 suggests Xn intermediate conformations, where Xn corresponds to intermediate unfolded states that occur in the process of Mb unfolding prior to reaching the rod-like chain characteristic of the UA state. Here the Xn notation refers to conformations that are not characteristic of previously identified transitions (i.e., N → MG → UA) and are, therefore, intermediates. Previous studies have identified specific structure intermediates [7, 57, 58]. Our experimental observations suggest that the process of Mb unfolding occurs via several transitions, N → MG → Xn → UA, where n refers to several conformational transitions as illustrated by the apoMb CCS distribution. That is, in the absence of the heme, it is expected that Mb will exist in several intermediate denatured states before complete unfolding to the UA form. Conformers corresponding to lowest charge state of the apoMb form, [M + 9H]+9, are then most likely attributed to the MG state or intermediate unfolded states that can occur via the MG → UA transition. The progressive increase in CCS with ion charge state suggests that unfolding is the result of the loss of the stabilizing intramolecular protein interactions via the absence of the porphyrin heme that have been reported as essential to both maintaining and allowing refolding of Mb [2]. CCS increase with charge state is also expected since it may/can be accompanied by stronger Coulombic repulsion from the protonated residue side chains. For higher charge states of apoMb, [M + 14H]+14–[M + 19H]+19, the number of IMS bands decreases, suggesting that further protein unfolding is not energetically favorable and/or the unfolding intermediates adopt comparable conformation, resulting in similar CCS values. This trend may be related to a point of stability in the Xn → UA transition; that is, the protonation sites are distant and minimize the Coulombic repulsion that was the driving force resulting in an increase in CCS. UA structures have been reported when harsher conditions are employed, but the corresponding CCS is much larger (>4300 Å2) than experimentally obtained in the current study [7].
H/D back exchange experiments were conducted under the same conditions as the CCS measurements. The number of IMS bands did not show any variation over time; however, in a 5–2000 ms time frame, the large number of collisions in the TIMS cell permitted the study of the H/D back exchange rate (Figure 2 and Table 1). No major changes were observed for the holoMb and apoMb [M + 9H]+9 HDX distribution (rate and number of exchanges), which suggests that despite the absence of the heme, no major changes in the protein conformation occurs; that is, the porphyrin cavity is preserved. There is, however, a general expansion of the protein following the loss of the porphyrin heme and/or a loosening of the porphyrin heme cavity, confirmed by the increase in CCS. This result can be interpreted as the transition from holoMb to apoMb resulting in the adoption of the MG conformation from the N state. That is, this change in conformation does not yield a difference in the extent of the unfolding [9] and, therefore, should not be accompanied by additional hydrogen-deuterium exchange. This information was used in the candidate assignment of the [M + 9H]+9 apoMb potential candidates (see Supplemental Material, which includes both compact and partially unfolded conformations).
A comparison in the HDX exchange observed for the higher apoMb charge states, [M + 10H]+10–[M + 19H]+19 (Figure 2), yields similar rates, suggesting a commonality in the unfolding pathway. The increase in the number of exchanges for each of the charge states can be related to an unfolding of the protein exposing additional inner cavity amino acid residues.
Candidate conformers have been proposed for all IMS bands (see Figure 3 for the most stable conformation for each identified IMS band). Inspection of the candidate structures pertaining to the protein unfolding observed in Mb suggest that unfolding occurs via the N → MG → Xn transition. That is, several intermediates (Xn) are observed in the [M + 10H]+10 – [M + 19H]+19 apoMb charge states, which lack a native tertiary structure as a result from the loss of the interior cavity structure (shown in red in Figure 3) with further unfolding accompanying each additional protonation. Theoretical CCS calculations also suggest that the experimental conditions (e.g., charge state distribution) utilized here are not harsh enough to populate the UA state (e.g., pH 2 would yield unfolding to the rod-like UA state). Close similarity between the candidate structures for the [M + 14H]+14 –[M + 19H]+19 apoMb charge states suggests that intramolecular Coulombic repulsion aides in stabilizing the conformational motifs presented (Figure 4). This can be corroborated by the pairwise RMSD comparisons of conformations that occur initially via the N → MG → Xn transition (i.e., [Mholo + 9H]+9 → [MApo + 9H]+9 → [MApo + 10H]+10]). For example, the increase in RMSD observed for these transitions in comparison to RMSD within a charge state infers larger differences in the three-dimensional structure that is accompanied by the loss of important protein stabilizing interactions. This can be further corroborated by a comparison of the secondary structure composition of the conformations; an increase in charge state results in an increase in random coil contribution in place of α-helix content (see Supplemental Information for more details).
Comparison of the most stable candidate structures corresponding to each identified conformation from Figure 1 for both the holo and apo forms of myoglobin. Heme cavity is depicted in red
Pathway of protein unfolding following the transition from holo to apomyoglobin. Initial loss of the heme results in a loosening of the heme cavity, while additional unfolding requires the loss of essential interactions between the alpha helices. The alpha helices have been depicted in purple (A, residues 4–19), blue (B, residues 21–37), cyan (C, residues 40–44), green (D, residues 52–59), light green (E, residues 61–78), yellow (F, residues 83–97), orange (G, residues 101–120), and red (H, residues 127–151)
Experimental and theoretical results did not result in compact denatured structures as previously reported for Mb intermediate states [7]. On the contrary, our results suggest that only unfolded, elongated intermediates correspond to the [M + 10H]+10–[M + 19H]+19 apoMb states with varying degrees of unfolding related to the loss of the interactions between helices A and G that are important to maintaining the globular shape [59]. This loss takes place upon the MG → Xn transition for the [M + 10H]+10 apoMb charge state. Helices A, G, and H become stable early in the protein folding process [60, 61] and the loss of the interaction of the helices may reflect the existence of multiple tertiary structures (or structural diversity by the lack of constraints).
Conclusion
The potential of simultaneous interrogation of protein conformations using IMS combined with HDX has been applied to the study of Mb kinetic intermediates. In particular, the unique capability of TIMS to sample the molecular ions after their formation over a large time scale (1–2000 ms) and simultaneously acquire CCS and HDX data permits a better screening of candidate structures. This translates into the possibility to postulate or confirm folding pathways and intermediates during Mb protein unfoldings: N → MG → Xn → UA. That is, results confirm the existence of intermediate denatured states, Xn, which occur prior to completely unfolding to UA. It was shown that the Xn intermediates are a consequence of the destabilization of the interactions between A and G helices. The ability to simultaneously study multiple conformation (using HDX and IMS) using a TIMS analyzer offers a unique advantage for time-resolved conformational studies of biomolecules.
References
Kendrew, J.C., Bodo, G., Dintzis, H.M., Parrish, R.G., Wyckoff, H., Phillips, D.C.: A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature 181, 662–666 (1958)
Jennings, P.A., Wright, P.E.: Formation of a molten globule intermediate early in the kinetic folding pathway of apomyoglobin. Science 262, 892–896 (1993)
Eliezer, D., Yao, J., Dyson, H.J., Wright, P.E.: Structural and dynamic characterization of partially folded states of apomyoglobin and implications for protein folding. Nat. Struct. Biol. 5, 148–155 (1998)
Scott, E.E., Gibson, Q.H., Olson, J.S.: Mapping the pathways for O-2 entry into and exit from myoglobin. J. Biol. Chem. 276, 5177–5188 (2001)
Ji, H.F., Shen, L., Grandori, R., Muller, N.: The effect of heme on the conformational stability of micro-myoglobin. FEBS J. 275, 89–96 (2008)
Olson, J.S., Phillips, G.N.: Myoglobin discriminates between O-2, NO, and CO by electrostatic interactions with the bound ligand. J. Biol. Inorg. Chem. 2, 544–552 (1997)
Kamatari, Y.O., Ohji, S., Konno, T., Seki, Y., Soda, K., Kataoka, M., Akasaka, K.: The compact and expanded denatured conformations of apomyoglobin in the methanol–water solvent. Protein Sci. 8, 873–882 (1999)
Kamei, T., Oobatake, M., Suzuki, M.: Hydration of apomyoglobin in native, molten globule, and unfolded states by using microwave dielectric spectroscopy. Biophys. J. 82, 418–425 (2002)
Eliezer, D., Wright, P.E.: Is apomyoglobin a molten globule? Structural characterization by NMR. J. Mol. Biol. 263, 531–538 (1996)
Koeniger, S.L., Merenbloom, S.I., Clemmer, D.E.: Evidence for many resolvable structures within conformation types of electrosprayed ubiquitin ions. J. Phys. Chem. B. 110, 7017–7021 (2006)
Kahsai, A.W., Rajagopal, S., Sun, J.P., Xiao, K.H.: Monitoring protein conformational changes and dynamics using stable-isotope labeling and mass spectrometry. Nat. Protoc. 9, 1301–1319 (2014)
Akke, M.: Conformational dynamics and thermodynamics of protein-ligand binding studied by NMR relaxation. Biochem. Soc. T. 40, 419–423 (2012)
Wales, T.E., Engen, J.R.: Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 25, 158–170 (2006)
Dobo, A., Kaltashov, I.A.: Detection of multiple protein conformational ensembles in solution via deconvolution of charge-state distributions in ESI MS. Anal. Chem. 73, 4763–4773 (2001)
Suckau, D., Shi, Y., Beu, S.C., Senko, M.W., Quinn, J.P., Wampler III, F.M., McLafferty, F.W.: Coexisting stable conformations of gaseous protein ions. Proc. Natl. Acad. Sci. U. S. A. 90, 790–793 (1993)
Horn, D.M., Breuker, K., Frank, A.J., McLafferty, F.W.: Kinetic intermediates in the folding of gaseous protein ions characterized by electron capture dissociation mass spectrometry. J. Am. Chem. Soc. 123, 9792–9799 (2001)
Hewitt, D., Marklund, E., Scott, D.J., Robinson, C.V., Borysik, A.J.: A hydrodynamic comparison of solution and gas phase proteins and their complexes. J. Phys. Chem. B. 118, 8489–8495 (2014)
Konermann, L., Douglas, D.J.: Equilibrium unfolding of proteins monitored by electrospray ionization mass spectrometry: distinguishing two-state from multi-state transitions. Rapid Commun. Mass Spectrom. 12, 435–442 (1998)
Hopper, J.T.S., Oldham, N.J.: Collision induced unfolding of protein ions in the gas phase studied by ion mobility-mass spectrometry: the effect of ligand binding on conformational stability. J. Am. Soc. Mass Spectrom. 20, 1851–1858 (2009)
Wang, F., Tang, X.J.: Conformational heterogeneity and stability of apomyoglobin studied by hydrogen deuterium exchange and electrospray ionization mass spectrometry. Biochemistry 35, 4069–4078 (1996)
Shvartsburg, A.A., Li, F., Tang, K., Smith, R.D.: Characterizing the structures and folding of free proteins using 2-D gas-phase separations: observation of multiple unfolded conformers. Anal. Chem. 78, 8575 (2006)
Shvartsburg, A.A., Smith, R.D.: High-resolution differential ion mobility spectrometry of a protein. Anal. Chem. 85, 10–13 (2013)
Jurneczko, E., Barran, P.E.: How useful is ion mobility mass spectrometry for structural biology? The relationship between protein crystal structures and their collision cross sections in the gas phase. Analyst 136, 20–28 (2011)
Bush, M.F., Hall, Z., Giles, K., Hoyes, J., Robinson, C.V., Ruotolo, B.T.: Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology. Anal. Chem. 82, 9557–9565 (2010)
Ruotolo, B.T., Benesch, J.L.P., Sandercock, A.M., Hyung, S.-J., Robinson, C.V.: Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 3, 1139–1152 (2008)
Valentine, S.J., Counterman, A.E., Clemmer, D.E.: A database of 660 peptide ion cross sections: use of intrinsic size parameters for bona fide predictions of cross sections. J. Am. Soc. Mass Spectrom. 10, 1188–1211 (1999)
Tao, L., McLean, J.R., McLean, J.A., Russell, D.H.: A collision cross-section database of singly-charged peptide ions. J. Am. Soc. Mass Spectrom. 18, 1232–1238 (2007)
Fernandez-Lima, F.A., Blasé, R.C., Russell, D.H.: A study of ion-neutral collision cross-section values for low charge states of peptides, proteins, and peptide/protein complexes. Int. J. Mass Spectrom. 298, 111–118 (2010)
Covey, T., Douglas, D.J.: Collision cross sections for protein ions. J. Am. Soc. Mass Spectrom. 4, 616–623 (1993)
Shi, H.L., Clemmer, D.E.: Evidence for Two new solution states of ubiquitin by IMS-MS analysis. J. Phys. Chem. B. 118, 3498–3506 (2014)
Wyttenbach, T., Pierson, N.A., Clemmer, D.E., Bowers, M.T.: Ion mobility analysis of molecular dynamics. Annu. Rev. Phys. Chem. 65, 175–196 (2014)
Li, H., Bendiak, B., Siems, W.F., Gang, D.R., Hill Jr., H.H.: Ion mobility-mass correlation trend line separation of glycoprotein digests without deglycosylation. Int. J. Ion Mobility Spectrom. 16, 105–115 (2013)
Zilch, L.W., Kaleta, D.T., Kohtani, M., Krishnan, R., Jarrold, M.F.: Folding and unfolding of helix-turn-helix motifs in the gas phase. J. Am. Soc. Mass Spectrom. 18, 1239–1248 (2007)
Niu, S., Rabuck, J.N., Ruotolo, B.T.: Ion mobility-mass spectrometry of intact protein–ligand complexes for pharmaceutical drug discovery and development. Curr. Opin. Chem. Biol. 17, 809–817 (2013)
Purves, R.W., Barnett, D.A., Ells, B., Guevremont, R.: Investigation of bovine ubiquitin conformers separated by high-field asymmetric waveform ion mobility spectrometry: cross section measurements using energy-loss experiments with a triple quadrupole mass spectrometer. J. Am. Soc. Mass Spectrom. 11, 738–745 (2000)
Rand, K.D., Pringle, S.D., Murphy III, J.P., Fadgen, K.E., Brown, J., Engen, J.R.: Gas-phase hydrogen/deuterium exchange in a traveling wave ion guide for the examination of protein conformations. Anal. Chem. 81, 10019–10028 (2009)
Fernandez-Lima, F.A., Kaplan, D.A., Park, M.A.: Note: Integration of trapped ion mobility spectrometry with mass spectrometry. Rev. Sci. Instr. 82, 126106 (2011)
Fernandez-Lima, F., Kaplan, D., Suetering, J., Park, M.: Gas-phase separation using a trapped ion mobility spectrometer Int. J. Ion Mobility Spectrom. 14, 93–98 (2011)
Hernandez, D.R., DeBord, J.D., Ridgeway, M.E., Kaplan, D.A., Park, M.A., Fernandez-Lima, F.: Ion dynamics in a trapped ion mobility spectrometer. Analyst 139, 1913–1921 (2014)
Silveira, J.A., Ridgeway, M.E., Park, M.A.: High resolution trapped ion mobility spectrometery of peptides. Anal. Chem. 86, 5624–5627 (2014)
Schenk, E.R., Ridgeway, M.E., Park, M.A., Leng, F., Fernandez-Lima, F.: Isomerization kinetics of AT hook decapeptide solution structures. Anal. Chem. 86, 1210–1214 (2014)
Molano-Arevalo, J.C., Hernandez, D.R., Gonzalez, W.G., Miksovska, J., Ridgeway, M.E., Park, M.A., Fernandez-Lima, F.: Flavin adenine dinucleotide structural motifs: from solution to gas-phase. Anal. Chem. 86, 10223–10230 (2014)
Schenk, E., Nau, F., Fernandez-Lima, F.: Theoretical predictor for candidate structure assignment from IMS data of biomolecule-related conformational space. Int. J. Ion Mobility Spectrom., submitted (2014)
McDaniel, E.W., Mason, E.A.: Mobility and Diffusion of Ions in Gases. John Wiley and Sons, Inc., New York, NY pp. 276 (1973)
Evans, S.V., Brayer, G.D.: High-resolution study of the three-dimensional structure of horse heart metmyoglobin. J. Mol. Biol. 213, 885–897 (1990)
Fernandez-Lima, F.A., Wei, H., Gao, Y.Q., Russell, D.H.: On the structure elucidation using ion mobility spectrometry and molecular dynamics. J. Phys. Chem. A. 113, 8221–8234 (2009)
Bogatyreva, N.S., Ivankov, D.N.: The relationship between the solvent-accessible surface area of a protein and the number of native contacts in its structure. Mol. Biol. 42, 932–938 (2008)
Kaltashov, I.A., Mohimen, A.: Estimates of protein surface areas in solution by electrospray ionization mass spectrometry. Anal. Chem. 77, 5370–5379 (2005)
Susa, A.C., Mortensen, D.N., Williams, E.R.: Effects of cations on protein and peptide charging in electrospray ionization from aqueous solutions. J. Am. Soc. Mass Spectrom. 25, 918–927 (2014)
Onufriev, A., Case, D.A., Bashford, D.: Structural details, pathways, and energetics of unfolding apomyoglobin. J. Mol. Biol. 325, 555–567 (2003)
Mesleh, M.F., Hunter, J.M., Shvartsburg, A.A., Schatz, G.C., Jarrold, M.F.: Structural information from ion mobility measurements: effects of the long-range potential. J. Phys. Chem. 100, 16082–16086 (1996)
Shvartsburg, A.A., Jarrold, M.F.: An exact hard-spheres scattering model for the mobilities of polyatomic ions. Chem. Phys. Lett. 261, 86–91 (1996)
Shelimov, K.B., Jarrold, M.F.: Conformations, unfolding, and refolding of apomyoglobin in vacuum: an activation barrier for gas-phase protein folding. J. Am. Chem. Soc. 119, 2987–2994 (1997)
Pringle, S.D., Giles, K., Wildgoose, J.P., Williams, J.P., Slade, S.E., Thalassinos, K., Bateman, R.H., Bowers, M.T., Scrivens, J.H.: An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave IMS/oa-ToF instrument. Int. J. Mass Spectrom. 261, 1--12 (2007)
Eliezer, D., Jennings, P.A., Wright, P.E., Doniach, S., Hodgson, K.O., Tsuruta, H.: The radius of gyration of an apomyoglobin folding intermediate. Science 270, 487–488 (1995)
Ohgushi, M., Wada, A.: Molten-globule state—a compact form of globular-proteins with mobile side-chains. Febs Lett. 164, 21–24 (1983)
Fink, A.L., Oberg, K.A., Seshadri, S.: Discrete intermediates versus molten globule models for protein folding: characterization of partially folded intermediates of apomyoglobin. Fold. Design 3, 19–25 (1998)
Tcherkasskaya, O., Ptitsyn, O.B.: Molten globule versus variety of intermediates: influence of anions on pH-denatured apomyoglobin. FEBS Lett. 455, 325–331 (1999)
Nishimura, C., Dyson, H.J., Wright, P.E.: Identification of native and non-native structure in kinetic folding intermediates of apomyoglobin. J. Mol. Biol. 355, 139–156 (2006)
Yao, J., Chung, J., Eliezer, D., Wright, P.E., Dyson, H.J.: NMR structural and dynamic characterization of the acid-unfolded state of apomyoglobin provides insights into the early events in protein folding. Biochemistry 40, 3561–3571 (2001)
Lecomte, J.T.J., Sukits, S.F., Bhattacharya, S., Falzone, C.J.: Conformational properties of native sperm whale apomyoglobin in solution. Protein Sci. 8, 1484–1491 (1999)
Acknowledgments
This work was supported by the National Institute of Health (grant no. R00GM106414) and a FFL Bruker Daltonic, Inc. fellowship. The authors acknowledge the Instructional and Research Computing Center (IRCC) at Florida International University for providing high performance computing resources that have contributed to the results reported within this research.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
Supplemental Information: Supporting material includes experimental mass spectrometry data of the myoglobin protein at different pH values (Supplemental Figure S1), all conformations identified for each charge state distribution (Figures S2–S76). Tables characterizing Mb conformations (Supplemental Tables S1 and S2) as well as a table depicting the secondary structure composition of all conformations have been included. PDB files for all candidate structures are included. (PDF 27386 kb)
Rights and permissions
About this article
Cite this article
Schenk, E.R., Almeida, R., Miksovska, J. et al. Kinetic Intermediates of Holo- and Apo-Myoglobin Studied Using HDX-TIMS-MS and Molecular Dynamic Simulations. J. Am. Soc. Mass Spectrom. 26, 555–563 (2015). https://doi.org/10.1007/s13361-014-1067-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-014-1067-9