Abstract
The COVID-19 pandemic has elicited a rapid response from the scientific community with significant advances in understanding the causative pathogen (SARS-CoV-2). Mechanisms of viral transmission and pathogenesis, as well as structural and genomic details, have been reported, which are essential in guiding containment, treatment, and vaccine development efforts. Here, we present a concise review of the recent research in these domains and an exhaustive analysis of the genomic origins of SARS-CoV-2. Particular emphasis has been placed on the pathology and disease progression of COVID-19 as documented by recent clinical studies, in addition to the characteristic immune responses involved therein. Furthermore, we explore the potential of nanomaterials and nanotechnology to develop diagnostic tools, drug delivery systems, and personal protective equipment design within the ongoing pandemic context. We present this as a ready resource for researchers to gain succinct, up-to-date insights on SARS-CoV-2.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The recent COVID-19 outbreak has generated considerable interest in medical and research circles around the world. This highly contagious disease poses severe challenges in clinical settings, with respiratory distress and immune dysregulation serving as chief hallmarks. The novel virus, identified as Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), presents many interesting similarities with viruses that were associated with earlier outbreaks such as the MERS-CoV and SARS-CoV. Notable attempts to develop vaccine candidates and therapeutic solutions have been initiated, and pioneering efforts have helped produce a wealth of knowledge on the virus in question [83]. However, the rapid pace of development has also resulted in an overwhelming spread of inaccurate information about the viral origin and infectivity. Thus, we aim to provide comprehensive coverage of the SARS-CoV-2 virology, transmission modes, associated clinical pathogenesis—in particular, cytokine storm responses and acute respiratory distress syndrome (ARDS). Besides, we also discussed the promising potential of nanotechnology-based tools to combat the disease. A concise, schematic summary of viral pathogenesis and transmission has been provided in Fig. 1.
Transmission and Pathogenesis of SARS-CoV-2. The SARS-CoV-2 is an enveloped, single-stranded RNA virus. The primary hosts are conjectured to be bats, from which the virus may have transmitted to a human host via intermediate carriers. An infected human host may readily transmit the pathogen to other individuals following zoonosis. Green boxes indicate the major events of SARS-CoV-2 infection including respiratory discomfort, renin-angiotensin system (RAS) dysfunction, and dysregulation of the immune system. Unless contained by proper medical intervention, these may culminate in potentially lethal conditions like ARDS and cytokine storm responses, as denoted by the red boxes
Origin and evolution of SARS-CoV-2
An outbreak of a novel coronavirus, originating from Wuhan, China, was declared in December 2019. Phylogenetic analysis of viral genome isolated from the infected patients and the market environment suggested that the virus could be clustered with a group that includes the SARS/SARS-like coronavirus, belonging to the genus Betacoronavirus and was thus named as SARS-CoV-2 [65, 109]. Whole-genome comparison of SARS-CoV-2 with other related coronaviruses indicated 79%, 50%, 87.23%, and 87.99% similarity to SARS-CoV, MERS-CoV, bat-SL-CoVZXC21, and bat-SL-CoVZC45, respectively [32, 56]. Interestingly, a remarkable sequence similarity of 96.2% was found between the bat CoV RaTG13 (Rhinolophus affinis) and SARS-CoV-2 [119]. Further understanding of host selection revealed that the bat CoV RaTG13 forms an inner joint neighbor with bat-SL-CoVZXC21 and bat-SL-CoVZC45 as the closest relatives. However, the bat HKU9 coronaviruses form an immediate outgroup sharing a common ancestor with the SARS-CoV-2 group (Fig. 2). Since most of the outer and inner joint neighbors of SARS-CoV-2 have bats as their natural host, these observations are suggestive that bats would be the convenient native hosts of SARS-CoV-2 [77, 109, 119].
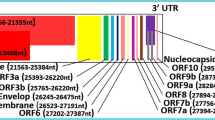
Structure, Genome organization, and Phylogenetic analysis of SARS-CoV-2. The upper panel figures depict the schematic structure of SARS-COV-2 virion and its genomic organization. The phylogenetic tree in the lower panel describes the evolutionary history of SARS-CoV-2 by the Neighbor-joining method [79]. Whole-genome sequence alignment was done by ClustalW (MEGA 7) with default settings [47], and an optimal tree with the sum of branch length = 2.24553015 is shown here. The alphacoronavirus HCoV-NL63 and HCoV-229E sequence were considered as the outgroup. The evolutionary distances were calculated using the Maximum Composite Likelihood method [88]. The tree is constructed to scale with units in the number of base substitutions per site. The analysis involved 14 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 24,863 positions in the final dataset
It is interesting to note that the seafood market in question also sold non-aquatic animals such as wild mammals, frogs, and snakes; however, bats were neither on sale nor found in the market during the outbreak [102]. Despite high sequence similarity, SARS-CoV-2 and bat SARS-like RaTG13 differ in several key genomic features. One of them is the single insertion of a polybasic cleavage site (Pro-Arg-Arg-Ala) at the S1/S2 junction of the SARS-CoV-2’s spike (S) glycoprotein, which was speculated to increase the infectivity of the virus [17, 113, 116]. The phylogenetic analysis also showed a long branch in between SARS-CoV-2 and the close relatives bat-SL-CoVZXC21 and bat-SL-CoVZC45 with sequence similarity less than 90%, suggesting that bat-SL-CoVZXC21 and bat-SL-CoVZC45 may not be the direct ancestors of SARS-CoV-2 (Fig. 2). This raises the possibility that another mammal may have served as an intermediate host from which the insertion was acquired, following which the spill-over event to infect humans may have occurred [32, 56]. A similar occurrence was also noted in the case of related MERS-CoV and SARS-CoV, with dromedary camels and palm civet being the intermediate hosts, respectively [2, 31].
Prior to the onset of the epidemic, in late October 2019, a SARS-CoV-like coronavirus was detected in the rescued dead Malayan pangolins’ lung samples (Manis javanica) from Guangdong province [53]. The whole-genome sequence alignment suggested Pangolin CoV to be the second closest relative of SARS-CoV-2 (91.02% similarity) after batCoV RaTG13 (90.55% similarity). A similar result was found from the amino acid phylogenetic analysis of S1 protein. However, unlike SARS-CoV-2, Pangolin-CoV does not have the furin polybasic cleavage site at S1/S2 junction [114]. Both sequence and structural comparisons of the S protein’s receptor-binding domain (RBD) suggests that similar to the RBD of SARS-CoV; the SARS-CoV-2’s RBD is also suitable for binding to the human Angiotensin-Converting Enzyme—2 (ACE-2) receptor [51, 56, 100]. Virus infectivity study in HeLa cells proved that the ACE2 receptor is required explicitly for SARS-CoV-2’s cellular entry, unlike that of other coronavirus receptors (dipeptidyl peptidase 4 or aminopeptidase N) [119]. The computational analysis suggests that five critical amino acid residues in RBD—necessary for binding to the ACE2 receptor—are conserved between Pangolin-CoV and SARS-CoV-2; whereas, the bat CoV RaTG13 contains four amino acid mutations in the RBD. These results suggest the possibility of similar entry profiles of both Pangolin-CoV and SARS-CoV-2, with a higher probability of crossing the host barriers to infect humans [53, 114].
Biophysical assays suggest that the S protein of SARS-CoV-2 binds at least ten times more strongly to the human cellular ACE2 receptor compared to its relative SARS-CoV [100]. This increases the chances of SARS-CoV-2 having passed through more than one intermediate host in order to acquire such high binding efficiency. The evidence thus far suggests that in addition to pangolins (Pelodiscus sinensis), snakes and turtles (Chelonia mydas and Chrysemys pictabellii) may have been potential intermediate hosts for the SARS CoV-2, prior to it's transmission to humans [45, 53]. Apart from structural aspects, characteristic patterns of genomic evolution and transformation lend credibility to a zoonotic origin theory. A well-known correlation exists between the viral CpG dinucleotide concentration and the mammalian host selection. The interferon-mediated mammalian zinc-finger antiviral protein (ZAP) binds to the CpG dinucleotides and inhibits viral replication, thereby promoting viral genome degradation [23, 60]. Hence, to ensure its existence, the virus would preferably evolve with a low CpG deficiency index (ICpG) in a high ZAP expressing mammalian tissue [105]. Interestingly, BatCoV RaTG13 and SARS-CoV-2 have an extremely low ICpG compared to other β-CoVs from bat species, suggesting that their ancestors either have low ICpG or have evolved with low ICpG values. Studies found that only genomes from canine coronaviruses (CCoVs) have similarly low ICpG values as observed in SARS-CoV-2 and BatCoV RaTG13. Thus, there is also a possibility that before acquiring the ability to evade human ZAP-mediated immune responses, the ancestors of these two viruses infected the canids [105].
Based on the above findings, it appears obvious that SARS-CoV-2 may have originated in the natural environment, influenced by various selection pressures exerted by primary and intermediate hosts. This also discredits conjectures about its genesis as a result of manipulation within laboratory conditions. However, more detailed serological, metagenomic, and experimental evolution studies need to be performed to confirm this hypothesis indubitably and ascertain whether the natural selection in RBD can occur prior to zoonosis.
Genome organization and structural biology of the virus
SARS-CoV-2, like other β-coronaviruses, is enveloped, with a positive single-strand non-fragmented RNA genome of 29.0–30.2 kb [59] (Fig. 2). The name coronavirus derives from its corona solis-like appearance (corona in Latin means crown) in the electron micrographs caused by the protrusion of spike glycoproteins from the viral envelope. Each virion ranges approximately between 50 and 200 nm in diameter [14]. The novel virus genome has 5’ (265 nt) and 3’ (229 nt) terminal sequences with a gene order as 5’- replicase gene, spike (S), envelope (E), membrane (M), and nucleocapsid (N)-3’ [34, 101] (Fig. 2). Unlike lineage-A β-CoVs, SARS-CoV-2 lacks hemagglutinin esterase, facilitating viral entry into the host-cell mediated by the S glycoprotein [59]. The replicase gene contains two large open reading frames (ORFs), orf1ab and orf1a, encompassing two-thirds of the genome, with orf1ab as the largest gene. Orf1ab and orf1a encode polyprotein-1ab (pp1ab) and polyprotein-1a (pp1a), respectively, which are co-translationally cleaved into 16 non-structural proteins (nsps) required for facilitating the viral replication [111]. These cleavages are mediated by virally encoded papain-like proteases or chymotrypsin-like protease (3CLpro) or main protease (Mpro) at specific recognition sites within the ORFs [22, 71].
The next ORF encodes monomeric S protein (175 kDa), a type I fusion glycoprotein that trimerizes and facilitates the receptor attachment [7, 20, 49]. The S1 domain is located in the N-terminus of S protein, which contains the glycosylated NTD (N-terminal domain), an RBD, and an RBM (receptor-binding motif) that directly interacts with the host ACE2 receptor for viral attachment. On the other hand, the S2 domain acts as the fusion machinery containing two heptad repeats (HR1 and HR2) that brings the membranes of both viruses and host cell together to facilitate the membrane fusion [10, 51, 106, 107].
The E protein and the M protein are 8-12 kDa and 25-30 kDa glycoproteins, respectively, located in the viral envelope. Both E and M are transmembrane proteins containing a C-terminal endodomain and N-terminal ectodomain [73]. Relatively scarce E proteins have ion channel activity and were speculated to promote the viral pathogenesis; whereas, M proteins are abundant in the virus and interact with other structural proteins to give the virion its shape [19, 62].
The N proteins are heavily phosphorylated proteins present in the helical nucleocapsid of the virus that protects the viral genome. It is an abundant viral protein in infected cells, featuring a C-terminal domain (CTD) and N-terminal domain (NTD) that can bind to RNA molecules of both host and viral origin [13]. The N protein interacts with the transcription regulatory sites (TSGs), present at the beginning of every structural and accessory gene for transcriptional control. Its CTD contains the genomic packaging signal required to pack the viral genome into a ribonucleoprotein complex called the capsid [13, 48, 61, 85]. The N protein subsequently interacts with the M protein and nsp3, a component of the replicase complex, to produce viral particles by facilitating the tethering of encapsulated viral genome to the replicase-transcriptase complex [39, 86].
Transmission of SARS-CoV-2
SARS-CoV-2, similar to the MERS-CoV and SARS-CoV, spreads mainly from an infected individual through respiratory droplets released while coughing. These droplets are > 5–10 μm in diameter and can remain suspended in a closed, static environment for as long as 8–14 min or undergo airborne transmission from an infected person to a healthy individual [12, 70, 84, 91, 103, 104] (Fig. 1). The virus is also known to be transmitted to healthy individuals directly by kissing or via sexual contact with an infected person [41, 68]. Several studies have suggested modes of indirect transmission through fomites and fecal–oral routes [30, 41, 64, 66]. Once the infected host sheds the virus into the environment, it may survive with a half-life of 6.8 h on plastic, 5.6 h on stainless steel, 3.5 h on cardboard, and 1.1 h in aerosols. If a healthy individual touches these contaminated surfaces and then their eyes, mouth, or nose, the viral particles may gain entry into the host cells and initiate infection [93]. It was also demonstrated that the virus could retain viability on the skin surface for up to 9 h, a property that could accelerate the pandemic by increasing the possibility of contact transmission [35].
The third trimester of pregnancy presents an immunocompromised situation, as well. Studies were conducted to evaluate the possible potential for vertical-transmission of the virus from the infected mother to neonates. However, it remains unclear whether viral transmission could occur during normal delivery since cesarean section-assisted deliveries conducted during the pandemic and neonates were found to be negative for SARS-CoV-2. The only exception to this so far was reported by Alzamora et al. [3, 15, 57]. On the other hand, children present a similar risk of infection to that of the general population. Nevertheless, children were less likely to develop severe symptoms due to the lower viral load, milder immunological responses, and less immune damage compared to severely affected adults [8, 58].
Based on this understanding of the modes of viral transmission, the appropriate use of personal protective equipment with N95 respirator without expiration valves for frontline healthcare workers was recommended. Additionally, preventive measures like environmental disinfection, social distancing, and use of face masks, protected contact, and frequent handwashing practices have been emphasized for the general population [11, 16, 97, 98]. However, the overall outcomes of these measures may be limited by the extent of public adherence to these guidelines and lack of effective implementation.
Pathogenesis of SARS-CoV-2
The highly pathogenic HCoVs, including SARS-CoV-2, MERS-CoV, and SARS-CoV, infects the lower respiratory tract, lungs, kidneys, cardio vasculature, skin tissues, and small intestine [21]. This infection leads to severe pneumonia, ARDS, and organ failures [4, 6, 25, 46].
The novel CoV-2 utilizes the host ACE2 receptor (especially in alveolar cells of lungs, goblet cells of the nasal mucosa, and absorptive enterocytes in the gut) in concert with the host’s serine protease TMPRSS2 to infiltrate the host cells [36, 119, 122]. The priming of viral S glycoprotein triggers the cleavage of S protein by host furin-like proteases at the S1/S2 cleavage site and brings the membranes of both viruses and host cell close to facilitate the membrane fusion [1, 17, 94, 114]. Subsequently, the viral genome is released into the host cells and follows replication, translation, and packaging into progeny viral particles. The active replication and release of viral particles induce the infected cells to undergo an inflammatory form of programmed cell death known as pyroptosis [24, 117]. Upon cell entry by binding to the host ACE2 receptor, the virus downregulates the expression of ACE2, required for angiotensin-II (Ang II) cleavage to produce angiotensin-1-7 [Ang (1-7)]. Being the key effector of the renin-angiotensin system (RAS), a high concentration of Ang II deregulates the RAS signaling and results in multi-system inflammation and severe lung injury [38].
As a consequence of Ang II upregulation, the infected alveolar cells secrete NLRP3 specific inflammatory mediators, promoting the secretion of IL-18, IL-1β, and damage-associated molecular patterns (DAMPs). The release of these inflammatory cytokines and DAMPs results in cell death by pyroptosis. These molecules are also recognized by alveolar macrophages and nearby alveolar cells, which promote local inflammatory responses by secreting specific pro-inflammatory cytokines and chemokines. These include IFN-γ, IL-6, IP-10, and MCP1, which attract the T lymphocytes, macrophages, and monocytes to the site of injury from the bloodstream [26, 37, 76, 89, 108, 115]. The infiltration of immune cells into the airways thus promotes further inflammation and establishes a pro-inflammatory feedback loop, triggering a cytokine storm [37]. The resulting cytokine storm not only damages the lungs but also shows a ripple effect across several other organs, causing septic shock and multiple organ failure [78, 87, 120]. In addition to the direct damages associated with a viral infection, the inflammatory cell infiltration causes hyaline membrane formation, desquamation of alveolar cells, and pulmonary edema leading to ARDS [89, 108].
To mount antiviral immunity, the alveolar macrophages engulf the viruses, process them, and present antigenic epitopes on their surface via the MHC molecules—especially epitopes from the spike glycoproteins. These processed antigens are recognized by specific Cytotoxic T-lymphocytes (CTLs) and initiate adaptive immune responses mediated by virus-specific B cells and T cells [52]. Upon infection, the levels of virus-specific IgG and IgM antibody titers were elevated, as suggested by clinical studies on 285 patients. All patients achieved seroconversion of IgM and IgG, either sequentially or simultaneously within 20 days after clinical manifestations [55]. Similar to SARS-CoV’s infection, the virus-specific IgM antibody titer starts declining three weeks post-symptom onset; however, the elevated IgG antibody levels last for a more extended period, suggesting an increased protective role offered by IgG against viruses [50, 55]. Apart from humoral responses, both SARS-CoV and SARS-CoV-2 patients suffer from lymphocytopenia, where the numbers of cellular immune CD8+ and CD4+ T cells are significantly reduced in the blood [33, 108, 118]. Follow-up studies in SARS-CoV patients indicate that the memory CD8+ and CD4+ T cells specific to SARS-CoV were persistent for a more extended period [69, 110]. Thus, it is clear that CoV-specific T-cells are essential in eliminating viruses and controlling disease progression. However, it remains uncertain whether T cell responses alone can stem the progress of infection. Intriguingly, recent evidence suggests the possibility that reinfection with SARS-CoV-2 may present more severe clinical symptoms as compared to the initial infection, bringing into question the ability of pre-existing immune responses to protect during the second exposure. Furthermore, variations were observed between viral isolates from the primary infection and reinfection scenarios. This observation raises concerns over whether a single vaccine can confer both individual and herd immunity [18, 90]. Further studies along these lines may help better inform the rational design of vaccines against SARS-CoV-2.
Nanotechnology as a potential tool to fight SARS-CoV-2
Recently, Weiss et al. (2020) argued that nanotechnology could be used to tackle the problem generated by SARS-CoV-2 and other future viral infections by various means such as (1) the nanomaterials can be used for the targeted delivery of drugs and vaccines into the lungs; (2) rapid, sensitive, precise detection of pathogen or detection of immunity (serological tests); (3) application of superior filters for face masks or filtering of blood; (4) developing nano-based surface coatings to inactivate the virus; (5) enhancement of tools for contact tracing [96] (Fig. 3).
Potential role of theranostic nanoparticles against SARS-CoV-2. Graphical representation of the pivotal role of nanoparticles in drug delivery, diagnosis, and protective coatings. The nanoparticles are conjugated to the therapeutic agents such as specific siRNA, mRNA, peptides, or antibodies. The conjugated agents are then prepared as a solution or an emulsion and are delivered to the patients via nasal routes, such as using a nasal spray. Nanoparticles are also used to develop diagnostic tools such as electrochemical devices or immunochromatographic strip tests. Besides, they are used as antiviral coatings over PPE, textiles, surfaces, AC filters, among other applications leading to an efficient blocking of viral transmission
Nanotechnology offers a new approach called ‘Nanotheranostics,’ which deals with diagnosis and drug delivery [27, 99]. The benefits of combined application of detection and therapeutics of nanomaterials have been extensively used in cancer research in the past; however, their potential has not been realized in tackling infectious diseases caused by viruses, bacteria, and fungi. There exist great potential in dealing with new and emerging diseases like COVID-19 [80] with the help of nanomaterials.
Theranostic nanoparticles render a pivotal role in diagnosing and drug delivery of siRNA, mRNA, drugs, vaccines, biomolecules, and peptides at the infection site (Fig. 3). In viral diseases, intranasal delivery based on nanoparticles was proposed by researchers to deliver antiviral agents for the treatment of lung infections. This kind of nasal delivery ensures the required concentration of drugs compared to other means of delivery without harming the normal cells [43].
Two types of nanomaterials-organic and inorganic, can be used for drug delivery. Organic nanoparticles are usually biodegradable, while inorganic nanoparticles are not degradable. The nanoparticles of silver, gold, silver sulfide, titanium oxide, zirconium, grapheme, and polymeric compounds can be used as a carrier for vaccines, which have a tremendous capacity as compared to traditional antigen-based vaccines. However, further toxicity studies are required in order to the safe delivery of drugs [63]. The metal nanoparticles are not biodegradable, and their toxicity should be evaluated. The basic advantage of biodegradable nanomaterials such as polymers, liposomes, dendrimers, and other small molecules is the controlled release of cargo materials at a specific site while maintaining the integrity of nanoparticles at off-targets [96] (Fig. 4). In addition, nanoparticles are used for the detection of SARS-CoV-2, enhancing the sensitivity for the detection by colorimetry. This technique is low-cost, simple, and fast. For example, gold nanoparticles are employed for the immunochromatographic strip test to detect SARS-CoV-2 (Fig. 3).
The nanomaterials can also be used in electrochemical devices for detecting SARS-CoV-2 (Fig. 3). More recently, Qiu et al. (2020) developed a new promising tool for the detection of COVID-19 [72]. The authors used a dual-functional plasmonic biosensor blending with the localized surface plasmon resonance and plasmonic photothermal effect. Gold nanoislands of two-dimensional nature were functionalized with DNA, which can detect the sequences of SARS-CoV-2. This method is accurate, rapid, and cost-effective and has demonstrated greater sensitivity. More recently, the potential applications of nanomaterials in combating COVID-19 have been discussed in diagnosis, drug delivery, and treatment [74]. In this context, a nanoparticle-based lateral flow biosensor assay was recently developed for the diagnosis of COVID-19 [121]. Furthermore, the size of viruses and nanoparticles are at comparable scales, which further bolsters the suitability of nanotechnology for vaccine delivery applications. For instance, the mRNA vaccine produced by Moderna is based on lipid nanoparticles [67]. There are other examples of vaccine delivery using nano platform such as emulsions, dendrimers, polysaccharide-based nanoparticles [9, 40, 67, 81, 112].
In addition to the use of nanoparticles in diagnosis and drug delivery, they can be coated on Personal Protection Equipment (PPE) to inactivate viral particles present on surfaces like door handles, tables, beds, light switches, basins, and toilets. Surface contamination poses a severe problem and places frontline healthcare workers such as doctors and nurses at risk despite proper personal protection by using protective masks, lab-coats, etc. because the indoor environment of the hospital is contaminated with SARS-CoV-2. Sportelli et al. suggested that nanometals can be used for developing antiviral and antibacterial textiles, antimicrobial surfaces, AC filters, and packaging solutions, among other diverse applications [82] (Fig. 3). There are now commercially available masks that are impregnated with silver and copper nanoparticles. In a study, Balagna et al. tested the activity of silver nanocluster/silica composite coating of face mask against COVID-19 and found that it showed strong virucidal activity [5]. Moreover, previous reports also provide evidence that metal nanoparticles in general and silver nanoparticles, in particular, are broad-spectrum antimicrobials [75] and offer a protective shield against viruses [28, 75]. In a comparative study, melt-blown filters used in N95 face masks were compared with nanofiber filters used in effective mask filters to test their reusability. The authors evaluated the airflow rate, filtration efficiency, surface, and morphological characteristics after cleaning them with 75% ethanol and recommended a nanofiber filter for broader usage in masks [92].
Considering the applications mentioned above, nanomaterials can be used effectively for the diagnosis, drug delivery, and treatment of the COVID-19 after extensive experimental trials.
Concluding remarks
The recent SARS CoV-2 pandemic has severely strained medical infrastructure and facilities. The highly detrimental socio-economic impacts of this outbreak have placed an urgent emphasis on accelerated vaccine development, for which understanding the fundamental biology of SARS CoV-2 is indispensable. Through this review, we have summarized the existing knowledge about SARS-CoV-2. Firstly, SARS-CoV-2 genesis occurred in the natural environment, with bat CoVs being the most closely related virus. Before jumping to humans, the SARS-CoV-2 may have passed through several intermediate hosts to acquire mutations in the spike proteins in order to bind the human ACE2 receptor with higher affinity as compared to SARS-CoV. Secondly, detailed information about the virus genomic structure has enhanced our understanding of the viral protein function. The different viral transmission modes explained thus far have been instrumental in characterizing the immunopathogenesis of SARS-CoV-2 and immune responses elicited within the host against the virus. In extreme cases, the infection induces cytokine storms and causes ARDS and multiple organ failure, which may be fatal to the infected person. We have also highlighted the emerging potential of nanomaterials as powerful tools for the detection, management, and treatment of COVID-19.
Certain target areas remain of chief concern and relevance to both the ongoing pandemic and prospective ones. Multiple questions regarding the nature of the disease in children and older adults persist. Exact transmission dynamics and the molecular mechanisms of viral entry along with its replication inside the host cell need to be characterized in greater detail. There is a possibility that the infectivity and transmissibility of the virus may be increasing rapidly among human hosts. This may provide the basis for the development of therapeutic strategies in the future. Furthermore, asymptomatic carriers pose a major threat, as well as a puzzle, since the specific innate immune mechanisms conferring resistance are yet to be elucidated. Understanding the acquisition of immunity against the infection remains a paradox. While certain studies provided evidence that individuals exposed to the virus can acquire immunity even without manifesting clinical symptoms, others have reported the absence of acquired immunity even after the post-infection recovery [29, 42, 44, 54, 95]. Thus, it remains inconclusive whether exposure to the virus may confer immunity, which in turn raises the possible threat of reinfection and recurrent outbreaks.
References
Abraham S, Kienzle TE, Lapps W, Brian DA. Deduced sequence of the bovine coronavirus spike protein and identification of the internal proteolytic cleavage site. Virology. 1990;176(1):296–301. https://doi.org/10.1016/0042-6822(90)90257-R.
Alagaili AN, Briese T, Mishra N, Kapoor V, Lipkin WI. Middle east respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia | mBio. 2014. https://doi.org/10.1128/mBio.00884-14.
Alzamora MC, Paredes T, Caceres D, Webb CM, Valdez LM, La Rosa M. Severe COVID-19 during pregnancy and possible vertical transmission. Am J Perinatol. 2020. https://doi.org/10.1055/s-0040-1710050.
Am Z, van Boheemen S, Tm B, Ad O, Ra F. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–20. https://doi.org/10.1056/nejmoa1211721.
Balagna C, Perero S, Percivalle E, Nepita EV, Ferraris M. Virucidal effect against coronavirus SARS-CoV-2 of a silver nanocluster/silica composite sputtered coating. Open Ceram. 2020;1:100006. https://doi.org/10.1016/j.oceram.2020.100006.
Bauer TT, Ewig S, Rodloff AC, Müller EE. Acute respiratory distress syndrome and pneumonia: a comprehensive review of clinical data. Clin Infect Dis. 2006;43(6):748–56. https://doi.org/10.1086/506430.
Beniac DR, Andonov A, Grudeski E, Booth TF. Architecture of the SARS coronavirus prefusion spike. Nat Struct Mol Biol. 2006;13(8):751–2. https://doi.org/10.1038/nsmb1123.
Bi Q, et al. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet Infect Dis. 2020. https://doi.org/10.1016/S1473-3099(20)30287-5.
BioNTech and Pfizer announce regulatory approval from German authority Paul-Ehrlich-Institut to commence first clinical trial of COVID-19 vaccine candidates | BioNTech’. https://investors.biontech.de/news-releases/news-release-details/biontech-and-pfizer-announce-regulatory-approval-german/.
Bosch BJ, van der Zee R, de Haan CAM, Rottier PJM. The coronavirus spike protein Is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77(16):8801–11. https://doi.org/10.1128/JVI.77.16.8801-8811.2003.
CDC. Coronavirus Disease 2019 (COVID-19) Centers for Disease Control and Prevention. 2020. https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/cloth-face-cover-guidance.html.
CDC. Coronavirus Disease 2019 (COVID-19), centers for disease control and prevention. 2020. https://www.cdc.gov/coronavirus/2019-ncov/more/scientific-brief-sars-cov-2.html.
Chang C, et al. Modular organization of SARS coronavirus nucleocapsid protein. J Biomed Sci. 2006;13(1):59–72. https://doi.org/10.1007/s11373-005-9035-9.
Chen N, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–13. https://doi.org/10.1016/S0140-6736(20)30211-7.
Chen H, et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395(10226):809–15. https://doi.org/10.1016/S0140-6736(20)30360-3.
Coronavirus disease (COVID-19): Masks. https://www.who.int/news-room/q-a-detail/coronavirus-disease-covid-19-masks.
Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir Res. 2020;176:104742. https://doi.org/10.1016/j.antiviral.2020.104742.
de Araujo Torres D, do Carmo Bueno Ribeiro L, de Freitas Linhares Riello AP, Horovitz DDG, Pinto LFR, Croda J. Reinfection of COVID-19 after 3 months with a distinct and more aggressive clinical presentation: Case report. J Med Virol. 2020. https://doi.org/10.1002/jmv.26637.
de Haan CAM, Vennema H, Rottier PJM. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J Virol. 2000;74(11):4967–78. https://doi.org/10.1128/JVI.74.11.4967-4978.2000.
Delmas B, Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J Virol. 1990;64(11):5367–75.
Ding Y, He L, Zhang Q, Jiang S. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS‐CoV) in SARS patients: implications for pathogenesis and virus transmission pathways—Ding—2004. J Pathol. Wiley Online Library. 2004. https://doi.org/10.1002/path.1560.
Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. In: Maier HJ, Bickerton E, Britton P, editors. Coronaviruses: methods and protocols. New York, NY: Springer; 2015. p. 1–23.
Ficarelli M, et al. CpG dinucleotides inhibit HIV-1 replication through zinc finger antiviral protein (ZAP)-dependent and -independent mechanisms. J Virol. 2020. https://doi.org/10.1128/JVI.01337-19.
Fink SL, Cookson BT. Apoptosis pyroptosis and necrosis: mechanistic description of dead and dying eukaryotic cells. Infec Immun. 2005;73(4):1907–16. https://doi.org/10.1128/IAI.73.4.1907-1916.2005.
Franks TJ, et al. Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum Pathol. 2003;34(8):743–8. https://doi.org/10.1016/S0046-8177(03)00367-8.
Freeman TL, Swartz TH. Targeting the NLRP3 inflammasome in severe COVID-19. Front Immunol. 2020. https://doi.org/10.3389/fimmu.2020.01518.
Fusco L, et al. Graphene and other 2D materials: a multidisciplinary analysis to uncover the hidden potential as cancer theranostics. Theranostics. 2020;10(12):5435–88. https://doi.org/10.7150/thno.40068.
Gaikwad S, et al. Antiviral activity of mycosynthesized silver nanoparticles against herpes simplex virus and human parainfluenza virus type 3. Int J Nanomed. 2013;8:4303–14. https://doi.org/10.2147/IJN.S50070.
Gao G, et al. Absent immune response to SARS-CoV-2 in a 3-month recurrence of coronavirus disease 2019 (COVID-19) case. Infection. 2020. https://doi.org/10.1007/s15010-020-01485-6.
Gu J, Han B, Wang J. COVID-19: gastrointestinal manifestations and potential fecal-oral transmission. Gastroenterology. 2020. https://doi.org/10.1053/j.gastro.2020.02.054.
Guan Y et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China | Science. 2003. https://doi.org/10.1126/science.1087139.
Guo Y-R, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak—an update on the status. Mili Med Res. 2020. https://doi.org/10.1186/s40779-020-00240-0.
He Z, et al. Effects of severe acute respiratory syndrome (SARS) coronavirus infection on peripheral blood lymphocytes and their subsets. Int J Infect Dis. 2005;9(6):323–30. https://doi.org/10.1016/j.ijid.2004.07.014.
He M-L, et al. Kinetics and synergistic effects of siRNAs targeting structural and replicase genes of SARS-associated coronavirus. FEBS Lett. 2006;580(10):2414–20. https://doi.org/10.1016/j.febslet.2006.03.066.
Hirose R, et al. Survival of SARS-CoV-2 and influenza virus on the human skin: importance of hand hygiene in COVID-19. Clin Infect Dis. 2020. https://doi.org/10.1093/cid/ciaa1517.
Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271. https://doi.org/10.1016/j.cell.2020.02.052.
Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. https://doi.org/10.1016/S0140-6736(20)30183-5.
Huang Z, Jiang Y, Chen J, Zhou Y. Inhibitors of the renin–angiotensin system: the potential role in the pathogenesis of COVID-19. Cardiol J. 2020. https://doi.org/10.5603/CJ.a2020.0056.
Hurst KR, Koetzner CA, Masters PS. Characterization of a critical interaction between the coronavirus nucleocapsid protein and nonstructural protein 3 of the viral replicase-transcriptase complex. J Virol. 2013;87(16):9159–72. https://doi.org/10.1128/JVI.01275-13.
Iavarone C, O’hagan DT, Yu D, Delahaye NF, Ulmer JB. Mechanism of action of mRNA-based vaccines. Expert Rev Vaccin. 2017;16(9):871–81. https://doi.org/10.1080/14760584.2017.1355245.
Iorio-Morin C, et al. Letter: the risk of COVID-19 infection during neurosurgical procedures: a review of severe acute respiratory distress syndrome coronavirus 2 (SARS-CoV-2) modes of transmission and proposed neurosurgery-specific measures for mitigation. Neurosurgery. 2020. https://doi.org/10.1093/neuros/nyaa157.
Isho B, et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci Immunol. 2020;5(52):08. https://doi.org/10.1126/sciimmunol.abe5511.
Itani R, Tobaiqy M, Al Faraj A. Optimizing use of theranostic nanoparticles as a life-saving strategy for treating COVID-19 patients. Theranostics. 2020;10(13):5932–42. https://doi.org/10.7150/thno.46691.
Iyer AS, et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci Immunol. 2020;5(52):08. https://doi.org/10.1126/sciimmunol.abe0367.
Ji W, Wang W, Zhao X, Zai J, Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 2020;92(4):433–40. https://doi.org/10.1002/jmv.25682.
Kuiken T, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362(9380):263–70. https://doi.org/10.1016/S0140-6736(03)13967-0.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4. https://doi.org/10.1093/molbev/msw054.
Kuo L, Masters PS. Functional analysis of the murine coronavirus genomic RNA packaging signal. J Virol. 2013;87(9):5182–8192. https://doi.org/10.1128/JVI.00100-13.
Li F. Structure function and evolution of coronavirus spike proteins. Ann Rev Virol. 2016;3(1):237–61. https://doi.org/10.1146/annurev-virology-110615-042301.
Li G, Chen X, Xu A. Profile of specific antibodies to the SARS-associated coronavirus. N Engl J Med. 2003;349(5):508–9.
Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–8. https://doi.org/10.1126/science.1116480.
Li X, Geng M, Peng Y, Meng L, Lu S. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020;10(2):102–8. https://doi.org/10.1016/j.jpha.2020.03.001.
Liu P, Chen W, Chen J-P. ‘Viral metagenomics revealed sendai virus and coronavirus infection of Malayan Pangolins (Manis javanica). Viruses. 2019;11(11):979. https://doi.org/10.3390/v11110979.
Liu T, et al. Prevalence of IgG antibodies to SARS-CoV-2 in Wuhan - implications for the ability to produce long-lasting protective antibodies against SARS-CoV-2. medRxiv. 2020. https://doi.org/10.1101/2020.06.13.20130252.
Long Q-X, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020. https://doi.org/10.1038/s41591-020-0897-1.
Lu R, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–74. https://doi.org/10.1016/S0140-6736(20)30251-8.
Lu D, Sang L, Du S, Li T, Chang Y, Yang X-A. Asymptomatic COVID-19 infection in late pregnancy indicated no vertical transmission. J Med Virol. 2020. https://doi.org/10.1002/jmv.25927.
Ludvigsson JF. Systematic review of COVID-19 in children shows milder cases and a better prognosis than adults. Acta Paediatr. 2020;109(6):1088–95.
Luk HKH, Li X, Fung J, Lau SKP, Woo PCY. Molecular epidemiology, evolution and phylogeny of SARS coronavirus. Infect Genet Evol. 2019;71:21–30. https://doi.org/10.1016/j.meegid.2019.03.001.
Meagher JL, et al. Structure of the zinc-finger antiviral protein in complex with RNA reveals a mechanism for selective targeting of CG-rich viral sequences. Proc Natl Acad Sci USA. 2019;116(48):24303–9. https://doi.org/10.1073/pnas.1913232116.
Molenkamp R, Spaan WJ. Identification of a specific interaction between the coronavirus mouse hepatitis virus A59 nucleocapsid protein and packaging signal. Virology. 1997;239(1):78–86. https://doi.org/10.1006/viro.1997.8867.
Nieto-Torres JL, et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLOS Pathog. 2014. https://doi.org/10.1371/journal.ppat.1004077.
Nikaeen G, Abbaszadeh S, Yousefinejad S. Application of nanomaterials in treatment, anti-infection and detection of coronaviruses. Nanomedicine. 2020. https://doi.org/10.2217/nnm-2020-0117.
Nikolich-Zugich J, Knox KS, Rios CT, Natt B, Bhattacharya D, Fain MJ. SARS-CoV-2 and COVID-19 in older adults: what we may expect regarding pathogenesis, immune responses, and outcomes. GeroScience. 2020. https://doi.org/10.1007/s11357-020-00186-0.
Normile D. Mystery virus found in Wuhan resembles bat viruses but not SARS, Chinese scientist says, Science | AAAS, https://www.sciencemag.org/news/2020/01/mystery-virus-found-wuhan-resembles-bat-viruses-not-sars-chinese-scientist-says (2020).
Nouri-Vaskeh M, Alizadeh L. Fecal transmission in COVID-19: a potential shedding route. J Med Virol. 2020. https://doi.org/10.1002/jmv.25816.
Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines—a new era in vaccinology. Nat Rev Drug Disc. 2018;17(4):261–79. https://doi.org/10.1038/nrd.2017.243.
Patrì A, Gallo L, Guarino M, Fabbrocini G. Sexual transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a new possible route of infection? J Am Acad Dermatol. 2020. https://doi.org/10.1016/j.jaad.2020.03.098.
Peng H, et al. Human memory T cell responses to SARS-CoV E protein. Microbes Infect. 2006;8(9–10):2424–31. https://doi.org/10.1016/j.micinf.2006.05.008.
Prather KA, Wang CC, Schooley RT. Reducing transmission of SARS-CoV-2. Science. 2020. https://doi.org/10.1126/science.abc6197.
Prentice E, McAuliffe J, Lu X, Subbarao K, Denison MR. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J Virol. 2004;78(18):9977–86. https://doi.org/10.1128/JVI.78.18.9977-9986.2004.
Qiu G, Gai Z, Tao Y, Schmitt J, Kullak-Ublick GA, Wang J. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14(5):5268–77. https://doi.org/10.1021/acsnano.0c02439.
Rabaan AA, et al. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Infez Med. 2020;28(2):2.
Rai M, et al. Nanotechnology-based promising strategies for the management of COVID-19: current development and constraints. Expert Rev Anti-Infect Ther. 2020. https://doi.org/10.1080/14787210.2021.1836961.
Rai M, Kon K, Ingle A, Duran N, Galdiero S, Galdiero M. Broad-spectrum bioactivities of silver nanoparticles: the emerging trends and future prospects. Appl Microbiol Biotechnol. 2014;98(5):1951–61. https://doi.org/10.1007/s00253-013-5473-x.
Ratajczak MZ, Kucia M. SARS-CoV-2 infection and overactivation of Nlrp3 inflammasome as a trigger of cytokine “storm” and risk factor for damage of hematopoietic stem cells. Leukemia. 2020;34:1–4. https://doi.org/10.1038/s41375-020-0887-9.
Rehman SU, Shafique L, Ihsan A, Liu Q. Evolutionary trajectory for the emergence of novel coronavirus SARS-CoV-2. Pathogens. 2020;9(3):240. https://doi.org/10.3390/pathogens9030240.
Remy KE, et al. Immunotherapies for COVID-19: lessons learned from sepsis. Lancet Resp Med. 2020. https://doi.org/10.1016/S2213-2600(20)30217-4.
Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–25. https://doi.org/10.1093/oxfordjournals.molbev.a040454.
Shetty Y, Prabhu P, Prabhakar B. Emerging vistas in theranostic medicine. Int J Pharm. 2019;558:29–42. https://doi.org/10.1016/j.ijpharm.2018.12.068.
Smith TRF, et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat Commun. 2020. https://doi.org/10.1038/s41467-020-16505-0.
Sportelli MC, et al. Can Nanotechnology and materials science help the fight against SARS-CoV-2? Nanomaterials. 2020;10(4):802. https://doi.org/10.3390/nano10040802.
Sreepadmanabh M, Sahu AK, Chande A. COVID-19: Advances in diagnostic tools, treatment strategies, and vaccine development. J Biosci. 2020;45(1):148. https://doi.org/10.1007/s12038-020-00114-6.
Stadnytskyi V, Bax CE, Bax A, Anfinrud P. The airborne lifetime of small speech droplets and their potential importance in SARS-CoV-2 transmission. Proc Natl Acad Sci. 2020;117(22):11875–7.
Stohlman SA, Baric RS, Nelson GN, Soe LH, Welter LM, Deans RJ. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J Virol. 1988;62(11):4288–95.
Sturman LS, Holmes KV, Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J Virol. 1980;33(1):449–62.
Sun X, et al. Cytokine storm intervention in the early stages of COVID-19 pneumonia. Cytokine Growth Factor Rev. 2020. https://doi.org/10.1016/j.cytogfr.2020.04.002.
Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101(30):11030. https://doi.org/10.1073/pnas.0404206101.
Tian S, Hu W, Niu L, Liu H, Xu H, Xiao S-Y. Pulmonary pathology of early-phase 2019 novel coronavirus (COVID-19) pneumonia in two patients with lung cancer. J Thorac Oncol. 2020;15(5):700–4. https://doi.org/10.1016/j.jtho.2020.02.010.
Tillett RL, et al. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2020. https://doi.org/10.1016/S1473-3099(20)30764-7.
Transmission of SARS-CoV-2: implications for infection prevention precautions. https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions.
Ullah S, et al. Reusability comparison of melt-blown vs nanofiber face mask filters for use in the coronavirus pandemic. ACS Appl Nano Mater. 2020;3(7):7231–41. https://doi.org/10.1021/acsanm.0c01562.
van Doremalen N, et al. Aerosol and Surface Stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl J Med. 2020. https://doi.org/10.1056/NEJMc2004973.
Wang Q, Qiu Y, Li J-Y, Zhou Z-J, Liao C-H, Ge X-Y. A unique protease cleavage site predicted in the spike protein of the novel pneumonia coronavirus (2019-nCoV) potentially related to viral transmissibility. Virol Sin. 2020. https://doi.org/10.1007/s12250-020-00212-7.
Ward H, et al. Declining prevalence of antibody positivity to SARS-CoV-2: a community study of 365,000 adults. medRxiv. 2020. https://doi.org/10.1101/2020.10.26.20219725.
Weiss C, et al. Toward nanotechnology-enabled approaches against the COVID-19 pandemic. ACS Nano. 2020;14(6):6383–406. https://doi.org/10.1021/acsnano.0c03697.
West R, Michie S, Rubin GJ, Amlôt R. Applying principles of behaviour change to reduce SARS-CoV-2 transmission. Nat Hum Behav. 2020;4(5):5. https://doi.org/10.1038/s41562-020-0887-9.
WHO. Modes of transmission of virus causing COVID-19: implications for IPC precaution recommendations. 2020. https://www.who.int/news-room/commentaries/detail/modes-of-transmission-of-virus-causing-covid-19-implications-for-ipc-precaution-recommendations.
Wong XY, Sena-Torralba A, Álvarez-Diduk R, Muthoosamy K, Merkoçi A. Nanomaterials for nanotheranostics: tuning their properties according to disease needs. ACS Nano. 2020;14(3):2585–627. https://doi.org/10.1021/acsnano.9b08133.
Wrapp D, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–3.
Wu F, et al. Complete genome characterisation of a novel coronavirus associated with severe human respiratory disease in Wuhan, China. bioRxiv. 2020. https://doi.org/10.1101/2020.01.24.919183.
Wu F, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–9. https://doi.org/10.1038/s41586-020-2008-3.
www.ETHealthworld.com. Novel coronavirus can survive on skin for 9 hours: study—ET healthworld, ETHealthworld.com. https://health.economictimes.indiatimes.com/news/diagnostics/novel-coronavirus-can-survive-on-skin-for-9-hours-study/78587107.
www.ETHealthworld.com. WHO acknowledges “evidence emerging” of airborne spread of COVID-19—ET healthworld, ETHealthworld.com. https://health.economictimes.indiatimes.com/news/industry/who-acknowledges-evidence-emerging-of-airborne-spread-of-covid-19/76847993.
Xia X. Extreme genomic CpG deficiency in SARS-CoV-2 and evasion of host antiviral defense | molecular biology and evolution | Oxford Academic. 2020. https://doi.org/10.1093/molbev/msaa094.
Xia S, et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–55. https://doi.org/10.1038/s41422-020-0305-x.
Xia S, et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020;17:1–3. https://doi.org/10.1038/s41423-020-0374-2.
Xu Z, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–2. https://doi.org/10.1016/S2213-2600(20)30076-X.
Xu X, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63(3):457–60. https://doi.org/10.1007/s11427-020-1637-5.
Yang L-T, et al. Long-lived effector/central memory T-cell responses to severe acute respiratory syndrome coronavirus (SARS-CoV) S antigen in recovered SARS patients. Clin Immunol. 2006;120(2):171–8. https://doi.org/10.1016/j.clim.2006.05.002.
Ye Z-W, Yuan S, Yuen K-S, Fung S-Y, Chan C-P, Jin D-Y. Zoonotic origins of human coronaviruses. Int J Biol Sci. 2020;16(10):1686–97. https://doi.org/10.7150/ijbs.45472.
Zeng C, et al. Leveraging mRNAs sequences to express SARS-CoV-2 antigens in vivo. bioRxiv. 2020. https://doi.org/10.1101/2020.04.01.019877.
Zhang Y-Z, Holmes EC. A genomic perspective on the origin and emergence of SARS-CoV-2: Cell. 2020. https://doi.org/10.1016/j.cell.2020.03.035.
Zhang T, Wu Q, Zhang Z. Probable pangolin origin of 2019-nCoV associated with outbreak of COVID-19. Curr-Biol-D-20–00299. 2020.
Zhang C, Wu Z, Li J-W, Zhao H, Wang G-Q. The cytokine release syndrome (CRS) of severe COVID-19 and interleukin-6 receptor (IL-6R) antagonist tocilizumab may be the key to reduce the mortality. Int J Antimicrob Agents. 2020;55:105954. https://doi.org/10.1016/j.ijantimicag.2020.105954.
Zhang L, Shen F, Chen F, Lin Z. Origin and evolution of the 2019 novel coronavirus. Clin Infect Dis. 2020. https://doi.org/10.1093/cid/ciaa112.
Zhang Y, Gao Y, Qiao L, Wang W, Chen D. Inflammatory response cells during acute respiratory distress syndrome in patients with coronavirus disease 2019 (COVID-19). Ann Intern Med. 2020. https://doi.org/10.7326/L20-0227.
Zheng M, et al. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17(5):533–5. https://doi.org/10.1038/s41423-020-0402-2.
Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–3. https://doi.org/10.1038/s41586-020-2012-7.
Zhou F, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–62. https://doi.org/10.1016/S0140-6736(20)30566-3.
Zhu X, et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens Bioelectron. 2020;166:112437. https://doi.org/10.1016/j.bios.2020.112437.
Ziegler CGK, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020. https://doi.org/10.1016/j.cell.2020.04.035.
Acknowledgments
Authors thank Ankit Sahu, Minita Desai, Pallavi Gupta and all the lab members for their help at various stages during preparation of the draft. AKS and SM acknowledge fellowships from DST-INSPIRE and KVPY, respectively. AC is a recipient of the IYBA fellowship from the DBT. AC thanks the Department of Biotechnology (DBT), and the DBT/Wellcome Trust India Alliance for supporting various research programs in the lab. We are thankful to BioRender for providing us the platform to generate the figures.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sahu, A.K., Sreepadmanabh, M., Rai, M. et al. SARS-CoV-2: phylogenetic origins, pathogenesis, modes of transmission, and the potential role of nanotechnology. VirusDis. 32, 1–12 (2021). https://doi.org/10.1007/s13337-021-00653-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13337-021-00653-y