Abstract
Triticale (x Triticosecale Wittmack) is a relatively new cereal crop. In Poland, triticale is grown on 12 % of arable land (http://www.stat.gov.pl). There is an increasing interest in its cultivation due to lowered production costs and increased adaptation to adverse environmental conditions. However, it has an insufficient tolerance to the presence of aluminum ions (Al3+) in the soil. The number of genes controlling aluminum tolerance in triticale and their chromosomal location is not known. Two F2 mapping biparental populations (MP1 and MP15) segregating for aluminum (Al) tolerance were tested with AFLP, SSR, DArT, and specific PCR markers. Genetic mapping enabled the construction of linkage groups representing chromosomes 7R, 5R and 2B. Obtained linkage groups were common for both mapping populations and mostly included the same markers. Composite interval mapping (CIM) allowed identification of a single QTL that mapped to the 7R chromosome and explained 25 % (MP1) and 36 % (MP15) of phenotypic variation. The B1, B26 and Xscm150 markers were 0.04 cM and 0.02 cM from the maximum of the LOD function in the MP1 and MP15, respectively and were highly associated with aluminum tolerance as indicated by Kruskal–Wallis nonparametric test. Moreover, the molecular markers B1, B26, Xrems1162 and Xscm92, previously associated with the Alt4 locus that encoded an aluminum-activated malate transporter (ScALMT1) that was involved in Al tolerance in rye (Secale cereale) also mapped within QTL. Biochemical analysis of plants represented MP1 and MP15 mapping populations confirmed that the QTL located on 7R chromosome in both mapping populations is responsible for Al tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Triticale (x Triticosecale Wittmack) is a relatively new cereal crop (Mergoum et al. 2009). In Poland, triticale is grown on 12 % of arable land (http://www.stat.gov.pl). There is an increasing interest in its cultivation due to better adaptation to adverse environmental conditions, efficient utilization of nutrients as compared to wheat (González et al. 2005), and increased amount of lysine (Boros 2002). Triticale grain is a valuable component of poultry and swine fodder (Myer and Barnett 2004). It contains more protein than rye. Moreover digestibility of triticale grain is higher compared to barley and similar to wheat (van Barneveld 1999). These properties favor triticale for low input farming systems.
Acidic soils constitute at least 40 % of Earth’s arable land (Kochian 1995). The geological origin of the soil, farming practices, and industrial pollution are the most likely causes for acidification. Plant tolerance to the presence of aluminum ions (Al3+) is an essential trait that allows growth and reasonable grain yield under such stressful conditions.
Under alkaline or neutral pH, aluminum persists in virtually insoluble inorganic complex compounds; thus, it is not present in ionic form in the soil solution. These insoluble complexes are not harmful to plants. However, at pH < 5.0, Al3+ ions are released to the soil solution (Williams 1999). The soluble Al3+ ions are taken up by plant roots and interrupt cap cell division, which results in a reduction of cell wall extension growth and reduction of DNA replication caused by binding of Al3+ to phosphate groups (Foy 1992). This process results in the inhibition of root elongation and reduction of growth of the root system, which decreases crop yield (Matsumoto et al. 1996).
One of the mechanisms of plant tolerance involves immobilization of the Al3+ ions in the rhizosphere by exudation of chelating organic acids from roots. The organic acids can accumulate in root cells and be secreted to the soil (Pellet et al. 1996). Alternatively, the root system may increase the pH in the extracellular environment of root apices (Henderson and Ownby 1991; Dengenhardt et al. 1998).
Hexaploid triticale is a synthetic species with a complex genome that contains two wheat (AABB) and one rye (RR) genomes. Its tolerance to Al3+ ions is intermediate between wheat and rye (Anioł and Gustafson 1984). The combined functions of aluminum tolerance genes from rye and wheat control this trait in triticale. However, there are unknown factors in triticale that influence gene expression (Anioł and Gustafson 1984). Studies of substitution lines in hexaploid triticale resulted in assignment of the trait to the 3R chromosome (Ma et al. 2000; Budzianowski and Woś 2004). However, in octoploid triticale, it appears that the wheat genome is responsible for aluminum tolerance. The trait is associated with citrate exudation which is due to the origin of the species where wheat cv Carazinho was tolerant to the presence of Al3+ via this mechanism (Stass et al. 2008).
Studies with ditelosomic and nullitetrasomic lines of the wheat cultivar Chinese Spring assigned aluminum-tolerance genes to the 2D, 4D, and 5A chromosomes (Takagi et al. 1983). Similar results were obtained by Anioł (1990). Chromosomes 2A, 6A, 7A, 3B, 4B, 3D, and 7D code for minor Al-tolerance factors (Anioł and Gustafson 1984; Cai et al. 2008; Ryan et al. 2009). In rye, at least five different loci were identified that control aluminum tolerance, including Alt1, Alt2, Alt3, Alt4 and Alt5 which localized on chromosomes 6RS, 3RS, 4RL, 7RS and 7RS, respectively (Anioł and Gustafson 1984; Ma et al. 2000; Matos et al. 2005; Benito et al. 2010; Silva-Navas et al. 2011).
The known aluminum-tolerance genes code for organic acid transporters that are either aluminum-activated malate transporters (ALMT) (Sasaki et al. 2004; Hoekenga et al. 2006; Ligaba et al. 2006; Fontecha et al. 2007) or multidrug and toxin efflux (MATE) proteins involved in the transport of citrate activated by aluminum (Furukawa et al. 2007; Magalhaes et al. 2007; Silva-Navas et al. 2011). In wheat, as well as in rye, both genes were detected in mapping populations (Sasaki et al. 2004; Ryan et al. 2009; Silva-Navas et al. 2011). The wheat malate transporter gene TaALMT (Sasaki et al. 2004) was mapped to chromosome 4DL (Raman et al. 2005). Its rye ortholog (ScALMT) was mapped to Alt4 locus on the 7RS chromosome (Fontecha et al. 2007; Collins et al. 2008). Gene responsible for Al induced citrate efflux (AACT1) was located on chromosome 4BL (Ryan et al. 2009) in wheat and on 7RS in rye (Silva-Navas et al. 2011). The ScAACT1 gene co-segregated with Alt5 locus localized 25 cM apart from the ScALMT1 gene (Silva-Navas et al. 2011). Silva-Navas et al. (2011) suggested that ALMT and MATE family genes may have been controlled by a gene localized on 3R chromosome. Its DNA sequence exhibited homology to the Arabidopsis STOP1 transcriptional factor (Liu et al. 2009; Silva-Navas et al. 2011) that regulates expression of the Al tolerant genes while being phosphorylated.
The availability of genetic maps is immensely useful for any molecular breeding program. Such maps exist for rye (Korzun et al. 2001; Khlestkina et al. 2004; Matos et al. 2007; Bolibok-Brągoszewska et al. 2009; Milczarski et al. 2011) and wheat (Gale et al. 1995; Röder et al. 1998; Somers et al. 2004; Akbari et al. 2006; Crossa et al. 2007). González et al. (2005) developed the genetic map of triticale saturated with AFLPs, RAPDs, RAMPs, and SSRs. Recently, Tyrka et al. (2011) elaborated a DArT, SSR, and AFLP based genetic map of the species. Later Alheit et al. (2011) presented triticale map based exclusively on DArT markers. Using association mapping and indirect mapping we showed that Al tolerance in triticale is encoded by several QTLs assigned to the 3R, 4R, 6R and 7R chromosomes (Niedziela et al. 2012). However, a lack of detailed information on the location of aluminum-tolerance genes in triticale motivated further work to evaluate biparental mapping populations and their saturation with different marker systems. We also used markers linked to the ALMT gene family that participates in the expression of the trait in cereals (Miftahudin et al. 2004, 2005) and is known to be involved in Al tolerance to verify whether those genes also play the same role in triticale.
The aim of the study was to identify molecular markers linked to and/or associated with Al-tolerance in triticale using specially designed biparential mapping populations and to verify which gene is responsible for the expression of the trait in the given case.
Materials and methods
Plant material
Two biparental mapping populations, namely MP1 and MP15 were derived via crossing individual double haploid plants originating from several individuals of cv. Bogo. The parental forms of the mapping populations differed in aluminum tolerance. In both cases, the tolerant plant was used as the pollinator. The F1 plants were isolated to prevent cross pollination. The F2 seeds collected from seedlings were used for phenotyping, following their cultivation in greenhouse. Each of the two populations comprised 94 individuals.
Aluminum tolerance test
The physiological test of aluminum tolerance described by Anioł (1984) was used. Sterilized seeds were placed on moist filter paper in Petri dishes and kept at 10 °C for 24 h. Germinated seedlings were transferred onto a polyethylene net supported by polystyrene foam, and placed in a tray filled with basic medium (2.0 mM CaCl2, 3.25 mM KNO3, 1.25 mM MgCl2, 0.5 mM (NH4)2SO4, and 0.2 mM NH4NO3, pH 4.5). The seedlings were maintained under a controlled environment (25 °C, a 12 h day/night photoperiod, and light intensity 40 Wm−2) in a growth cabinet (POL-EKO-APARATURA, ST500 B40 FOT10). After three days, seedlings were transferred for 24 h onto the same medium containing Al3+ ions (16 ppm, 0.59 mM) in the form of AlCl3. The plants were washed in water and then placed again in the basic medium for 48 h. Aluminum tolerance was evaluated by measuring root regrowth visualized after root staining with 0.1 % Eriochrome Cyanine R for 10 min.
Al treatment leads to irreversible damage of apical meristem of seedlings. Eriochrome Cyanine R binds to those regions resulting in dark purple color of the damaged roots. Tolerant and moderately tolerant seedlings preserve their ability to root grow after the Al3+ is removed from the solution. The newly growing root parts are not stained with Eriochrome Cyanine R. The length of the regrowth measured in millimeters reflects the level of plant tolerance to the presence of toxic Al3+ ions.
Genotyping
DNA isolation
Isolation of genomic DNA was performed using fresh leaves of 14-days old seedlings and the Plant DNeasy Mini Kit (Qiagen), followed by spectrophotometric quantification (NanoDrop ND-1000) and integrity testing on a 1.0 % agarose gel.
Amplified fragment length polymorphism (AFLP), simple sequence repeats (SSR), diversity arrays technology (DArT), and specific markers were used to profile the F2 mapping populations.
Amplified fragment length polymorphism analysis
The AFLP procedure followed that described previously by Vos et al. (1995). Genomic DNA (500 ng) representing individual plants was digested with EcoRI/MseI endonucleases (37 °C for 3 h; 70 °C for 15 min) followed by ligation to respective adapters (20 °C for 12 h). Pre-selective [(94 °C for 30 s; 56 °C for 60 s; 72 °C) × 19 cycles] and selective amplifications using various combinations of EcoRI (E) and MseI (M) primers (Table 1) with three selective nucleotides were performed. Reaction mixture volumes were 10 μl and contained 1.5 μl of diluted pre-amplification products, 0.025 μl HotStart DNA Polymerase, 0.5 μl MseI primer, 0.5 μl 32P-labeled EcoRI primer, 0.5 μl 10 mM dNTPs, 1.0 μl 25 mM MgCl2, 1.0 μl 10× PCR buffer (Qiagen), and distilled water up to the required volume. PCR reactions were performed using a PTC-225 Peltier Thermal Cycler (MJ Research) with the following thermal profiles: [95 °C, 15 min] [94 °C, 30 s; 65 °C, 30 s (Ramp 0.7 °C/cycle); 72 °C, 60 s] × 12 [94 °C, 30 s; 56 °C, 30 s; 72 °C, 60 s] × 29 [72 °C, 10 min]. The PCR products then underwent denaturation and separation on a 7 % denaturing polyacrylamide gel followed by overnight exposure to X-ray films at −35 °C.
Simple sequence repeats analysis
SSR analysis included 157 rye markers (SCM) from the BAZ Database of Secale cereale microsatellites (http://www.bafz.de.) provided by the Federal Centre for Breeding Research on Cultivated Plants, GrosLusewitz; 20 rye-specific SSRs related to aluminum tolerance (SCM) obtained by Matos et al. (2007); 36 rye expressed microsatellite site (REMS) sequences made available to us by Dr. V. Korzun (Lochow Petkus GmbH, Bergen, Germany); and 46 Gatersleben wheat microsatellites (GWM) (Röder et al. 1998). These markers were amplified on DNA matrixes of the F2 mapping populations. Genomic DNA was amplified in 10 μl containing 10 ng temple DNA, 5.0 pmole of each primer labeled with 32P, 2.5 mM dNTPs, 1.5 mM MgCl2, 1× reaction buffer, and 0.25 u of HotStart DNA Polymerase. The thermal profiles followed those described in previous reports (Röder et al. 1998; Khlestkina et al. 2004; Matos et al. 2007).
Sequence-specific markers
Several PCR markers flanking Alt4 locus in rye (B1, B4, B11, B25, B26, B27, BCD1230) were tested on both mapping populations (Miftahudin et al. 2004, 2005; Benito et al. 2010). In total, 10 μl of the reaction mixture consisted of 10 ng of genomic DNA, 5.0 pmole of each primer labeled with 32P, 2.5 mM dNTPs, 1.5 mM MgCl2, 1× reaction buffer, and 0.25 u of HotStart DNA Polymerase. The thermal profiles followed those described in previous reports (Miftahudin et al. 2004, 2005). In addition, PCR products underwent separation on a 7 % denaturing polyacrylamide gel.
Diversity arrays technology
DArT profiling was performed by Triticarte Pty Ltd., Yarralumla, ACT, Australia with the current triticale assay (http://www.triticarte.com.au/).
Sequencing ladder
The marker length was evaluated using the sequencing ladder with the SequiTherm EXCELTM II (EPICENTRE® Biotechnology) sequencing kit.
Linkage analysis and QTL mapping
Data preparation
Molecular markers were transformed into binary (presence/absence) matrix for both mapping populations. Markers with a minor allele frequency of less than or equal to 5 % were removed from the analyses. Redundant markers were preliminary eliminated using agglomeration analysis (UPGMA) and Dice genetic distance in PAST software (Hammer et al. 2001). Markers separated by genetic distance lower than or equal to 5 % (formed marker assembly) were merged. Missing data was completed using information from the redundant markers of the given contiguous assembly. Only one representative of the given redundant marker assembly was retained, and information on the removed markers retained for further analysis. Such prepared data were loaded to Rcran software with installed qtl package (Broman 2010). All analysis were performed according to manual (Broman 2010). Individuals and markers with missing data below 70 % were omitted from further analysis using function subset.cross() and drop.markers(), respectively. The next step was to study the segregation patterns of the markers using function geno.table. Markers that show significant distortion at the 5 % level, after a Bonferroni correction for the multiple tests were removed from future analysis.
Genetic mapping
R/qtl package was used for linkage analysis (Broman 2010). Duplicated individuals, makers with numerous missing data were removed from further analysis prior to construct linkage groups (LGs). LGs were obtained using logarithm of odds (LOD) set to 15 and maximum recombination fraction (rf) equal to 0.35. The best order of markers was determined with orderMarkers function with the following parameters: Kosambi mapping function, error probability = 0.005, tolerance for determining convergence = 1e-6 and maximum number of EM (expectation–maximization) iterations = 10,000. To explore alternate orders “countxo” and “likelihood” methods were applied with window size up to 7. The “countxo” option determines the minimum number of crossing overs while the “likelihood” method calculates the post probable marker order within a given window. The linkage groups with the lowest number of crossing overs, maximum likelihood order, the lowest map length and the minimum gap size were used for future analysis. The marker order was verified visually using plotRF r/qtl function. Genotypic error was estimated and singletons were removed within r/qtl package according to the manual (Broman 2010).
Map comparison
To align genetics maps the marker order was considered and the maps were juxtaposed visually.
QTL detection
To perform composite interval mapping missing data were approximated via fill.geno function using Haley-Knott regression method implemented in the r/qtl package (Broman et al. 2003). Missing data were approximated via fill.geno function using Haley-Knott regression method. Mapping of quantitative trait loci (QTLs) was performed using function cim() with Haley-Knott regression method, Kosambi map function, error probability equal to 0.001. A 1000-permutations was used to evaluate empirical LOD thresholds for QTL significance determination at 0.05 alpha-value level. Explained phenotyping variance for a single QTL model was assessed using fitqtl() function.
Association between molecular markers and Al tolerance was tested using the non-parametric Kruskal-Wallis test implemented in MapQTL 5.0 software (van Ooijen 2004).
Organic acid assays
The eight F3 plants representing four Al non-tolerant (15/69, 15/85, 1/88, 1/24) and four tolerant lines (15/62, 15/44, 1/74, 1/81) originated from the F2 biparental mapping populations (MP1 and MP15) were used to perform malate and citrate efflux from excised root apices. The tests followed assays described by Delhaize et al. (1993). Sterilized seeds were placed on filter paper in Petri dishes and kept at 5 °C for 48 h. Germinated seedlings were transferred onto a polyethylene net supported by polystyrene foam, and placed in a tray filled with sterile 0.2 mM CaCl2 solution (pH 4.3) which was gently aerated with a pump. The seedlings were grown under a controlled environment (25 °C, a 12 h day/night photoperiod, and light intensity 40 Wm−2) in a growth cabinet (POL-EKO-APARATURA, ST500 B40 FOT10) for 7 days. Ten existed root apices (5 mm long) were placed in a 2 ml sample tube with 1 ml control solution (0.2 mM CaCl2, pH 4.3) to remove any malate and citrate released from the wounded tissue. Then the apices were resuspended in 1 ml treatment solution (0.2 mM CaCl2 with or without 100 μM AlCl3, pH 4.3) and incubated on a shaker (300 rpm) for 18 h. Malic acid in treatment solution was assayed using modifications of previously described enzymatic methods (Delhaize et al. 1993). Sample 0.68 ml was incubated in 0.75 ml of glycine buffer (0.5 M Gly, 0.4 M hydrazine hydrate, pH 9.0) and 50 μl 40 mM NAD+ solution. The reaction mixture was preincubated for 30 min to obtain a stable A 340 reading before the addition of 5 μl of malic dehydrogenase (2.2 mg/ml, Sigma-Aldrich). The amount of NADH formed in the reaction pathway is stoichiometric with the amount of malate in the sample.
Citric acid in treatment solution was analyzed using commercially available assay kit (K-CITR 07/11, Megazyme International Ireland Ltd.). All assays were conducted for the four-replicated treatments.
Statistics
The normal distribution of scores was verified by Shapiro–Wilk test to validate the use of parametric tests. Broad sense heritability coefficient (h2) for aluminum tolerance was estimated as the ratio of genotypic to the phenotypic variance. Variance components estimated of REML method were used to compute statistics (SAS 2009).
Statistical analysis of biochemical data was performed using the SAS ®System (SAS 2009). Bartlett’s test (Bartlett 1937) was performed first to determine the equality of variances among the specified groups. If significant differences were found with Bartlett’s test, then the Satterthwaite t-test (Satterthwaite 1946) was performed (SAS 2009).
Results
Aluminum tolerance test
Physiological tests revealed that parental forms of the mapping populations exhibited the lowest and the highest regrowth values in the presence of Al3+ ions, whereas the F2 individuals showed segregation of the trait. The observed root regrowth ranged from 0 to 2.0 cm. No perfect normal distribution in case of both mapping populations was observed.
Marker analysis
Depending on the mapping population and selective primers, the number of AFLPs ranged from 33 to 86 (Table 1). In total, 1077 (MP1) and 1060 (MP15) AFLPs were amplified on genomic DNAs of the mapping populations. Fifteen and 14 markers were polymorphic in the MP1 and MP15 populations, respectively (Tables 1 and 2). Out of 259 SCM, REMS, and GWM markers tested, 10.4 % appeared to be polymorphic between parental forms of the populations (Table 2). There were 53 polymorphisms in MP1 and 51 in MP15. They segregated as dominant and codominant (22) markers. DArT technology resulted in at least 153 (MP1) and 130 (MP15) polymorphisms (Table 2). The DNA sequences of two BAC clones converted by the others (Benito et al. 2010; Miftahudin et al. 2005) to the specific PCR-based markers (B1 and B26) segregated in both populations (Table 2). In total, there were 226 (MP1) and 200 (MP15) polymorphisms useful for construction of the linkage groups.
Data preparation for genetic mapping
After removal of redundant markers, their number dropped from 226 to 128 (MP1) and from 200 to 131 (MP15), respectively. Additionally, in MP1 population there were two individuals missing more than 50 % of genotypic data and three markers that missed about 70 % of markers shared among the individuals. In MP15 one individual with more than 60 % and eight markers missing for 70 % of genotypes were removed from further analysis. Moreover 29 and 23 markers in MP1 and M15 exhibited significant segregation distortion and were also omitted during mapping steps. In a consequence of preliminary analysis MP1 population consisted of 92 individuals and 96 makers, whereas MP15 of 93 individuals and 99 markers.
Genetic mapping
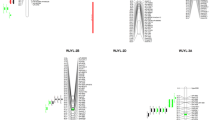
Linkage analysis allowed identification of three groups assigned to 5R, 7R and 2B chromosomes for both mapping populations (Table 3). Localization of the linkage groups was possible due to the known chromosomal assignment of DArTs and SSRs mapped in triticale (Tyrka et al. 2011), rye (Bolibok-Brągoszewska et al. 2009; Milczarski et al. 2011), and wheat (Akbari et al. 2006; Crossa et al. 2007; Francki et al. 2009). The linkage groups encompassed 4, 9, and 63 markers, and spanned over 10, 11, and 84 cM, respectively, for MP1 (Table 3). For MP15, linkage groups consisted of 4, 9, and 62 markers, and spanned over 10, 9, and 117 cM, respectively. Markers assigned to chromosome 5R occupied just two positions and were located at a distance of 10 cM in both populations (Fig. 1). The average density of markers assigned to chromosome 2B was one marker per 1.6 cM in the MP1 and 1.3 cM in the MP15 populations (Fig. 2). Markers representing linkage groups that mapped to chromosome 7R were present every 2.4 cM (MP1) and 3.0 cM (MP15), with the largest gap spanning over 6.4 cM for both mapping populations. The distribution of the markers was not uniform between the genomes, with 29.6 % (MP1) and 33.0 % (MP15) of markers mapping to rye, and 4.0 % (MP1) and 4.5 % (MP15) of markers mapping to wheat B genomes. We did not identify linkage groups representing the wheat A genome. Genotypic error was estimated to be equal to 0.03.
The alignment of the 5R linkages of the MP1 and MP15 mapping populations and 5R chromosome map published by Alheit et al. (2011). Common markers are indicated in red
The alignment of the 2B linkages of the MP1 and MP15 mapping populations 2B chromosome map published by Alheit et al. (2011). Common markers are indicated in red
Alignment of linkage groups for MP1 and MP15 populations with genetic maps of triticale and rye
Linkage groups assigned to 5R and 2B chromosomes and mapped in the MP1 and MP15 populations exhibited nearly perfect matching of marker order (Figs. 1 and 2). Comparison of the two linkage groups with the triticale genetic maps of chromosome 5R published by Alheit et al. (2011) demonstrated the presence of only one common marker (rPt-401561) (Fig. 1). Similarly, the order of DArTs also was congruent in the 2B linkage group (Fig. 2), with some shifts of a few markers within a short genetic distance. Alignment of the 2B linkage groups with maps by Alheit et al. (2011) showed the presence of one common marker (wPt-1294). In case of both mapping populations there were no common markers for chromosome 5R and 2B with the map published by Tyrka et al. (2011).
The linkage group mapped to the 7R chromosome in the MP1 population consisted of 37 markers. The same linkage group evaluated for the MP15 population had 39 markers. The number of markers increased almost double after adding their redundant counterparts and was 63 and 62 for MP1 and MP15 respectively (Table 3). There were at least 55 common markers shared between the two linkage groups. When comparing linkage group of MP1 with the 7R chromosome published by Tyrka et al. (2011) and by Alheit et al. (2011), 16 and 31 common markers were detected, respectively (Fig. 3). In the case of MP15, 14 and 28 markers common with Tyrka et al. (2011) and Alheit et al. (2011) maps were detected, respectively. Most of the markers exhibited colinearity.
QTL detection
Composite interval mapping (CIM) allowed identification of a single QTL that mapped to the 7R chromosome in both mapping populations (Table 3, Figs. 4 and 5). The maximum of the LOD function exceeded values of 6.1 (MP1) and 9.0 (MP15) (Table 4), which passed the permutation test with LOD cut-off values equal to 3.2 and 5.5, respectively. The 7R QTL showed an additive effect and explained 25.3 % (MP1) and 35.9 % (MP15) of phenotypic variance.
QTL composite interval mapping (CIM) profile of the 7R linkage group for the average length of root regrowth in the MP1 population. The LOD threshold is that obtained in R/qtl package (R-CRAN) after 1000 permutations (PT). CIM-LOD(max)) states for LOD maximum while QTL maximum reflects its position on genetic map
QTL composite interval mapping (CIM) profile of the 7R linkage group for the average length of root regrowth in the MP15 population. The LOD threshold is that obtained in R/qtl package (R-CRAN) after 1000 permutations (PT). CIM-LOD(max)) states for LOD maximum while QTL maximum reflects its position on genetic map
The B1, B26 and Xscm150 markers were 0.04 cM and 0.02 cM away from the LOD function maximum in the MP1 and MP15 populations, respectively (Table 4). The above-mentioned alleles originated from an aluminum-tolerant parent. Markers linked with the aluminum-tolerance QTL had positive additive effect (Table 4). Alignment of the linkage groups for chromosome 7R in the QTL region demonstrated that the markers were in the same order (Fig. 3) and the maximums of the LOD functions overlapped.
The analysis using Kruskal-Wallis test revealed that all markers located on chromosome 7R were significantly associated with aluminum tolerance in the case of both mapping populations. The highest significance level (p = 0.0001) and statistic value was observed in the case of B1_299; B26_287; Xscm150_213, EM5_156, Xrems1162_315, Xrems1234_278, Xrems1188_177, rPt-507379 and Xscm092_295 markers (Table 5). All those markers were assembled in the QTL function maximum. Markers located on remaining chromosomes were not associated with the trait.
Organic acid assays
The test for the presence of malate efflux in the root apices of triticale F3 tolerant and non-tolerant plants indicated that the samples treated with Al3+ ions differed according to their tolerance in the MP15 (t = 10.86, p < 0.001) and MP1 (t = 59.63, p < 0.0001) mapping populations. No differences were observed when the samples representing the same MP15 (t = 2.24, p = 0.2677) and MP1 (t = 0.45, p = 0.7323) lines were not treated with Al3+.
Similar tests for the presence of citric acid efflux stimulated by the Al3+ treatment revealed its absence in both mapping populations.
Heritability coefficient
Broad sense heritability coefficient (h2) for aluminum tolerance equaled to 0.996 (95 % confidence interval (CI), 0.995, 0.996) and 0.994 (95 % confidence interval (CI), 0.993, 0.995) in the case of MP1 and MP15 populations, respectively.
Discussion
The development of the first DArT-based genetic map of triticale (Tyrka et al. 2011) facilitated new research to map agricultural traits without the necessity of constructing saturated genetic maps of the species. Assuming that DArTs usually identify several thousand segregating polymorphisms (Wenzl et al. 2006; Crossa et al. 2007; Bolibok-Brągoszewska et al. 2009; Tyrka et al. 2011), the mapping populations used in the study should be capable of detecting enough markers to obtain saturated linkage groups. In the present work, a limited number of polymorphisms were detected with DArTs or other marker platforms. This could be due to the choice of parental forms derived via androgenesis from plants of the cultivar ‘Bogo’. We cannot exclude that the parental forms were somaclones, which were selected based on differences in Al tolerance. This would explain why few linkages were detected and why they were poorly saturated with markers (a homozygous genetic background within the genome regions that was different from that encoding Al tolerance) except the map of chromosome 7R. The parental forms seem to differ in their Al tolerance due to chromosomal rearrangements that are typical for triticale and predominantly affect the R genome (Oleszczuk et al. 2011). The somaclones, and the rearrangements affecting the Al tolerant locus but not the other genome regions, should limit the number of polymorphisms and linkages identified.
The majority of all mapped markers come from the R genome, whereas saturation of the wheat one was the poorest. A heterogeneous distribution of markers among the different triticale genomes (González et al. 2005; Alheit et al. 2011; Tyrka et al. 2011) as well as problems with mapping wheat genome in triticale (Francki et al. 2009) were reported previously. This phenomenon was explained by different polymorphic natures of the triticale genomes (González et al. 2005; Alheit et al. 2011; Badea et al. 2011; Tyrka et al. 2011). Higher saturation of the R genome than the wheat genome relates to the allogamous nature of the reproductive system of rye (González et al. 2005), or to the fact that it undergoes more frequent rearrangements than the wheat genome. This is due to the difference in the size of the rye and wheat chromosomes, which results in delayed replication of the R genome (Oleszczuk et al. 2011).
Alignment of DArT markers encompassing the linkage groups identified in the F2 mapping populations of MP1 and MP15, and those described for genetic maps of triticale (Alheit et al. 2011; Tyrka et al. 2011) and rye (Bolibok-Brągoszewska et al. 2009), demonstrated nearly perfect matching of the data. Thus, the order of the markers is consistent with previous results (Alheit et al. 2011; Tyrka et al. 2011) and confirms proper assignment of the linkage groups to the triticale chromosomes. In the case of the largest linkage group mapped to chromosome 7R, additional information on precise chromosomal assignment came from analyses with sequence-specific and microsatellite markers directed to Alt4. Benito et al. (2010) demonstrated that B1 and B26 sequence-specific markers and microsatellite Xscm92 are mapped to the chromosome 7R in rye. These markers also mapped on 7R in the MP1 and MP15 populations. The average density of markers forming linkage groups in the MP1 and MP15 populations (2.4 cM and 3.0 cM) was similar to that obtained by Alheit et al. (2011) and Tyrka et al. (2011). This marker density is sufficient for QTL mapping (Piepho 2000). However, our phenotypic data did not followed normal distribution. Deviation of a trait from normal distribution has been observed by the others (Singh et al. 2009; Septiningsih et al. 2003). The problem of skewed distribution could be theoretically overcome by transformation of the data or by using the non-parametric Kruskal–Wallis test. The test usually provides further confirmation of the marker trait association, and indicates that the results of the QTL analysis were not influenced by segregation distortion or non-normal distribution of certain traits (Singh et al. 2009). Considering such a problem in the case of our populations we correlated the genotypic data with aluminum tolerance using Kruskal-Wallis nonparametric test and succeeded in identification of highly associated markers. All those markers mapped exclusively to the chromosome 7R with the most associated markers spanning over 3.0 cM reflecting the presence of the QTL that was revealed by us using composite interval mapping.
The QTL analyses revealed that the locus that accounts for up to 25 % and 36 % of explained phenotypic variance were mapped on chromosome 7R for the F2 mapping populations of MP1 and MP15. The QTL location is in agreement with our previous results based on association mapping (at least in the case of the QTL on 7R), which showed that only the rye genome appears to be involved in Al tolerance in triticale (Niedziela et al. 2012). Thus, either Al tolerance in our populations is encoded exclusively by the R genome, or the known QTL responsible for the trait and mapped by the others (Sasaki et al. 2004) to wheat genome, might have explained only minor phenotypic variance in our mapping populations and were masked by the main QTL or were missing in the given plant materials. The presence of only a single QTL within our mapping populations seems to be supported by the lack of QTL located on chromosomes 3R (Alt2), 4RL (Alt3), 6RS (Alt1), and 7RS (Alt4, Alt5) (Anioł and Gustafson 1984; Gallego and Benito 1997; Miftahudin et al. 2002; Matos et al. 2005; Benito et al. 2010; Silva-Navas et al. 2011) in rye or that such QTL could be masked in triticale. Obviously, detection of such QTL based on the MP1 and MP15 populations might have been impossible due to small population sizes (or) and a limited number of polymorphic markers available for the ‘A’ and ‘B’ genomes, but above all without the availability of a genetic linkage map for the chromosomes of interest, except chromosome 7.
The aluminum-tolerance QTL mapped on chromosome 7R of triticale in this work occupies the same locus as Alt4 in rye (Miftahudin et al. 2005; Benito et al. 2010), therefore we suggest the gene encoding aluminum tolerance should be the same, with possibly different alleles in the two species. A confirmation of this hypothesis was obtained with the analyses of the sequence-specific markers (Miftahudin et al. 2004, 2005) and SSRs (Benito et al. 2010). The B26 marker mapped to the 7RS chromosome with a distance of 0.05 cM from the Alt4 locus in rye, whereas the B1 marker had a distance of 0.4 cM to Alt4 (Miftahudin et al. 2005). The B1 and B26 markers segregated in the MP1 and MP15 populations and linked to the aluminum-tolerance QTL of the chromosome 7R in triticale. The B1 marker was 0.04 cM from the QTL in the MP1 population and 0.02 cM from the QTL in the MP15 population. Marker B26 mapped in the same position as B1 for both populations. Moreover, the Xscm92 marker that segregates in rye and maps about 17.2 cM from the Alt4 locus (Benito et al. 2010) was identified in the 7R linkage group of the MP1 population, where it maps 0.04 cM away from the QTL maximum in MP1 and 20.3 cM in MP15. Moreover, our study demonstrated that the microsatellites Xrems1188, Xscm019 and Xscm150 previously localized on 7R (Hackauf and Wehling 2001; Khlestkina et al. 2004) also were linked to the B1 and B26 markers. These B1 and B26 sequence-specific markers are associated with the Alt4 locus that encodes the ScALMT1 (Fontecha et al. 2007), which is known to be involved in aluminum tolerance in rye via malate efflux mechanisms. Biochemical analysis of our data confirmed presence of malic acid in treatment solution containing Al3+ ions. Thus, we suggest that in rye and triticale, the same locus that is located on chromosome 7RS may control at least part of the aluminum tolerance response. By assuming that the markers linked with the aluminum-tolerance QTL had a positive additive effect, and considering the way that the mapping populations were arranged, the most tightly linked markers should prevail among tolerant individuals.
Considering the very few markers mapped on chromosome 3R we failed to detect a QTL on chromosome 3R in both mapping populations. However, this QTL was found by us (Niedziela et al. 2012) and the others (Ma et al. 2000; Budzianowski and Woś 2004) using association mapping. If detected it usually explains most of the available phenotypic variance of the trait (Ma et al. 2000; Budzianowski and Woś 2004). Recently, Silva-Navas et al. (2011) have suggested that it could be responsible for the expression of a putative transcriptional factor regulating Al tolerant genes located on 7R. Possibly the lack of this QTL may explain why only a single Al tolerance QTL mapped to the chromosome 7R was detected by us. As in both mapping populations Al tolerance was related to malic acid exudation one may speculate that the transcriptional factor activates gene coding for the expression of citric efflux.
The traits under control of additive genes have usually high heritability (Manal 2009; Ajmal et al. 2009). It was demonstrated that Al tolerance in triticale could be inherited in 85–89 % (Schlegel 1996; Zhang et al. 2001). Our results clearly support those data demonstrating that the heritability in triticale could be even greater reaching 99 %. Such great values of Al tolerance heritability were also published for Arabidopsis thaliana (Kobayashi and Koyama 2002). Broad sense heritability can be controlled by several factors (Ukai 2000). In case of aluminum tolerance such high values are undoubtedly connected with accurate determination of the phenotypes and slight environment effect due to control conditions of experiment. The results of phenotypic variance (25 % and 36 %) and heritability coefficient obtained by us may suggest that triticale selection for aluminum tolerance would be effective in MP1 and MP15 populations involving crosses of sensitive and tolerant parents and the QTL identified in this study could be efficiently transferred to another population by marker-assisted backcrossing procedures.
Conclusions
The results of the present study indicate that aluminum tolerance in triticale is related to the QTL located on chromosome 7R. This proposal is consistent with association mapping studies in which numerous markers assigned to chromosome 7R were strongly associated with the trait (Niedziela et al. 2012). By considering the location of the B1 and B26 markers and the known location of microsatellite markers (Benito et al. 2010), it is also likely that the QTL is on the short arm of chromosome 7R. Moreover, based on the biochemical tests performed by us, linkage group of the markers specific for the QTL identified in the present study and in previous studies (Miftahudin et al. 2005; Benito et al. 2010), the documented presence of the ScALMT1 gene in the locus, and the linkage group of markers with the aluminum-tolerance QTL in triticale, our data confirm that malate efflux mechanisms play important roles in the aluminum tolerance of triticale.
Abbreviations
- AFLP:
-
Amplified fragment length polymorphism
- ALMT:
-
Aluminum-activated malate transporter
- DArT:
-
Diversity arrays technology
- CIM:
-
Composite interval mapping
- DH:
-
Doubled haploid
- GWM:
-
Gatersleben wheat microsatellites
- LOD:
-
Logarithm of odds
- MATE:
-
Multidrug and toxin efflux
- QTL:
-
Quantitative trait locus
- PCR:
-
Polymerase chain reaction
- REMS:
-
Rye expressed microsatellite sites
- SCM:
-
Secale cereale microsatellites
- SSR:
-
Simple sequence repeat
References
Ajmal SU, Zakir N, Mujahid MY (2009) Estimation of genetic parameters and character association in wheat. J Agric Biol Sci 1(1):15–18
Akbari M, Wenzl P, Caig V, Carling J, Xia L, Yang S, Uszynski G, Mohler V, Lehmensiek A, Kuchel H, Hayden MJ, Howes N, Sharp P, Vaughan P, Rathmell B, Huttner E, Kilian A (2006) Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor Appl Genet 113(8):1409–1420
Alheit KV, Reif JC, Maurer HP, Hahn V, Weissmann EA, Miedaner T, Würschum T (2011) Detection of segregation distortion loci in triticale (x Triticosecale Wittmack) based on a high-density DArT marker consensus genetic linkage map. BMC Genomics 12:380
Anioł A (1984) Induction of aluminium tolerance in wheat seedlings by low doses of aluminium in the nutrient solution. Plant Physiol 75:551–555
Anioł A (1990) Genetics of tolerance to aluminium in wheat (Triticum aestivum L.). Plant Soil 123:223–227
Anioł A, Gustafson JP (1984) Chromosome location of genes controlling aluminium tolerance in wheat, rye, and triticale. Can J Genet Cytol 26:701–705
Badea A, Eudes F, Salmon D, Tuvesson S, Vrolijk A et al. (2011) Development and assessment of DArT markers in triticale. Theor Appl Genet 122:1547–1560
Bartlett MS (1937) Properties of sufficiency and statistical tests. Proc R Soc Lond 160:268–282
Benito C, Silva-Navas J, Fontecha G, Hernández-Riquer MV, Eguren M, Salvador N, Gallego FJ (2010) From the rye Alt3 and Alt4 aluminum tolerance loci to orthologous genes in other cereals. Plant Soil 327:107–120
Bolibok-Brągoszewska H, Heller Uszyńska K, Wenzl P, Uszyński G, Kilian A, Rakoczy-Trojanowska M (2009) DArT markers for the rye genome—genetic diversity and mapping. BMC Genomics 10:578
Boros D (2002) Physico-chemical quality indicators suitable in selection of triticale for high nutritive value. In: Proceedings of the 5th International Triticale Symposium: 2002; Radzikow, Poland, p. 239–244
Broman KW (2010) Genetic map construction with R/qtl. University of Wisconsin-Madison, Department of Biostatistics and Medical Informatics, Technical Report # 214, 4 November 2010, Revised 21 March 2012
Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Budzianowski G, Woś H (2004) The effect of single D-genome chromosomes on aluminium tolerance of triticale. Euphytica 137:165–172
Cai S, Bai G-H, Zhang D (2008) Quantitative trait loci for aluminum resistance in Chinese wheat landrace FSW. Theor Appl Genet 117:49–56
Collins NC, Tardieu F, Tuberosa R (2008) Quantitative trait loci and crop performance under abiotic stress: where do we stand? Plant Physiol 147:469–486
Crossa J, Burgueño J, Dreisigacker S, Vargas M, Herrera-Foessel SA, Lillemo M, Singh RP, Trethowan R, Warburton M, Franco J, Reynolds M, Crouch JH, Ortiz R (2007) Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics 177:1889–1913
Delhaize E, Craig S, Beaton CD, Bennet RJ, Jagadish VC, Randall PJ (1993) Aluminum tolerance in wheat (Triticum aestivum L.) II. Aluminum-stimulated excretion of malic acid from root apices. Plant Physiol 103:695–702
Dengenhardt J, Larsen PB, Howell SH, Kochian LV (1998) Aluminum resistance in the Arabidopsis mutant alr-104 is caused by an aluminum-induced increase in rhizosphere pH. Plant Physiol 117:19–27
Fontecha G, Silva-Navas J, Benito C, Mestres MA, Espino FJ, Hernández-Ríquer V, Mestres MA, Gallego FJ (2007) Candidate gene identification of an aluminum-activated organic acid transporter gene at the Alt4 locus for aluminum tolerance in rye (Secale cereale L.). Theor Appl Genet 114:249–260
Foy CD (1992) Soil chemical factors limiting plant root growth. In: Hatfield JL, Stewart BA (eds) Advances in soil science: limitation to plant root growth, vol 19. Springer, New York, pp 97–149
Francki MGWE, Crawford AC, Broughton S, Ohm HW, Barclay I, Wilson RE, McLean R (2009) Comparison of genetic and cytogenetic maps of hexaploid wheat (Triticum aestivum L.) using SSR and DArT markers. Mol Genet Genomics 281:181–191
Furukawa J, Yamaji N, Wang H, Mitani N, Murata Y (2007) An aluminum-activated citrate transporter in barley. Plant Cell Physiol 48:1081–1091
Gale MD, Atkinson MD, Chinoy CN, Harcourt RL, Jia J et al. (1995) Genetic maps of hexaploid wheat. In: Li ZSXZ (ed) Proceedings of the 8th international wheat genet symposium. China Agricultural Scientech Press, Beijing, pp 29–40
Gallego FJ, Benito C (1997) Genetic control of aluminium tolerance in rye (Secale cereale L.). Theor Appl Genet 95:393–399
González JM, Muñiz LM, Jouve N (2005) Mapping of QTLs for androgenetic response based on a molecular genetic map of × Triticosecale Wittmack. Genome Res 48:999–1009
Hackauf B, Wehling P (2001) Identification of microsatellite polymorphisms in an expressed portion of the rye genome. Plant Breed 121:17–25
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron 4:1–9
Henderson M, Ownby JD (1991) The role of root cap mucilage secretion in aluminum tolerance in wheat. Curr Top Plant Biochem Physiol 10:134–141
Hoekenga OA, Maron LG, Piñeros MA, Cancado GMA, Shaff J, Kobayashi Y, Ryan PR, Dong B, Delhaize E, Sasaki T, Matsumoto H, Yamamoto Y, Koyama H, Kochian LV (2006) AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc Natl Acad Sci U S A 103(25):9738–9743
Khlestkina EK, Than MHM, Pestsova EG, Röder MS, Malyshev SV, Korzun V, Börner A (2004) Mapping of 99 new microsatellite-derived loci in rye (Secale cerale L.) including 39 expressed sequence tags. Theor Appl Genet 109(4):725–732
Kobayashi Y, Koyama H (2002) QTL analysis of Al tolerance in recombinant inbred lines of Arabidopsis thaliana. Plant Cell Physiol 43(12):1526–1533
Kochian LV (1995) Cellular mechanisms of aluminium toxicity and resistance in plants. Annu Rev Plant Physiol Plant Mol Biol 46:237–260
Korzun V, Malyshev S, Voylkov AV, Börner A (2001) A genetic map of rye (Secale cerale L.) combining RFLP, isozyme, protein, microsatellite and gene loci. Theor Appl Genet 102:709–717
Ligaba A, Katsuhara M, Ryan PR, Shibasaka M, Matsumoto H (2006) The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiol 142(3):1294–1303
Liu X, Magalhaes JV, Shaff J, Kochian LV (2009) Aluminum-activated citrate and malate transporters from MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant Physiol 123:1537–1543
Ma JF, Taketa S, Yang ZM (2000) Aluminium tolerance genes on the short arm of chromosome 3R are linked to organic acid release in triticale. Plant Physiol 122:687–694
Magalhaes JV, Liu J, Guimaraẽs CT, Lana UGP, Alves VM, Wang Y-H, Schaffert RE, Hoekenga OA, Shaff JE, Pineros MA, Klein PE, Kochian LV (2007) A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet 39:1156–1161
Manal HE (2009) Estimation of heritability and genetic advance of yield traits in wheat (Triticum aestivum L.) under drought condition. Int J Genet Mol Biol 1:115–120
Matos M, Camacho MV, Pérez-Flores V, Pernaute B, Pinto-Carnide O (2005) A new aluminium tolerance gene located on rye chromosome arm 7RS. Theor Appl Genet 111:360–369
Matos M, Pérez-Flores V, Camacho MV, Pernaute B, Pinto-Carnide O, Benito C (2007) Detection and mapping of SSRs in rye ESTs from aluminium-stressed roots. Mol Breeding 20(2):103–115
Matsumoto H, Senoo Y, Kasai M, Maeshima M (1996) Response of the plant root to aluminum stress: analysis of the inhibition of the root elongation and changes in membrane function. J Plant Res 109:99–105
Mergoum M, Singh PK, Peña RJ, Lozano-del Río AJ, Cooper KV, Samon DF, Gómez Macpherson H (2009) Triticale: a ‘‘new’’ crop with old challenges. In: Carena MJ (ed) Cereals. Springer, New York, pp 267–286
Miftahudin T, Chikmawati T, Ross K, Scoles GJ, Gustafson JP (2005) Targeting the aluminium tolerance gene Alt3 region in rye, using rice/rye micro-colinearity. Theor Appl Genet 110:906–913
Miftahudin T, Scoles GJ, Gustafson JP (2002) AFLP markers tightly linked to the aluminium-tolerance gene Alt3 in rye (Secale cereale L.). Theor Appl Genet 104:626–631
Miftahudin T, Scoles GJ, Gustafson JP (2004) Development of PCR-based codominant markers flanking the Alt3 gene in rye. Genome 47:231–238
Milczarski P, Bolibok-Bragoszewska H, Myśków B, Stojałowski S, Heller-Uszyńska K, Góralska M, Brągoszewski P, Uszyński G, Kilian A, Rakoczy-Trojanowska M (2011) A high density consensus map of rye (Secale cereale L.) based on DArT markers. PLOS One 6(12)
Myer R, Barnett, RD (2004) Triticale grain in swine diets. Available: http://www.edisifasufledu/an032: 37 Accessed 2008 March 2004
Niedziela A, Bednarek P, Cichy H, Budzianowski G, Kilian A, Aniol A (2012) Aluminum tolerance association mapping in triticale. BMC Genomics 13(67). doi:10.1186/1471-2164-1113-1167
Oleszczuk S, Rabiza-Swider J, Zimny J, Łukaszewski AJ (2011) Aneuploidy among androgenic progeny of hexaploid triticale (X triticosecale Wittmack). Plant Cell Rep 30(4):575–586
Pellet D, Papernik LA, Kochian LV (1996) Multiple aluminum-resistance mechanisms in wheat. Roles of root apical phosphate and malate exudation. Plant Physiol 112:591–597
Piepho HP (2000) Optimal marker density for interval mapping in a back cross population. Heredity 84(4):437–440
Raman H, Zhang K, Cakir M, Appels R, Garvin DF, Maron LG, Kochian LV, Moroni JS, Raman R, Imtiaz M, Drake-Brockman F, Waters I, Martin P, Sasaki T, Yamamoto Y, Matsumoto H, Hebb DM, Delhaize E, Ryan PR (2005) Molecular characterization and mapping of ALMT1, the aluminium-tolerance gene of bread wheat (Triticum aestivum L.). Genome 48:781–791
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier M-H, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Ryan PR, Raman H, Gupta S, Horst WJ, Delhaize E (2009) A second mechanism for aluminum resistance in wheat relies on the constitutive efflux of citrate from roots. Plant Physiol 149:340–351
SAS I, Inc. (2009) SAS/STAT 9.2 User’s Guide, Second Edition, Ed, Vol SAS Publishing, SAS Institute Inc., Cary, NC, USA
Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ (2004) A wheat gene encoding an aluminium-activated malate transporter. Plant J 37:645–653
Satterthwaite FE (1946) An approximate distribution of estimates of variance components. Biom Bull 2:110–114
Schlegel R (1996) In: Guedes-Pinto H, Darvey N, Carnide VP (eds) Triticale: today and tomorrow. Kluwer, Dordrecht, p. 21–31
Septiningsih EM, Trijatmiko KR, Moeljopawiro S, McCouch SR (2003) Identification of quantitative trait loci for grain quality in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor Appl Genet 107:1433–1441
Silva-Navas J, Benito C, Téllez-Robledo B, El-Moneim A, Gallego FJ (2011) The ScAACT1 gene at the Qalt5 locus as a candidate for increased aluminium tolerance in rye (Secale cereale L.). Mol Breed. doi:10.1007/s11032-011-9668-5
Singh R, Tan SG, Panandam JM, Rahman RA, Ooi LCL, Low E-TL, Sharma M, Jansen J, Cheah S-C (2009) Mapping quantitative trait loci (QTLs) for fatty acid composition in an interspecific cross of oil palm. BMC Plant Biol 9:114
Somers D, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Stass A, Smit I, Eticha D, Oettler G, Horst JH (2008) The significance of organic-anion exudation for the aluminium resistance of primary triticale derived from wheat and rye parents differing in aluminium resistance. J Plant Nutr Soil Sci 171(4):634–642
Takagi H, Namai H, Murakami K (1983) Exploration of aluminium tolerant genes in wheat. In: Proceeding of the 6th International Wheat Genetics Symposium: 1983; Kyoto, Japan; 1983: 143
Tyrka M, Bednarek PT, Kilian A, Wędzony M, Hura T, Bauer E (2011) Genetic map of triticale compiling DArT, SSR, and AFLP markers. Genome 54:391–401
Ukai Y (2000) Genetic analysis at the genomic level: map and QTL. University of Tokyo Press, Tokyo
van Barneveld RJ (1999) Chemical and physical characteristics of grains related to variability in energy and amino acid availability in pigs: a review. Aust J Agric Res 50(5):667–687
van Ooijen JW (2004) MapQTL_5 Software for the mapping of quantitative trait loci in experimental populations. Kyazma BV, Wageningen
Vos P, Hogers R, Bleeker M, Rijans M, Van de Lee T, Hormes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Website: http://www.bafz.de. BAZ Database of Secale cerale Microsatellites (SCM) Federal Centre for Breeding Research on Cultivated Plants
Website: http://www.stat.gov.pl/cps/rde/xbcr/gus/PUBL_rl_uzytkowanie_gruntow_r_2011.pdf Central Statistical Office
Website: http://www.triticarte.com.au/
Wenzl P, Li H, Carling J, Zhou M, Raman H, Paul E, Hearnden P, Maier C, Xia L, Caig V, Ovesná J, Cakir M, Poulsen D, Wang J, Raman R, Smith KP, Muehlbauer GJ, Chalmers KJ, Kleinhofs A, Huttner E, Kilian A (2006) A high-density consensus map of barley linking DArT markers to SSR, RFLP and STS loci and agricultural traits. BMC Genomics 7:206
Williams RJP (1999) The J.D. Birchall memorial lecture. What is wrong with aluminium? J Inorg Biochem 76:81–88
Zhang XG, Jessop RS, Ellison F, Evans P (2001) Assessment of aluminium stress tolerance of triticale breeding lines in hydroponics “Science and Technology: Delivering Results for Agriculture?” Rowe B, Donaghy D, Mendham N (eds). Proceedings of the 10th Australian Agronomy Conference, January 2001, Hobart, Tasmania
Acknowledgments
This research was funded by the Ministry of Science and Higher Education projects no. PBZ-MNiSW 2/3/2006. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors’ contributions
AN carried out the physiological tests and molecular genetic studies, performed the statistical analyses and wrote the manuscript, PTB conceived the study, participated in its design and coordination, participated in running statistics, and participated in writing the manuscript, ML carried out biochemical tests, DM carried out statistical analysis of the biochemical results, heritability and test of normality, AA provided intellectual input during the experiments and revised the manuscript. All authors read and approved the final manuscript.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Niedziela, A., Bednarek, P.T., Labudda, M. et al. Genetic mapping of a 7R Al tolerance QTL in triticale (x Triticosecale Wittmack). J Appl Genetics 55, 1–14 (2014). https://doi.org/10.1007/s13353-013-0170-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-013-0170-0