Abstract
The novel Coronavirus disease (COVID-19), which first appeared at the end of December 2019, continues to spread rapidly in most countries of the world. Respiratory infections occur primarily in the majority of patients treated with COVID-19. In light of the growing number of COVID-19 cases, the need for diagnostic tools to identify COVID-19 infection at early stages is of vital importance. For decades, chest X-ray (CXR) technologies have proven their ability to accurately detect respiratory diseases. More recently, with the availability of COVID-19 CXR scans, deep learning algorithms have played a critical role in the healthcare arena by allowing radiologists to recognize COVID-19 patients from their CXR images. However, the majority of screening methods for COVID-19 reported in recent studies are based on 2D convolutional neural networks (CNNs). Although 3D CNNs are capable of capturing contextual information compared to their 2D counterparts, their use is limited due to their increased computational cost (i.e. requires much extra memory and much more computing power). In this study, a transfer learning-based hybrid 2D/3D CNN architecture for COVID-19 screening using CXRs has been developed. The proposed architecture consists of the incorporation of a pre-trained deep model (VGG16) and a shallow 3D CNN, combined with a depth-wise separable convolution layer and a spatial pyramid pooling module (SPP). Specifically, the depth-wise separable convolution helps to preserve the useful features while reducing the computational burden of the model. The SPP module is designed to extract multi-level representations from intermediate ones. Experimental results show that the proposed framework can achieve reasonable performances when evaluated on a collected dataset (3 classes to be predicted: COVID-19, Pneumonia, and Normal). Notably, it achieved a sensitivity of 98.33%, a specificity of 98.68% and an overall accuracy of 96.91%
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The novel Coronavirus (COVID-19), which originated in Wuhan, China at the end of 2019, has become a serious threat worldwide to public health [1]. Accordingly, this pandemic stands as a global health emergency. The continuing spread of the COVID-19 pandemic, which claims a large number of victims and serious infections every day, affecting several territories, such as the United States, Italy, Spain, etc., makes its treatment increasingly challenging. According to the World Health Organization (WHO), COVID-19 has a considerable impact on human beings. Till November 2020, the number of confirmed COVID-19 cases in the affected countries reached 58,900,547, of which 1,393,305 have died, reported to WHO. The majority of infected patients suffer from mild to moderate respiratory issues, the severity of which changes over time [2]. The disease is highly contagious because it can spread rapidly from infected people to healthy people through micron-size droplets from the nose and oral cavity or close contact between infected and uninfected people; COVID-19 has a reproduction rate of 3 or more, meaning that on average 3 or more people can be infected per COVID-19 case [3].
Currently, the most recognized strategy to fight the pandemic involves intensified screening for the infection [4]. To slow down its spread, millions of people still need to be screened over time. The real-time reverse transcriptase-polymerase chain reaction (RT-PCR) test can detect and screen the presence of the virus with a high-level of sensitivity. This kind of test has often been used as the main screening process for COVID-19 by directly identifying the existence of the virus. Despite this advantage, RT-PCR suffers from some drawbacks. For example, some centers have a limited number of RT-PCR kits available and there is a high risk of receiving false-negative RT-PCR results over time [5]. Moreover, it is very time-consuming and expensive especially when the collected specimens need to be processed by external specialized laboratories. Hence, this makes it very difficult to perform RT-PCR testing for a large number of suspected patients in as short a time as possible.
To address these shortcomings, researchers have developed an important complement to RT-PCR tests by detecting COVID-19 from chest computed tomography (CT) scans [6,7,8,9]. This will help reduce treatment delays and patient isolation. In [6], Ai et al. concluded that chest CT scans were more sensitive for the diagnosis of COVID-19 than traditional RT-PCR, although it should be noted this work was published very early in the pandemic. In [7], Dangis et al. showed that the chest CT scanner has considerable performance in diagnosing COVID-19 in terms of speed, specificity, and sensitivity. A comparative study of 51 patients showed that chest CT scans were very sensitive (50/51 patients) for screening for COVID-19 disease [8]. In a study by Bai et al. [9], a total of 219 COVID-19 positive patients detected by both RT-PCR and chest CT were conducted. The researchers concluded quantitatively that the distinction of COVID-19 from other types of pneumonia on CT chest scans was more sensitive than RT-PCR tests. Regardless of these strengths, CT scans still have some limitations. Among these is the fact that screening for COVID-19 generally takes much longer to perform. Moreover, high-quality scanners are very expensive and patients have to deal with much higher radiation doses [10].
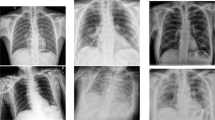
In contrast, chest X-rays (CXRs) are one of the most common large-scale medical imaging techniques that have proven to be effective in speeding-up and facilitating the screening of the pandemic [11, 12]. These techniques allow the early detection and tracking of the virus. Figure 1 shows CXRs of an older man patient from Wuhan, China [13]. These samples indicate the progression of lung consolidation from days 0, 4, and 7, respectively. However, COVID-19 patients have some clinical features and symptoms that make it distinctive from other types of viral pneumonia [14, 15]. For example, Zhao et al. [14] compared the difference between COVID-19 pneumonia and other cases of pneumonia. They found that the patients’ features were very similar and the most common symptoms were fever and cough. Generally, the early symptoms that make COVID-19 infection different from other viruses were fever, dry cough, heavy sweating, and weakness. Consequently, it was difficult to distinguish COVID-19 pneumonia when compared to other cases of pneumonia.
Temporal evolution of CXR outcomes in an older male patient with three consolidation changes in the lung periphery [13]
AI-based methods have been widely used in many real-world applications, including healthcare, medical imaging, and so on. For example, AI-based CT scans were introduced to distinguish COVID-19 features from other diseases [16]. Currently, the development of new diagnostic methods using deep learning algorithms is underway.
Convolutional neural networks (CNNs) have been one of the deep neural networks (DNNs) used to screen for COVID-19 from patient data, leading to impressive results [17]. The availability of large-scale CT and CXR image datasets for deep model training and testing purposes has encouraged the deep learning community to develop intelligent systems for the screening of coronavirus disease [17,18,19]. For instance, the results obtained in [17] showed that the use of large CT datasets (618 samples in total) for the training model led to reliable COVID-19 diagnostic outcomes. Since the beginning of the pandemic, many researchers have proposed a range of deep learning-based methods for COVID-19 detection and diagnosis from X-ray scans [20,21,22,23,24,25,26]. Narin et al. [20] developed a deep model based on CXR data with a high accuracy of up to 98%. The network parameters were loaded from a pre-trained ResNet-50 [27] model. Similarly, Apostolopoulos and Bessiana [21] used the transfer learning technique with CNNs to speed-up the detection of COVID-19 on X-ray samples. They achieved an accuracy, sensitivity, and specificity of 96.78%, 98.66%, and 96.46%, respectively. In [22], Karim et al. proposed a screening model based on ensemble learning techniques, called “DeepCOVIDExplainer”, which was used to predict the COVID-19 pandemic from X-ray features (16,995 samples in total). Basu et al. [23] introduced a binary classification model (i.e. 2 classes to be predicted: COVID-19 or non-COVID-19, respectively) using ResNet-18 as the backbone network. The backbone network was used to extract visual patterns from CXR images following the transfer learning paradigm. They reached a sensitivity of 96.00% and a specificity of 70.65% on 1531 X-ray images. In [24], Hemdan et al. developed a powerful deep learning framework, called COVIDX-Net, based on CXR images and deep CNN models (e.g. VGG, ResNet, etc.). The best-proposed model received F1-scores of 89% (normal cases) and 91% (COVID-19 cases), respectively. Ozturk et al. [25] introduced a deep learning-based algorithm for booth binary and multi-class classification to discriminate COVID-19 patients from other diseases in CXR images. The model was produced with an accuracy of 98.08–87.02% for binary and multi-class scenarios. In [26], Toğaçar et al. introduced a novel screening system based on fuzzy color and social mimic optimization techniques for automated classification of CXR images. The overall classification rate of 99.27% was the outcome of the model. Das et al. [28] proposed the Truncated Inception Net to detect COVID-19 positive cases from other non-COVID cases. It should be noted that all these methods were based on 2D CNN architectures.
Most computer vision tasks, such as image classification, semantic segmentation, object recognition, etc., are based on 2D CNN architectures [29]. However, only a small number of methods using 3D CNNs have been adopted, mainly because of their complexity and their inability to perform well for classes with similar visual appearances across several bands [30]. In fact, the main characteristic that distinguishes 2D CNNs from 3D CNNs is their ability to identify contextual structures from high-dimensional spaces. In particular, 2D CNNs can only handle 2D datasets by capturing visual 2D information (i.e. width and height) from still images. Conversely, their 3D counterparts can efficiently process 3D input data by locally stacking nodes in three dimensions (i.e. width, height, and depth). Regarding applications, these complex models are capable of processing spatial and contextual cues from video sequences [31] and are most commonly used to analyze medical imaging (e.g. CT scans, MRI, etc.) for detection, screening, and development of patient-specific models [32]. Despite the aforementioned strengths, there is a potential problem when deep 3D CNN architectures are learned end-to-end, especially after stacking 3D convolution layers, maximizing the expansion of feature dimensions and parameter space. Accordingly, the motivation behind the proposal of a hybrid 2D/3D CNN model is to fill these gaps based on the transfer learning paradigm. An adaptive combination of a pre-trained deep 2D CNN and a shallow 3D CNN allows better use of the dynamic synergies between the multidimensional activation maps, generating more informative patterns and, thus, enabling more powerful learning techniques for the whole network. For instance, we have previously combined 2D–3D CNNs based on the transfer learning paradigm to ensure a better compromise between accuracy and efficiency [33].
In this paper, a faster and more accurate deep learning system has been developed for screening the COVID-19 pandemic based on the combination of a deep 2D CNN pre-trained on ImageNet (i.e. VGG16 [34]) with a shallow 3D CNN, a depth-wise separable convolution [35], and a spatial pyramid pooling module [36], using CXR samples. More specifically, our idea is to transfer the 2D parameters learned from a 2D CNN and integrate them into a shallow 3D CNN, so that extra contextual information gets provided and the training process is faster.
The main innovation presented in this paper is the introduction of a novel hybrid 2D/3D CNN-based framework to efficiently and effectively diagnose COVID-19 from CXR images. To our knowledge, we are the first to propose a hybrid 2D/3D CNN network for the screening of COVID-19 using CXR images.
Methodology
Collections
In practice, collecting and annotating medical image collections is a very challenging task. For reasons of data confidentiality, only a few large CXR datasets are publicly available. Although these datasets can be used for classification purposes, there are only a few large datasets associated with COVID-19. In this work, the main sources of CXR images are derived from three publicly available collections: COVID-19 collection (C1) [18], COVID-19 Radiography collection (C2) [37], and Pneumonia collection (C3) [19]. In collection C1, the data collected included updated items for patients with COVID-19 (535 CXRs) and other viral and bacterial Pneumonia (MERS, SARS, and ARDS). Here, only COVID-19 positive posterior–anterior CXRs are taken into account. Collection C2 consists of CXR scans of COVID-19 positive cases (219 CXRs) with normal (1341 CXRs) and viral Pneumonia (1345 CXRs). The third collection C3 consists of a total of 5863 CXR scans of patients. It includes 1583 normal (i.e. without lung infection) cases and 4280 viral and bacterial Pneumonia cases. Figure 2 illustrates an example of CXR images from Normal cases (Fig. 2a), COVID-19 patients (Fig. 2b), and Pneumonia cases (Fig. 2c).
For our purposes, a total of 754 COVID-19 positive CXRs from the first and second collections (C1 and C2), a total of 1341 normal CXRs from the second and third collections (C2 and C3), and only a total of 1345 Pneumonia positive CXRs from the third collection (C3) are considered (3440 in total). Table 1 displays the distribution of data collected for each category.
The details of the experiments conducted will be discussed and explained in “Experiments” section.
Proposed method
This section describes the proposed transfer learning-based method for end-to-end screening of COVID-19 infected patients from CXR images.
Motivation and description
In computer vision, transfer learning, commonly known as cross-domain adaptation [38], aims to strengthen the generalization capacity of the target model by loading knowledge from the source one. Moreover, this paradigm can effectively contribute to overcoming the lack of available CXR datasets. Up to now, no attempt has been made to use 3D CNNs to learn spatial and contextual cues for effective recognition of COVID-19 using CXR data. Despite their promising capabilities, 3D CNNs are not as efficient for still image recognition as their 2D counterparts. This may be due to the complexity of feature mapping and the enormous memory costs associated with stacking multiple 3D convolution layers. Therefore, we propose a transfer learning-based hybrid 2D/3D CNN, called Hybrid-COVID, that integrates a pre-trained deep 2D CNN with a series of 3D convolutions to generate and learn more complex and informative representations while reducing model complexity, when modeling spatio-contextual information at each processing stage.
Figure 3 show the workflow diagram of the proposed framework, which is composed mainly of six blocks, namely, respectively, a pre-trained 2D CNN (VGG16 network), an up-sampling step, a depth-wise separable convolution, a shallow 3D CNN (building block), a spatial pyramid pooling (SPP) module, followed by the classification scheme (dense layers).
Inspired by the work of [33], the VGG16 model is used as a backbone network for extracting generic features from a huge dataset (i.e. ImageNet). This pre-trained model has already achieved high performance for the image classification task. The up-sampling step plays a crucial role, as it is useful to reconstruct the loaded feature maps without skipping any spatial information. The depth-wise separable convolution ensure that useful features are preserved while reducing the model’s computational cost. The computational complexity of stacking multiple layers of 3D convolution layers could be overcome by fine-tuning a pre-trained model, making it easier to learn deeper and more informative feature maps. This involves significantly reducing network parameters as well as retaining 2D spatial information that is otherwise missed by the 2D CNN. A systematic integration of 2D and 3D convolutions can be achieved by reshaping their output activations through merging operations. An SPP module is then suitable for extracting multi-scale representations from intermediate levels.
In the following subsections, we will describe in detail each block of the proposed Hybrid-COVID model.
Hybrid-COVID
Hybrid-COVID is a novel transfer learning-based model architecture that aims to ensure a better compromise between accuracy and computational complexity while integrating both 2D and 3D CNNs. Table 2 shows the output shape for each level of our architecture.
In the following paragraphs, each component of the model will be described in more detail.
2D CNN in computer vision, 2D CNN models have been one of the most popular deep models for image classification and understanding tasks [39]. They allow the dynamic extraction of discriminating features directly from raw data (e.g. still images, etc.) or from an intermediate level of the network. Visual Geometry Group Network, VGG [34] is one of the deep CNN architectures that have proven their performance in extracting generic features from large datasets. To reduce the network parameters and the computational complexity, the VGG network was used a cascaded convolution layers with \(3 \times 3\) filter sizes and 1 strides. This pre-trained network can be taken as a feature extractor by exploiting only its convolutional side. To date, this network is available in two versions, VGG16 and VGG19, which differ in their depth and hyper-parameters. In this work, we used VGG16 since it was lighter and cheaper than VGG19 for model training. Originally, the VGG16 network consists of several hidden layers (i.e. the convolutional side), including 13 convolution layers, and 3 visible layers (i.e. dense layers).
Up-sampling step structurally, the VGG16 architecture applies a series of 2D convolutions to the input data to generate sparse features while reducing their spatial dimensions through max-pooling layers. An up-sampling operation [40] is then performed, which upsamples the pooled representation, resulting in dense feature maps that can subsequently be used as input for the next processing step. Technically, this operator is the inverse transformation of the down-sampling (pooling) layer. In this work, we used only one UpSampling2D layer with 2 scale factors and bicubic interpolation. This step can contribute to reducing the number of unnecessary calculations without wasting any semantic information.
Depth-wise separable convolution our entire architecture is based on a depth-wise separable convolution operation [35]. Indeed, it is a variant form of the regular convolution operation. In regular convolution, the filter window is applied to multiple input channels, allowing the channels to be mixed separately to produce an output feature map. In contrast, the depth-wise separable convolution operation convolves each input channel separately, after which a point-wise convolution will be performed to combine the resulting output channels. In other words, a 2D convolution with a filter size of \(3\times 3\) will be factorized into a depth-wise convolution of \(3\times 3\) and a point-wise convolution of \(1\times 1\), each capturing spatial and cross-channel correlations. Formally, consider a standard convolutional kernel K of size W \(\times \) W \(\times \) M\( \times \) N, where W is the spatial dimension of the kernel, M is the number of input channels and N is the number of output channels, respectively. After its application on the input representation I, the output feature map F will be
The shape of the input and output representation is S \(\times \) S \(\times \) M and S \(\times \) S \(\times \) N, where S stands for the spatial dimension, and M and N for the number of input and output channels, respectively. In depth-wise separable convolution, we first extract a spatial representation from depth-wise convolution (\({\tilde{K}}\)) of size W \(\times \) W,
we then apply the point-wise convolution (\({\hat{K}}\)) of size 1 \(\times \) 1 \(\times \) M across the channel to mix the output of the depth-wise convolution. Thus, the output of the depth-wise separable convolution will be
Figure 4 displays the steps of the depth-wise separable convolution operation. In this study, we used only a single layer with a kernel size of \(3\times 3\). The use of this operation makes it possible to reduce the network parameters and speed-up the convergence rate of the model.
3D CNN in this work, a set of 3D convolution kernels is applied to the reshaped feature maps (i.e. 3D data cubes) to capture any inherited contextual patterns in the data stream. This is accomplished by using a reshaping operation that ensures the spatial matching of the input and output shapes while keeping the sliced data unchanged. Structurally, the proposed 3D CNN consists of stacking three convolution layers (i.e. a shallow network) applied with kernels of different sizes [(\(3\times 3\times 3\)), (\(2\times 2\times 2\)), and (\(1\times 1 \times 1\)), respectively]. Note that the padding and stride hyper-parameters remain at 1 for simplicity. Also, in order to reduce the number of trainable parameters and save resources, we only set the number of filters in each layer to 5. Compared to 2D CNN, 3D CNN shows better modeling capacity of contextual information by performing 3D convolutions on the space axes without loss of relevant features [41]. Technically, a 3D convolution can represent the multi-dimensional input information as a 4D tensor of the shape [D \(\times \) H \(\times \) W \(\times \) C], where D, H, W, C refers to the depth, height, width and number of slices, respectively. In this sense, a 3D CNN network can handle features at different scales, with one path for spatial information and another path for broader contextual information.
Unlike an ordinary 3D CNN model that takes multiple input video images [31] or hierarchical 3D medical image information [32], the proposed 3D building block fine-tunes the spatial representations of the previous stage for improved sharing of knowledge and computation across domains.
By sliding a 3D convolution kernel across the spatial and contextual dimensions, the output representation at the position (h, w, d) on the jth feature tensor of the ith layer can be computed as Eq. 4.
where \(\varPhi \) is the activation function, also called non-linearity function, \(w_{i,j,n}^{l,k,t}\) and \(b_{i,j}\) are the network parameters, n is the index of all feature maps. \(L_{i}\) and \(K_{i}\) designate respectively the spatial dimensions of the 3D convolution kernel, while \(T_{i}\) designates the contextually-ordered-feature axis. Figure 5 depicts a 3D convolution layer with h \(\times \) w \(\times \) d inputs and l \(\times \) k \(\times \) t kernel.
Spatial pyramid pooling in this paper, we integrated the spatial pyramid pooling (SPP) module [36] in our model architecture to extract multi-scale feature maps from spatial dimensions. Technically, SPP is the dynamic variant of the pooling operation, which consists of extracting a fixed-length feature map from representations of arbitrary size by concatenating their spatial outputs. In particular, the SPP technique is used in this work to produce feature vectors of fixed dimensions for X-ray features of varying scales. This can reduce scale-variance and avoid the risk of over-fitting concern. The proposed structure of the SPP block is shown in Fig. 6. The input of the SPP block is a set of reshaped feature maps of the last 3D convolution layer. The final classification is carried out by dense layers applied on SPP-features.
Hybridization process
The goal of the hybridization process is to learn multi-domain features from multi-dimensional data cube using both 2D and 3D CNNs. The proposed Hybrid-COVID model mainly consists of two correlated sides: the 2D CNN side and the 3D CNN side. After removal of the last dense layer, the 2D CNN side (i.e. a pre-trained VGG16 network) is used to load discriminating and abstract representations that are learned from a huge dataset (i.e. 1000K-ImageNet). Furthermore, the 3D CNN side is fine-tuned by the 2D CNN one to learn high-level contextual features from the reshaped data. Thus, the Hybrid-COVID model is produced by integrating both 2D CNN and 3D CNN through reshaping operations. This operation can be formulated as follows:
where \(R_{2D}^{l}\) and \(R_{3D}^{l}\) represent the learned activations from 2D and 3D CNN sides, respectively. f(.) is a function that allows the input tensors to be reshaped into a fixed-size tensor with the same rank values. As shown in Table 2, after performing the first 3D convolution layer (Conv3D) on the input tensor (reshaped feature map) with a learned 3D kernel (width, height, and channel), 2 spatial dimensions (\(3\times 3\)) and 2 feature dimensions (\(512\times 5\)) are generated, forcing the first of these dimensions to learn features that are in a meaningful sequence by running the convolution over them.
Classification scheme
The classification scheme is performed at the end of the SPP block to categorize the input CXR image into either a positive COVID-19 case, a positive Pneumonia, or a Normal case. For this purpose, we first added a dense layer of 64 neurons, followed by the rectified linear unit (ReLU) activation function. Indeed, ReLU is one of the fastest and most effective learning activation functions that has proven its effectiveness in avoiding the problem of vanishing gradient [42]. A last dense layer of three neurons, followed by the Softmax activation function (i.e. a multi-class predictor) is then included to perform the final prediction.
Experiments
In this subsection, we discuss three different experiments conducted to test and validate the performance of the proposed method. Note that in all these experiments, our Hybrid-COVID model is trained with the same hyper-parameters as those described in “Training implementation” section.
Baseline experiment
In this experiment, a total of 3440 CXR images are collected from the 3 collections (C1 to C3) and splitted into 1341 normal cases (i.e. no pulmonary complications at the time of acquisition), 754 COVID-19 confirmed cases, and 1345 Pneumonia cases (both bacterial and viral Pneumonia). The collected dataset is referred to as Dataset-0. For this experiment, we compare our results with a range of deep learning methods, which are described in detail in “Comparison with state-of-the-art methods” section. Since the benchmark models are trained with different data distributions, we re-trained each one of them with the same training/testing set to ensure comparability of the results of each method (see Table 5). The evaluation of classification performance is covered in “Performance evaluation” section.
The visualization of some activation maps generated by our Hybrid-COVID model allows a better interpretation of the hybridization scheme. Besides, identifying regions where the disease is present is a vital task for further screening. The Grad-CAM [43] is used in this work as a tool to predict the class-discriminative localization map of an affected region in a seemingly unreasonable way. A detailed qualitative analysis of the performance of our model is provided in “Visualization” section.
Experiment 1
The collections (C1 to C3) already mentioned in “Collections” section are acquired on different scanners with different parameters under different protocols, which means that they are derived from different data domains.
Therefore, quantitative data from different sources can be combined to demonstrate the robustness of our model in detecting COVID-19 positive cases in order to clearly identify the heterogeneity of each database and ensure that they are free of bias. Hence, an additional experiment must be conducted to further validate and evaluate the predictive performance of our architecture. The motivation behind this policy is to make sure that the discrimination capacity of the models is not distorted by the presence of sample artifacts and outlier observations in the collected data. Specifically, we modified the original dataset by splitting it into subsets (sub-datasets) to examine the performance of the model in discriminating the clinical features of COVID-19 disease from other pneumonia cases. The following are the 15 different combinations of datasets (Datasets-1 to -15):
-
Dataset-1 includes 535 COVID-19 positive CXR images and 670 normal CXRs from collections C1 and C2, respectively.
-
Dataset-2 includes 535 COVID-19 positive CXR images and 671 normal CXRs from collections C1 and C3, respectively.
-
Dataset-3 includes 535 COVID-19 positive CXR images from the collection C1 and 1341 normal CXRs, combined from collections C2 and C3.
-
Dataset-4 includes 535 COVID-19 positive CXR images, 1345 Pneumonia CXRs, and 671 normal CXRs from collections C1 and C3, respectively.
-
Dataset-5 includes 535 COVID-19 positive CXR images and 2686 non-COVID-19 CXRs (including 1345 Pneumonia positive and 1341 normal CXRs).
-
Dataset-6 includes 219 COVID-19 positive CXR images and 670 normal CXRs from the collection C2.
-
Dataset-7 includes 219 COVID-19 positive CXR images and 671 normal CXRs from collections C2 and C3, respectively.
-
Dataset-8 includes 219 COVID-19 positive CXR images from the collection C2 and 1341 normal CXRs, combined from collections C2 and C3.
-
Dataset-9 includes 219 COVID-19 positive CXR images, 1345 Pneumonia CXRs, and 671 normal CXRs from collections C2 and C3, respectively.
-
Dataset-10 includes 219 COVID-19 positive CXR images and 2686 non-COVID-19 CXRs (including 1345 Pneumonia positive and 1341 normal CXRs).
-
Dataset-11 includes 754 COVID-19 positive CXR images (combined from collections C1 and C2) and 670 normal CXRs from the collection C2.
-
Dataset-12 includes 754 COVID-19 positive CXR images (combined from collections C1 and C2) and 671 normal CXRs from the collection C3.
-
Dataset-13 includes 754 COVID-19 positive CXR images (combined from collections C1 and C2) and 1341 normal CXRs, combined from collections C2 and C3.
-
Dataset-14 includes 754 COVID-19 positive CXR images (combined from collections C1 and C2) and 1345 Pneumonia CXRs and 671 normal CXRs from the collection C3.
-
Dataset-15 includes 754 COVID-19 positive CXR images and 2686 non-COVID-19 CXRs (including 1345 Pneumonia positive and 1341 normal CXRs).
Table 3 shows the distribution of CXRs across categories in the constructed datasets. Details of the quantitative results will be outlined and analyzed in “Impact of data and device heterogeneity” section.
Experiment 2
To examine the impact of the hybridization pipeline on classification performance, we conducted another experience under the following three scenarios:
-
Scenario 1 increase the number of 3D convolutions by a factor of 2 [i.e. make the 3D building block deeper by stacking a set of three 3D convolution layers with kernel sizes of \((3\times 3\times 3)\), \((2\times 2\times 2)\) and \((1\times 1\times 1)\) respectively].
-
Scenario 2 replace 3D convolutions with 2D counterparts (i.e. 2D CNN). Here, the reshaping operations are not necessary for space mapping. Specifically, the 2D convolution layers have kernel sizes of \((3\times 3)\), \((2 \times 2)\) and \((1\times 1)\), respectively.
-
Scenario 3 in this scenario, we redesigned the hybrid architecture by removing the 2D and 3D building blocks in which only the 2D parameters of VGG16 are fine-tuned. Specifically, the VGG16 model is used as a feature extractor, and its output parameters are fed into the input of the target classifier.
In “Impact of the hybridization pipeline” section, we will discuss the classification performance obtained under all scenarios.
Training implementation
During the training process, the network parameters will be adjusted using the Back-propagation algorithm, which iteratively decreases the loss function until convergence. In this work, the categorical cross-entropy loss is employed as the loss function. Besides, we used the Adam optimizer [44] for a faster convergence of the algorithm. We fixed the maximum epochs at 30, the batch size at 16, and the initial learning rate at 0.0003. Each training sample is resized to a spatial dimension of \(112\times 112\) pixels. The one-hot encoding scheme is then applied to convert the labeled data into numerical values. For model regularization, the dropout [45] and data augmentation [46] techniques are used to reduce imbalance in data distribution, improve generalizability, and prevent the overfitting problem when training our network. The conventional data augmentation technique is applied as a pre-processing step by rotating each CXR image by up to \(20^{\circ }\). The details of the algorithm of the proposed Hybrid-COVID model are summarized in Algorithm 1. Note that X is a set of CXR images, while Y represents the corresponding set of class labels.
The training process is carried out with tenfold cross-validation procedure. The implementation of the proposed model is achieved by using Python 3 and the publicly available Keras library with the TensorFlow 2.0 backend on Nvidia Tesla K80 GPUs with about 12 GB of memory.

Metrics
The proposed model shows acceptable experimental findings in terms of accuracy and efficiency on a collected dataset. To quantitatively prove this performance, therefore, standard measures are required for empirical analysis. These metrics include Precision (PREC), F1-Score (F-SCORE), Sensitivity (SEN)/Recall (REC), Specificity (SPE), and Accuracy (ACC). The PREC measures the correct prediction rate.
The F-SCORE is the harmonic mean of precision and recall, which aims to strike a balance between the two. The SEN and SPE are used to measure true positive and true negative rates, respectively. The ACC determines the percentage of correctly predicted labels of total tests. These measures can be calculated as follows:
Note that TP, FP, TN, and FN, respectively, stand for true positives, false positives, true negatives, and false negatives. In addition to the above measures, the confusion matrix is also included in our experiments to statistically evaluate the classification performance of the trained model by plotting the relationships between predicted values and ground-truth values.
The obtained results will be analysed and discussed in the “Results” and “Discussion” sections of this study.
Results
The main objective of the proposed deep learning-based framework is to categorize CXR images into three categories: COVID-19, Normal, or Pneumonia. We emphasize that the proposed Hybrid-COVID is a 2D/3D hybrid architecture designed to detect patients suspected of having COVID-19 by analyzing CXR images for early screening of the disease. This section assesses the proposed model on the collected dataset and compares it with state-of-the-art methods.
Performance evaluation
In this study, the proposed Hybrid-COVID model is trained and evaluated on CXR samples. It seeks to ensure a better compromise between accuracy and complexity by reducing the false negative rate as much as possible (i.e. mis-classification cost of diagnosed COVID-19 cases) as well as the computational time. The screening performance of the model is assessed using a set of previously mentioned classification metrics, including confusion matrix.
Nevertheless, we first assess the accuracy and loss of training and validation over epochs. These two indicators have been most often used in the literature to assess the performance of deep learning classifiers. Figure 7 shows the evolution of training and validation accuracy and loss. As can be seen, the validation accuracy increases to a certain level of stability and reaches 95.45%, while the validation loss values decrease more rapidly to 14%.
Figure 8 displays the confusion matrix of our model. As we can see, the screening model predicted 260 of the COVID-19 cases as true positives, 150 and 260 of the Normal and Pneumonia classes as true negatives, respectively. Table 4 shows the class-wise classification performance of the model; we obtained a reasonable result for the COVID-19 diagnosis using our method with 99.77% PREC, 99.67% REC, and 99.13% F-SCORE with a spatial resolution of \(112\times 112\).
Comparison with state-of-the-art methods
The proposed screening system has proven its performance over state-of-the-art methods:
-
DarkCovidNet [25]: a framework is introduced to automatically diagnose COVID-19 infection in CXR images based on the DarkNet model [47].
-
VGG19 + Transfer learning [21]: a transfer learning-based framework is proposed for the early detection of COVID-19 cases. VGG19 was the best model based on experimental results.
-
CoroNet [48]: an architecture based on a pre-trained Xception network is proposed to discriminate the features of COVID-19 patients from other diseases.
-
CapsNet [49]: a deep learning-based architecture is introduced, allowing the detection of COVID-19 infection using CXR images and capsule networks [50].
Subsequently, the achieved performance is reported and compared with the aforementioned benchmark methods.
DarkCovidNet [25] achieved an ACC rate of 89.20%, a SPE of 93.20%, and a SEN of 88.10%, respectively. In [21], a screening model based on the transfer learning technique and the VGG19 network was reached an ACC rate of 89.20%. The authors of [48] obtained 94.17% SEN, 96.20% SPE, and 93.89% ACC, which represents less than 4.16%, 2.48 %, and 3.02% of our results, respectively. CapsNet [49] reached 86.27% ACC for multi-class classification purposes.
Table 5 shows the superiority of our method compared to the aforementioned methods. As seen, our method, which combines several components (i.e. a 2D CNN network, a building block of 3D convolutions, etc.), surpasses the best competing models by an ACC rate of 1.2%, a SPE of 2.43%, and a SEN of 4.16%, respectively.
Impact of data and device heterogeneity
Table 6 reports the quantitative results obtained in terms of SEN, SPE, and ACC for each of the fifteen different datasets (Datasets-1 to -15). As can be seen, the proposed Hybrid-COVID classifier shows significantly improved performance on the majority of datasets. For example, our proposed Hybrid-COVID model achieves good binary classification performance on Dataset-5 (COVID-19 vs. non-COVID-19) with 97.23% SEN, 100% SPE and 98.94% ACC, respectively. Similarly, our classification model performs well in the range of 98.03% to 96.68% SEN, 100% to 98.91% SPE, and 99.32% to 97.47% ACC on Dataset-10 (COVID-19 vs. non-COVID-19) and Dataset-15 (COVID-19 vs. non-COVID-19), respectively. In addition, it reaches a SEN, SPE and classification ACC of 97.08%, 100% and 98.90%, respectively, on Dataset-4 (COVID-19 vs. Pneumonia vs. Normal); 97.39%, 99.84% and 99.04%, respectively, on Dataset-9 (COVID-19 vs. Pneumonia vs. Normal); and 96.04%, 98.83% and 97.01%, respectively, on Dataset-14 (COVID-19 vs. Pneumonia vs. Normal). In Table 6, the overall ACC, SEN, and SPE for the two classes (COVID-19 vs. Normal) are also reported.
Impact of the hybridization pipeline
Table 7 displays the results obtained from the baseline scenario and the three other ones. Firstly, despite the increase in the depth of the network, we point out that the total number of parameters (i.e. trainable and non-trainable parameters) remains approximately the same. That is one of the reasons why transfer learning techniques make sense when combining 2D and 3D CNNs. As expected, the total number of parameters has decreased from 17M to 14M, as a result of replacing 3D convolutions with 2D counterparts. Compared to 2D CNN, the number of parameters required for each 3D convolutional kernel is comparatively high. The overall ACC of the classification of the different scenarios is also given in Table 7, which shows that the baseline scenario performs better than the other three. Consequently, the three scenarios yield different results. However, the baseline scenario gives a good outcome with an ACC rate of 96.91%, which is higher than the first (a gap of 0.85%), second (a gap of 2.13%), and third (a gap of 4.1%) scenarios.
Visualization
In the supervised learning mode, end-to-end learning of a deep CNN is regarded as a black box. To date, visualizing and interpreting how CNNs interact has been a vital challenge for the computer vision community [51]. Figure 9 shows how Hybrid-COVID trained by visualizing some of the feature maps generated by the 3D CNN output (building block). As can be seen, the representations learned are complex, rich, and informative. In general, the extracted features have become increasingly sophisticated at several levels of abstraction and representation [42]. Therefore, the patterns learned with our Hybrid-COVID model pay more attention to contextual cues as well as the spatial aspect of the input image.
Figure 10 shows a visualization of two X-ray images classified by our framework. As can be seen, the proposed Hybrid-COVID model gives a more accurate visual localization of the disease region. We can observe here that the model focuses mainly on the anterior thoracic region, where the discriminating features of COVID-19 are well classified.
Discussion
The proposed transfer learning-based framework consists of a hybrid 2D/3D CNN architecture that allows the automatic detection of COVID-19 using CXR modality. The implemented Hybrid-COVID model classifies each sample in a multi-class manner by predicting a class label for each testing example according to whether it belongs to the COVID-19 class, Normal, or Pneumonia.
In this paper, Hybrid-COVID exploits the potential synergy between the pre-trained VGG16 model (i.e. a 2D CNN) and a shallow 3D CNN for high-level contextual feature learning for each data sample. The proposed hybridization pipeline shows its proficiency in capturing both visual appearance and semantic detail from multi-dimensional feature space. The pipeline mainly consists of two correlated branches: the 2D CNN branch and the 3D CNN branch. On the one hand, the 2D CNN branch encodes features along the spatial axes of the data based on the transfer learning paradigm. On the other hand, the 3D CNN branch takes advantage of pre-trained parameters to capture and extract any inherent contextual information within the boundaries of SEN and SPE.
In practice, the high SEN and SPE rates of a screening test are crucial for many reasons. For instance, a false negative result can lead to a critical situation where a patient with COVID-19 will not be isolated based on this test. However, this can lead to the widespread transmission of the virus to other healthy people, both indoors and outdoors. Compared to recent work in the literature, the experimental results produced by the Hybrid-COVID model show promising findings in terms of ACC, SPE, and SEN (Table 5). As shown in Fig. 8, from the confusion matrix, the model can correctly classify most of the samples, allowing to focus on the visual features that help distinguish COVID-19 cases from non-COVID-19 cases (i.e. Normal and Pneumonia cases).
Based on the results in Table 6, we showed that different domains of data could have a significant impact on the automatic detection of COVID-19 from CXRs and thus on the model’s ability to discriminate COVID-19 from other similar diseases. From Table 7, we can see that the integration of a pre-trained deep 2D CNN with a shallow 3D CNN is the best scenario to obtain better classification performance. The results summarized in Table 7 show that the hybridization scheme is quite convincing in terms of classification accuracy. In other words, 2D or 3D CNNs alone cannot learn very informative and contextual cues compared to hybrid 2D/3D CNNs. Here, it is important to note that the number of parameters has a significant impact on the computational efficiency of our hybrid architecture, as a decrease in the number of parameters can affect the decision-making capacity of the classifier. Also, the experimental outcomes obtained for the baseline scenario are obtained by considering the computational complexity of the pre-trained VGG16 model and the 3D CNN network. Compared to the 2D/2D network, the adaptive combination of 2D/3D CNN networks proved to be the most suitable for accurate and efficient classification of CXRs.
In addition to quantitative analyses, we qualitatively demonstrated the discrimination power of Hybrid-COVID using localization maps where infected regions are visualized on X-ray images (Fig. 10). Besides, as shown in the generated feature maps of the 3D convolution layer (Fig. 9), the model shows robust and complex patterns of CXRs, which are then mapped to the classification scheme using a dense layer for final prediction. Moreover, this research went beyond the limited availability of training data, as further modeling generally requires much more samples to achieve satisfactory results.
Nevertheless, despite these promising results, the proposed Hybrid-COVID network still needs to be improved to meet the performance improvements expected by radiologists and health experts. Besides, it should be noted that screening for COVID-19 from medical imaging modalities remains an open research topic and requires special attention in the future. Given that a recent retrospective study of [5] indicated that more than 50% of patients had no changes in CT scans at the early stages of symptom onset. As a result, this would severely limit the clinical application of any imaging-based screening method.
Conclusion
In this paper, a transfer learning-based hybrid 2D/3D CNN architecture has been developed to automatically detect and classify COVID-19 from CXR data. We performed an in-depth analysis to find a compromise between accuracy and efficiency by integrating both 2D and 3D CNNs into a hybrid design, called Hybrid-COVID. The findings of the proposed method clearly show its robustness compared to recent studies by up to 96.91% accuracy, 98.68% specificity, and 98.33% sensitivity, respectively. We also showed that the integration of a depth-wise separable convolution and a spatial pyramid pooling module could produce high-level representations from multi-scale X-ray information. In the clinical practice, this high level of performance could help radiologists in making more informed and faster decisions to detect COVID-19 in the early stages. Finally, we consider that the proposed architecture can be further enhanced with larger datasets in the future. We also plan to redesign the architecture of the Hybrid-COVID model and integrate new datasets. Furthermore, we will rethink the computational burden of our method by proposing a new lightweight network for COVID-19 screening with CXR and other modalities.
Data availability
The data used to support the findings of this study are already publicly available and referenced.
References
World Health Organization (2020) Coronavirus. https://www.who.int/health-topics/coronavirus. Accessed 25 April 2020
Bai Y, Yao L, Wei T et al (2020) Presumed asymptomatic carrier transmission of COVID-19. JAMA 323(14):1406–1407. https://doi.org/10.1001/jama.2020.2565
Grech V (2020) Unknown unknowns—COVID-19 and potential global mortality. Early Hum Dev 144:105026
Ji T et al (2020) Detection of COVID-19: a review of the current literature and future perspectives. Biosens Bioelectron 166:112455
Bernheim A et al (2020) Chest CT findings in coronavirus disease-19 (COVID-19): relationship to duration of infection. Radiology 295:200463
Ai T et al (2020) Correlation of chest CT and RT-PCR testing in Coronavirus Disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. https://doi.org/10.1148/radiol.2020200642
Dangis A et al (2020) Accuracy and reproducibility of low-dose submillisievert chest CT for the diagnosis of COVID-19. Radiol Cardiothorac Imaging 2(2):e200196. https://doi.org/10.1148/ryct.2020200196
Fang Y et al (2020) Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. https://doi.org/10.1148/radiol.2020200432
Bai HX et al (2020) Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. https://doi.org/10.1148/radiol.2020200823
Willemink MJ, Noël PB (2019) The evolution of image reconstruction for CT-from filtered back projection to artificial intelligence. Eur Radiol 29(5):2185–2195. https://doi.org/10.1007/s00330-018-5810-7
Jacobi A, Chung M, Bernheim A et al (2020) Portable chest X-ray in coronavirus disease-19 (COVID-19): a pictorial review. Clin Imaging 64:35–42. https://doi.org/10.1016/j.clinimag.2020.04.001
Brady Z et al (2020) Technique, radiation safety and image quality for chest X-ray imaging through glass and in mobile settings during the COVID-19 pandemic. Phys Eng Sci Med. https://doi.org/10.1007/s13246-020-00899-8
Ng M-Y et al (2020) Imaging profile of the COVID-19 infection: radiologic findings and literature review. Radiol Cardiothorac Imaging 2(1):e200034. https://doi.org/10.1148/ryct.2020200034
Zhao D et al (2020) A comparative study on the clinical features of COVID-19 pneumonia to other pneumonias. Clin Infect Dis. https://doi.org/10.1093/cid/ciaa247
Fan L et al (2020) Progress and prospect on imaging diagnosis of COVID-19. Chin J Acad Radiol 3(1):4–13. https://doi.org/10.1007/s42058-020-00031-5
Jin S et al (2020) AI-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system in four weeks. medRxiv. https://doi.org/10.1101/2020.03.19.20039354
Butt C, Gill J, Chun D et al (2020) Deep learning system to screen coronavirus disease 2019 pneumonia. Appl Intell. https://doi.org/10.1007/s10489-020-01714-3
Cohen JP, Morrison P, Dao L (2020) COVID-19 image data collection. arXiv: 2003.11597
Chest X-ray images (pneumonia) (2020) https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia. Accessed 25 June 2020
Narin A, Kaya C, Pamuk Z (2020) Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. arXiv: 2003.10849
Apostolopoulos ID, Bessiana TA (2020) COVID-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med 43:1–6
Karim MR, Döhmen T, Rebholz-Schuhmann D et al (2020) DeepCOVIDExplainer: explainable COVID-19 predictions based on chest X-ray images. arXiv: 2004.04582
Basu S, Mitra S, Saha N (2020) Deep learning for screening COVID-19 using chest X-ray images. arXiv:2004.10507
Hemdan EE-D, Shouman MA, Karar ME (2020) COVIDX-Net: a framework of deep learning classifiers to diagnose COVID-19 in X-ray images. arXiv:2003.11055
Ozturk T et al (2020) Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med 121:103792
Toğaçar M, Ergen B, Cömert Z (2020) COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput Biol Med 121:103805
He K et al (2016) Deep residual learning for image recognition. In: IEEE conference on computer vision and pattern recognition (CVPR), pp 770–778. https://doi.org/10.1109/CVPR.2016.90
Das D, Santosh KC, Pal U (2020) Truncated inception net: COVID-19 outbreak screening using chest X-rays. Phys Eng Sci Med. https://doi.org/10.1007/s13246-020-00888-x
Li Z et al (2020) A survey of convolutional neural networks: analysis, applications, and prospects. arXiv:2004.02806
Singh RD, Mittal A, Bhatia RK (2019) 3D convolutional neural network for object recognition: a review. Multimed Tools Appl 78:15951–15995
Ji S et al (2013) 3D convolutional neural networks for human action recognition. IEEE Trans Pattern Anal Mach Intell 35:221–231
Singh SP et al (2020) 3D deep learning on medical images: a review. Sensors 20:5097
Bayoudh K et al (2020) Transfer learning based hybrid 2D–3D CNN for traffic sign recognition and semantic road detection applied in advanced driver assistance systems. Appl Intell. https://doi.org/10.1007/s10489-020-01801-5
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556
Chollet F, Xception (2017) Deep learning with depthwise separable convolutions. In: CVPR, pp. 1800–1807. https://doi.org/10.1109/CVPR.2017.195
He K et al (2014) Spatial pyramid pooling in deep convolutional networks for visual recognition. In: ECCV, pp. 346–361
COVID-19 Radiography Database (2020). https://www.kaggle.com/tawsifurrahman/covid19-radiography-database. Accessed 25 June 2020
Zhang L (2019) Transfer adaptation learning: a decade survey. arXiv:1903.04687
Khan A et al (2020) A survey of the recent architectures of deep convolutional neural networks. Artif Intell Rev 53:5455–5516
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: IEEE conference on computer vision and pattern recognition (CVPR), pp 3431–3440. https://doi.org/10.1109/CVPR.2015.7298965
Tran D et al (2015) Learning spatiotemporal features with 3D convolutional networks. In: IEEE international conference on computer vision (ICCV), pp 4489–4497. https://doi.org/10.1109/ICCV.2015.510
LeCun Y et al (2015) Deep learning. Nature 521(7553):436–444. https://doi.org/10.1038/nature14539azz
Selvaraju RR et al (2020) Grad-CAM: visual explanations from deep networks via gradient-based localization. Int J Comput Vis 128:336–359
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. arXiv:1412.6980
Srivastava N et al (2014) Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res 15(56):1929–1958
Shorten C, Khoshgoftaar TM (2019) A survey on image data augmentation for deep learning. J Big Data 6(1):60. https://doi.org/10.1186/s40537-019-0197-0
Redmon J, Divvala S, Girshick R, Farhadi A (2016) You only look once: unified, real-time object detection. In: IEEE conference on computer vision and pattern recognition (CVPR), pp 779–788. https://doi.org/10.1109/CVPR.2016.91
Khan AI, Shah JL, Bhat MM (2020) CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest X-ray images. Comput Methods Programs Biomed 196:105581
Toraman S, Alakus TB, Turkoglu I (2020) Convolutional CapsNet: a novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks. Chaos Solitons Fractals 140:110122
Kwabena Patrick M, Felix Adekoya A, Abra Mighty A, Edward BY (2019) Capsule networks—a survey. J King Saud Univ Comput Inf Sci. https://doi.org/10.1016/j.jksuci.2019.09.014
Zeiler MD, Fergus R (2013) Visualizing and understanding convolutional networks. In: Computer vision—ECCV, pp 818–833
Funding
This study did not receive external funding.
Author information
Authors and Affiliations
Contributions
All authors contributed equally.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent to publish
Not required.
Ethical approval
All human participant data in this study were sourced from publicly available datasets.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bayoudh, K., Hamdaoui, F. & Mtibaa, A. Hybrid-COVID: a novel hybrid 2D/3D CNN based on cross-domain adaptation approach for COVID-19 screening from chest X-ray images. Phys Eng Sci Med 43, 1415–1431 (2020). https://doi.org/10.1007/s13246-020-00957-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-020-00957-1