Abstract
Since December 2019, the Coronavirus Disease (COVID-19) pandemic has caused world-wide turmoil in a short period of time, and the infection, caused by SARS-CoV-2, is spreading rapidly. AI-driven tools are used to identify Coronavirus outbreaks as well as forecast their nature of spread, where imaging techniques are widely used, such as CT scans and chest X-rays (CXRs). In this paper, motivated by the fact that X-ray imaging systems are more prevalent and cheaper than CT scan systems, a deep learning-based Convolutional Neural Network (CNN) model, which we call Truncated Inception Net, is proposed to screen COVID-19 positive CXRs from other non-COVID and/or healthy cases. To validate our proposal, six different types of datasets were employed by taking the following CXRs: COVID-19 positive, Pneumonia positive, Tuberculosis positive, and healthy cases into account. The proposed model achieved an accuracy of 99.96% (AUC of 1.0) in classifying COVID-19 positive cases from combined Pneumonia and healthy cases. Similarly, it achieved an accuracy of 99.92% (AUC of 0.99) in classifying COVID-19 positive cases from combined Pneumonia, Tuberculosis, and healthy CXRs. To the best of our knowledge, as of now, the achieved results outperform the existing AI-driven tools for screening COVID-19 using the acquired CXRs, and proves the viability of using the proposed Truncated Inception Net as a screening tool.
Similar content being viewed by others
Introduction
Coronavirus Disease 2019 (COVID-19) is an infectious disease caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) [1]. The disease was first identified in 2019 in Wuhan, China, and has since spread globally, resulting in the 2019–2020 Coronavirus pandemic [2]. With more than 5.92 million confirmed cases of infection and 364,000 cases of death by the fifth month of its discovery (as on May 30, 2020), the SARS-CoV-2 continues to infect people worldwide [3]. The virus is primarily transmitted among individuals through respiratory droplets. Studies have also shown that the virus can persist on surfaces which an infected individual might have touched. As a consequence, by the end of March 2020, the spread of this virus had been described as exponential [3].
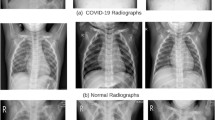
The gold standard for the diagnosis and detection of COVID-19 is the polymerase chain reaction (PCR). It can detect the SARS-CoV-2 RNA from respiratory specimens through nasopharyngeal or oropharyngeal swabs. Despite the high sensitivity and accuracy of the PCR technique, the method is highly time-consuming and resource-intensive. Therefore, considering the unprecedented spread rate of the virus across the globe and the rapid temporal progression of the disease throughout a subject’s body [4], a faster screening tool is necessary for COVID-19 outbreaks. As an alternative to the traditional PCR technique, researchers have proposed the use of radiography techniques such as Computed Tomography (CT) scans and chest X-rays (CXRs) for COVID-19 screening. Early studies of COVID-19 positive patients have shown that their CT scans and CXRs show identifiable abnormalities [5, 6], according to which COVID-19 pneumonia was more likely to have peripheral distribution, ground-glass opacity, fine reticular opacity, and vascular thickening, but less likely to have a central+peripheral distribution, pleural effusion and lymphadenopathy. The idea is further strengthened by observing the high correlation between the PCR and radiological results, as demonstrated in [7]. In [8, 9], authors establish that the sensitivity of CT scan imaging outperforms the conventional PCR technique. The possible reasons may be immature development of nucleic acid detection technology, low patient viral load or improper clinical sampling, as stated in [8]. According to [6, 10,11,12], the infestation of COVID-19 can primarily be characterized through radiographs by patches of Ground-Glass Opacity (GGO) and consolidation (See Fig. 1). Additionally, authors in [4, 13] have provided a deep insight into the statistical growth of radiological cues in COVID-19 positive patients and the temporal stages of the disease’s growth in a host’s body, respectively. According to these works, the disease can be temporally divided into four phases: early phase, progressive phase, severe phase, and dissipative phase. During this time, the CT scores and number of zones involved progress rapidly, which peak during illness days 6–11, followed by persistence of the high levels. The major pattern of abnormality found after symptom onset was ground-glass opacity, in accordance to the other contemporary works. These paved the way to a faster screening procedure than the PCR. Despite the radiological findings, there still exists a problem to use radiography as the primary screening tool for COVID-19. The problem being the lack of skilled radiologists across the globe. Ever since the advent of digital radiography, CT scans and CXRs have been used globally, but the final interpretations are required to be done by the experts, which could be time-consuming. Besides, authors in [5] have demonstrated through an experiment that sensitivity and specificity of screening COVID-19 positive CT images fluctuate significantly when done by radiologists. Therefore, for mass screening, automated or specifically AI-driven tools are necessary to be deployed across the globe, particularly in resource-constrained regions.
COVID-19 pneumonia is characterized primarily by patches of Ground-Glass Opacity (GGO) and consolidations. In these CXRs, the GGO areas, in early stages of COVID-19, are identified/annotated with white arrows. These annotations were made in the original dataset, which solely attribute the clinical implications
For all healthcare and/or (bio)medical problems, for more than a decade, deep learning has been a pinnacle in automation, especially in medical imaging. This motivates its use in the COVID-19 screening. Recently in [14], the author stated that to detect COVID-19, AI-driven tools are expected to have active-learning based cross-population train/test models that employ multitudinal and multimodal data. In this work [14], the use of deep learning and image data, such as CT scans and CXRs are addressed. Even though multimodal data can improve confidence in decision-making, for the COVID-19 case, such data are not available as of today. Due to lack of data, COVID-19 reveals the limits of AI-driven tools. As soon as the COVID-19 pandemic came into play, several systems have been released to automate the screening procedure. Alibaba released an AI-based system to screen COVID-19 infection from CT scans, with an accuracy of 96% [15]. Researchers in [16] proposed a Convolutional Neural Network (CNN) based technique to differentiate COVID-19 from Pneumonia and normal cases of CT scans, with a classification sensitivity of 0.90 for COVID-19. In [17], researchers have used a 3-dimensional deep learning model to segment infected regions from CT scans, followed by an attention driven network to classify COVID-19 from Influenza-A viral pneumonia and normal cases, and an accuracy of 86.7% was reported. Further, researchers in [18] have also proposed image segmentation schemes to detect lesions in CT scans. The prospective system is based on the popular UNet++ architecture, and it produces bounding boxes around lesion regions. The system achieved a result of 100% per patient and 94.34% per image sensitivity. Authors in [19, 20] have proposed deep learning models to classify COVID-19 positive CXRs from normal and Pneumonia cases, respectively. In [19], authors have investigated various standard CNN models, such as ResNet and AlexNet to extract deep features, which was followed by a Support Vector Machine (SVM) to classify COVID-19 positive cases. A maximum accuracy of 95.38% was reported using ResNet50 as the feature extractor. The latter work proposed a tailored CNN model using residual connections to achieve promising results of 80% positive predictive value (PPV)[20]. Additionally, authors in [21] have demonstrated the use of ResNet50, Inception Net V3, and Inception-Resnet V2 for identifying COVID-19 positive CXRs from healthy ones. Similar experiments have also been demonstrated in [22], which focuses on the importance of transfer learning for medical image classifications, in the context of COVID-19. On the whole, researchers found that the use of chest radiographs is better in terms of lung abnormalities screening [11, 14, 23,24,25,26]. With these, COVID-19 can be analyzed better using radiological image data [7, 8].
(Above) The original architecture of the Inception Net V3 model, which was implemented for classifying images of the ImageNet database [27]. (Below) The Truncated Inception Net model, which is proposed in our work for screening COVID-19 positive CXRs. The model retains 3 inception modules and 1 grid size reduction module from the original architecture (given above)
In this work, considering the fact that X-ray imaging systems are more prevalent and cheaper than CT scan systems, we use deep learning to screen COVID-19 using CXRs. We propose a CNN-based model, which we call Truncated Inception Net, solely based on the Inception Net V3 architecture [27] (Fig. 2). Using the limited number of COVID-19 positive CXRs, made available by Cohen [28], we analyse our model’s performance as a binary classifier, where additional datasets are also taken into account. As the COVID-19 dataset collection, alone, is not trivial, our experimental datasets that are composed of COVID-19, Pneumonia, Tuberculosis, and healthy cases, which are sufficient to validate COVID-19 positive cases. For this, several publicly available datasets, such as Pneumonia dataset [29], Tuberculosis datasets (Shenzhen, China, and Montgomery County, USA) [30] are used to create six different experimental tests. Such a varied dataset combinations for analyzing deep learning models on COVID-19 CXRs has not been demonstrated in the literature so far. On the whole, through this work, we demonstrate that the Truncated Inception Net deep learning model is a viable option for COVID-19 screening and it outperforms the state-of-the-art results for COVID-19 positive cases, on the obtained and manually combined datasets.
Proposed method
Given that COVID-19 shows patches of GGO and consolidation in CXRs [10], to detect COVID-19 positive cases, a multi-resolution analysis of the CXR images is deemed useful. This functionality of analyzing spatial data at multiple resolutions is possessed by the Inception module, which is the fundamental block of the popular Inception Net V3 [27]. Additionally, considering the fact that the number of data samples of COVID-19 positive CXRs is very scarce at present, a modified version of the Inception Net V3 model [27] is proposed, which we call Truncated Inception Net. The Truncated Inception Net is primarily designed to avoid possible overfitting due to the lack of COVID-19 positive samples. Further, the Truncated Inception Net is computationally efficient. Originally, the Inception Net model was used on the ImageNet database, consisting of more than 1.3 million images from 1000 different classes. In what follows, the different aspects of the Truncated Inception Net model architecture and implementation are discussed.
Block diagram: The block diagram presents the internal pipeline of an Inception module, which forms the building block of the InceptionNet. Multiple sized kernels (e.g. 3\(\times \)3 and 5\(\times \)5) are used to convolve with the input image, to extract features of varied spatial resolution. Finally, the activation maps obtained from the parallel computations are stacked depth-wise to form the output
Inception module
The multi-resolution analysis capability of the Inception module comes from its inherent architecture. In traditional CNN models, kernels of specific receptive field sizes are used in specific layers to capture features through the use of convolution. On the contrary, in an inception module, kernels of various receptive field sizes (1\(\times \)1, 3\(\times \)3, and 5\(\times \)5) are used in parallel to extract features of varying sizes. The extracted parallel features are then stacked depth-wise to form the output of the inception module. A 3\(\times \)3 max pooled version of the input is also stacked along with the previous feature maps. The combined output of the inception module provides rich feature maps of varying perspectives as inputs to the next convolutional layer of the CNN. Such a property of the Inception module explains its unique performance in medical imaging, and in our case, on the COVID-19 CXRs. For better understanding, the schematic representation of the Inception module is presented in Fig. 3.
Truncated architecture
Since the original Inception Net V3 model was built for the ImageNet database, the architectural complexity of the model is well justified. On the contrary, the COVID-19 dataset used in our work is immensely small compared to the ImageNet database. Therefore, a truncation of the model is necessary to reduce the model complexity and eventually the number of trainable parameters to prevent the model from overfitting issues. The model was truncated at a point, where it retained 3 Inception modules and 1 grid size reduction block from the beginning, followed by the cascading of a Max Pooling and a Global Average Pooling layer to reduce the output dimension. The point of truncation was chosen experimentally, that yielded the best classification results. Finally, a fully connected layer was cascaded to perform the classification task. The truncation of the model not only reduced training time and trainable parameters but also reduced the processing time while evaluating a CXR to detect COVID-19 positive cases. As a consequence, it facilitates mass screening at an efficient speed and accuracy. For more detailed information about computational efficiency, we refer to “Experimental setup” section. The architecture of the complete Truncated Inception Net can be visualized in Fig. 2.
Adaptive learning rate protocol
For training the Truncated Inception Net, a dynamic protocol was used to control the learning rate at each epoch because of the following reasons: (a) A constant learning rate of high value often leads to divergence of the weight vectors’ trajectory from global minimum in the loss function space during the optimization process, and (b) an arbitrarily chosen low value often takes longer period of training time. Therefore, a dynamic procedure was opted, where the learning rate was initialized with a value of 0.001 and then it was reduced by a factor of 2 every time the validation loss remained same or did not decrease for more than 3 epochs. In this case, the factor of 3 epochs is known as the patience factor. The process is well explained through the diagram in Fig. 4. This procedure yielded the behavior of reducing the velocity of approach when the weight vectors are close to the global minimum, to prevent overshooting. Note that, the initial value of 0.001, reduction factor of 2, and patience factor of 3 epochs were chosen after experimenting with multiple values and monitoring the classification performance. For the model and datasets considered in this work, the aforementioned values proved to have the best results. Additional optimization procedures like Grid Search, Particle Swarm Optimization, or Genetic Algorithm can be used for the optimization of the same.
Transfer learning
Deep learning models are inherently data intensive. However, since the size of the COVID-19 dataset is very small compared to standard datasets used in deep learning, the concept of transfer learning can be applied to augment the decision-making process. Transfer learning uses the concept of transferring knowledge from one domain to another by using trained weights from the previous domain. Traditionally in a CNN, the weight matrices of several layers from the beginning are frozen while training on the secondary domain, and only the remaining layers are fine-tuned. This process works well when both the domains share an overlapping region in the low-level features. In our case, since the ImageNet and the COVID-19 datasets belong to non-overlapping domains, the trained weights from the ImageNet dataset were used to initialize the weights of our model, but none of them were frozen. This kept all the layers initialized with relatively more meaningful weights than random initialization, and subject to learning during the training procedure.
Experimental setup
Datasets
Collecting COVID-19 dataset is not trivial. We, however, collect a number of CXR benchmark collections (C1 to C3) from the literature (See Table 1). They help to showcase/validate the usability and robustness of our model.
-
C1:
COVID-19 collection[28] is an open-source collection that is made available and maintained by Joseph Paul Cohen. At the time of the present study, it is composed of 162 COVID-19 positive CXRs, along with some other CXRs of diseases like MERS, SARS, and viral Pneumonia. For our purpose, only COVID-19 positive posteroanterior CXRs are considered.
-
C2:
Pneumonia collection[29] (Kaggle CXR collection) is composed of 5863 CXRs. Out of this, 1583 CXRs are normal or healthy CXRs and the remaining 4280 CXRs show various manifestations of viral and bacterial Pneumonia.
-
C3:
Two publicly available Tuberculosis (TB) collections[30] are considered: (a) Shenzhen, China and (b) Montgomery County, USA. These CXR benchmark collections were made available by the U.S. National Library of Medicine, National Institutes of Health (NIH). The Shenzhen, China collection is composed of 340 normal cases and 342 positive cases of TB. The Montgomery County, USA collection is composed of 80 normal CXRs and 58 TB positive CXRs.
A few samples from the aforementioned collections are visualized in Fig. 5. Using aforementioned collections, we constructed six different combinations of data to train and validate our model. As provided in Table 2, these six different combinations of datasets (D1 to D6) are enlisted below:
-
D1:
In dataset D1, 162 COVID-19 positive CXRs and 340 healthy CXRs from the Shenzhen, China collections are considered.
-
D2:
For this dataset D2, 162 COVID-19 positive CXRs and 80 healthy CXRs from the Montgomery County, USA are considered.
-
D3:
D3 consists of 162 COVID-19 positive CXRs and 1583 healthy CXRs from the Pneumonia collections are considered.
-
D2:
D4 contains 162 COVID-19 positive CXRs and 2003 healthy CXRs, combined from the Shenzhen, Montgomery and Pneumonia collections are considered.
-
D5:
In dataset D5, 162 COVID-19 positive CXRs, 4280 Pneumonia positive CXRs and 1583 healthy CXRs from the Pneumonia collections are considered.
-
D6:
In dataset D6, 162 COVID-19 positive CXRs and 6683 non-COVID CXRs (comprising of 4280 Pneumonia positive, 400 TB positive and 2003 healthy CXRs) are considered.
The primary motivation behind constructing the various data combinations (D1 to D6) is to show the robustness of the Truncated Inception Net to detect COVID-19 positive cases. Further, COVID-19 is believed to have a close relationship with traditional Pneumonia. Therefore, a separate dataset (D5) was constructed to show whether our proposed model is able to differentiate COVID-19 positive cases from those traditional Pneumonia positive cases. Besides, CXRs of Tuberculosis manifestation were also added in D6 to prove that our model is robust enough to identify COVID-19 from other diseases like TB, Pneumonia, and healthy CXRs. The robustness also lies in the way we collect data, where regional variation can be considered as a crucial element. In our datasets, the healthy CXRs in D1, D2, and D3 are collected from different regions of the world. Considering multiple combination of data from different places can help develop cross-population train/test models.Footnote 1
As an input to our model, CXR images were scaled down to the size of 224\(\times \)224\(\times \)3 to match the input dimensions of the Truncated Inception Net. Such a resizing can also reduce computational complexity. Since the pixels of the CXRs have bounded discrete values, the images were normalized using the min-max scaling scheme. The choice is further backed by the fact that standardization (zero-mean unit variance) assumes the data to always have a Gaussian distribution that might not always be the case. Additionally, pixel intensities of COVID-19 features like GGO patches and consolidation falls in the same range of bones in CXRs, as demonstrated by quick preliminary experiments. So histogram matching was also excluded as a normalization scheme, since it decreases the signal to noise ratio in this scenario.
Validation protocol and evaluation metrics
To validate our proposed model, a 10 fold cross-validation scheme was opted for training and testing purposes on all six datasets: D1–D6. The process of 10 fold cross-validation works in the following way: say there are 100 data samples in the total dataset. Then samples 1–10 are made a subset and labelled as fold-1, samples 11–20 are labelled as fold-2 and so on. These creates 10 disjoint subsets of the original dataset. Following this, the model to be tested is first trained on subsets 1–9 and tested on subset 10. Similarly, in the second trial the model is tested on subset 9 after being trained on the remaining subsets. This scheme ensures that the model’s performance is not biased by the presence of outlier data samples in the training or testing datasets. Following this strategy, each of the constructed datasets (D1–D6) was subdivided into 10 subsets of almost equal number of data samples. Then the model was trained on 9 subsets and tested on the remaining 1 subset. This process was repeated using each of the subsets as a test set for once. After the ten separate trials of training and testing, the result was averaged over the ten trials to assess the mean (and standard deviation) performance of the model on that dataset. This procedure can be well understood by observing the result pattern in Table 4, which tabulates the tenfold cross-validation performance of the model on dataset D6. For each of the 10 folds, six different evaluation metrics were employed: (a) Accuracy (ACC); (b) Area under the ROC curve (AUC); (c) Sensitivity (SEN); (d) Specificity (SPEC); (e) Precision (PREC); and (f) F1 score. These can be computed as follows:
where \(t_{p}\), \(f_{p}\), \(t_{n}\), and \(f_{n}\) are the total number of true positives, false positives, true negatives, and false negatives. The mean scores from all 10 folds were taken for each of the above metrics, to get the final results on a particular dataset.
In traditional deep learning tasks, a primary metric like accuracy is sufficient to judge the performance of a deep learning model as binary classifier. On the contrary, such an assumption does not work well when considering imbalanced datasets. In such cases (like, in medical datasets), the positive class to be predicted often has much lower data samples than the negative class. Therefore, accuracy would demonstrate a fairly high value even if the model labels all the test data to be negative. Therefore, special attention is given to metrics like Sensitivity/Recall, Precision, and F1 score here.
In the context of COVID-19, the Sensitivity metric plays a very crucial role when deploying a model for screening patients in the early stages of a pandemic. Sensitivity measures the likelihood that the model would not miss classifying COVID-19 positive samples/patients. This prevents the further spreading of the infection. Secondly, the precision measures the likelihood that a model would not make a mistake to classify normal patients as COVID-19 positive. This metric becomes very important in the later stages of a pandemic, when medical resources are limited, and they are available only to the patients that are in need. Besides, F1 score is used to extract the combined performance score of a model, which is the harmonic mean of the precision and sensitivity of a model.
Results and analysis
Before providing quantitative results, we first provide activation maps generated by our proposed model for a COVID-19 positive, Pneumonia positive, and TB positive CXR can be visualized in Fig. 6. It can be observed that in the preliminary layers (like Conv2D), the lung region is clearly visible in the activation map for normal CXR, while the clarity gradually decreases for pneumonia and further for COVID-19 CXR. This corresponds to the growth of GGO patches in COVID-19 positive CXRs. However, in the later layers of the model, the activation maps become more abstract, for which the terminal dense layer is used in the model to map these abstract feature representations to their corresponding labels (COVID+ or COVID−).
Following the validation protocol and evaluation metrics mentioned in the previous “Validation protocol and evaluation metrics” section, we present the mean scores that were achieved using tenfold cross-validation train-test scheme, on each of the six different datasets: D1–D6. The experimental results are well documented in Table 3. Also, standard deviation (\(\sigma \)) is reported in all cases, whose very low value proves the statistical robustness of our model. Our proposed Truncated Inception Net model achieves a classification ACC, AUC, SEN, SPEC, PREC, and F1 score of 99.96%, 1.0, 0.98, 0.99, 0.98, and 0.98, respectively, on the dataset: D5 (COVID-19 positive case detection against Pneumonia and healthy cases) and that of 99.92%, 0.99, 0.93, 1.0, 1.0, and 0.96, respectively, on the D6 dataset (COVID-19 positive case detection against Pneumonia, TB, and healthy CXRs). Since the custom datasets being used were highly imbalanced in terms of class representation, sensitivity and precision are the most significant metrics in our case, as said in “Validation protocol and evaluation metrics” section. Consequently, the proposed model achieves high sensitivity and precision on these datasets. For a better understanding of the results, six different ROC curves are shown in Fig. 7; one for each dataset, starting from D1 to D6.
Additionally, since for every dataset we computed tenfold cross-validation, for better understanding of how average scores and their standard deviation were computed, the results obtained from each fold on the dataset: D6 are provided in Table 4. Besides, the proposed Truncated Inception Net model performs \(2.3\pm 0.18\) times on an average faster than Inception Net V3 model. In Table 5, computational times (by taking 10 different CXR samples as input) are used to demonstrate the differences between them. The primary reason being the large number of parameters in the original Inception Net V3 model. Precisely, this model contains more than 21.7 million trainable parameters in contrast our model which contains only 2.1 million trainable parameters, making it a better choice for training on small datasets and also for active learning. Therefore, for mass screening in resource-constrained areas, employing a faster tool is the must.
Discussion
Since COVID-19 outbreak, very few pieces of works have been proposed/reported using CXRs to detect COVID-19 positive cases (see “Introduction” section): In our comparison, ResNet50 and SVM [19], COVID-Net [20], ResNet50 [21], and Inception Net V3 [21] are considered even though they are not peer-reviewed research articles. We have compared with these pieces works using exact same evaluation metrics (ACC in %, AUC, SEN, SPEC, PREC, and F1 score) and nature of dataset. Like other works, we take COVID-19 positive and healthy CXRs from Pneumonia dataset (D3 in our case), and used this result as a comparison to other works. Besides, since all models were based on deep learning models, we consider an essential element i.e., number of parameters in our comparison. Table 6 provides a complete comparative study. Not all the authors reported AUC, SPEC, and F1 score. Note that, our model was used as a binary classifier to screen a CXR as COVID+ or COVID-, while not all the stated works performed the same. The mentioned results belong to the COVID+ positive class, wherever multi-class classification was done instead of binary classification. On the whole, considering the number of parameters, the proposed Truncated Inception Net outperforms all. Note that, since our model is the derivative of Inception Net V3 model, it is worth to compare between them. We observe that, in both computational time (Table 5) and performance scores (Table 6), Truncated Inception Net performs better than Inception Net V3 [21]. For a better understanding, three different performance scores: poor, the best and average are considered from Table 3. This suggests that the Truncated Inception Net is not only more computationally effective in terms of training and usability, but also more flexible for the purpose of active learning [14].
Even though the performed experiments validate that the proposed deep learning model for screening COVID-19 positive CXRs, it is important to understand that the system relies completely on visual cues in the input data. Therefore, in the early stages of COVID-19, when the radiologically observable cues have not yet developed, the system might fail to perform as stated. A detailed study on this is a scope for future work, where the input data shall be additionally labeled with the stage of COVID-19 it depicts as well. However, data acquired for this work did not contain any explicit information regarding the stages of COVID-19 in the individual CXRs. Further, the system is limited by its capacity to localize the disease in the CXR. As seen in the activation maps of deeper layers (Fig. 6), the model develops an intrinsic representation of the CXR features rather than accurate spatial heat-map, which is then mapped to the output using a dense layer classifier. The mentioned goal can be achieved by using increased number of data or a deep learning model(s) that is/are pre-trained on a large number of CXRs of different diseases (like CheXNet [31]), which shall be our future goal.
Conclusion and future works
In this work, we have proposed the Truncated Inception Net deep learning model to detect COVID-19 positive patients using chest X-rays. For validation, experimental tests were done on six different experimental datasets by combining COVID-19 positive, Pneumonia positive, Tuberculosis positive, and healthy CXRs. The proposed model outperforms the state-of-the-art results in detecting COVID-19 cases from non-COVID ones. Besides, considering the number of parameters used in our proposed model, it is computationally efficient as compared to original Inception Net V3 model and other works proposed in the literature. It is important to note that the study has no clinical implications. Instead, we solely aimed to check whether the proposed Truncated Inception Net could be used in detecting COVID-19 positive cases using CXRs.
Observing the performance scores, the Truncated Inception Net can serve as a milestone for screening COVID-19 under active-learning framework on multitudinal/multimodal data [14]. It also motivates to work on cross-population train/test models. Integrating this model with CheXNet model [31] will be our immediate plan, since ChexNet is primarily employed to analyze CXRs.
Notes
Even though, our tests proved that the proposed model can be considered as a cross-population train/test model, it is beyond the scope of the paper.
References
World Health Organization (2020) Naming the coronavirus disease (covid-19) and the virus that causes it. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it
World Health Organization (2020) The continuing 2019-ncov epidemic threat of novel coronaviruses to global health - the latest 2019 novel coronavirus outbreak in wuhan, China. https://pubmed.ncbi.nlm.nih.gov/31953166/
World Health Organization (2020) Coronavirus disease (covid-2019) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports
Li M, Lei P, Zeng B, Li Z, Peng Y, Fan B, Wang C, Li Z, Zhou J, Shaobo H et al (2020) Coronavirus disease (covid-19): spectrum of ct findings and temporal progression of the disease. Acad Radiol 27:603
Bai HX, Hsieh B, Xiong Z, Halsey K, Choi JW, Tran Thi ML, Pan I, Shi L-B, Wang D-C, Mei J et al (2020) Performance of radiologists in differentiating covid-19 from viral pneumonia on chest ct. Radiology, pp 200823
Gross A, Thiemig D, Koch F-W, Schwarz M, Gläser S, Albrecht T (2020) CT appearance of severe, laboratory-proven coronavirus disease 2019 (covid-19) in a caucasian patient in Berlin, Germany. Georg Thieme Verlag KG, In RöFo-Fortschritte auf dem Gebiet der Röntgenstrahlen und der bildgebenden Verfahren
Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L (2019) Correlation of chest ct and RT-PCR testing in coronavirus disease, (covid-19) in China: a report of 1014 cases. Radiology 200642:2020
Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, Ji W (2020) Sensitivity of chest ct for covid-19: comparison to RT-PCR. Radiology. https://doi.org/10.1148/radiol.2020200432
Chung JH, Elicker BM, Ketai LH, Kanne JP, Little BP (2020) Essentials for radiologists on covid-19: an update-radiology scientific expert panel. Radiology 200527:27
Kong W, Agarwal PP (2020) Chest imaging appearance of covid-19 infection. Radiology 2(1):e200028
Huang C, Wang Y, Li X, Ren L, Zhao J, Yi H, Zhang L, Fan G, Jiuyang X, Xiaoying G et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. Lancet 395(10223):497–506
Ng M-Y, Lee EY, Yang J, Yang F, Li X, Wang H, Lui MM, Shing-Yen LC, Leung B, Khong P-L et al (2020) Imaging profile of the covid-19 infection: radiologic findings and literature review. Radiology 2(1):e200034
Wang Y, Dong C, Hu Y, Co Li, Ren Q, Zhang X, Shi H, Zhou M (2020) Temporal changes of CT findings in 90 patients with covid-19 pneumonia: a longitudinal study. Radiology 10:200843
Santosh KC (2020) Ai-driven tools for coronavirus outbreak: need of active learning and cross-population train/test models on multitudinal/multimodal data. J Med Syst 44(5):1–5
Technology org, ai algorithm detects coronavirus infections in patients from ct scans with 96% accuracy. https://www.technology.org/2020/03/01/ai-algorithm-detects-coronavirus-infections-in-patients-from-ct-scans-with-96-accuracy, 2020(accessed March 02, 2020)
Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q et al (2020) Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest CT. Radiology 284:574
Xu X, Jiang X, Ma C, Du P, Li X, Lv S, Yu L, Chen Y, Su J, Lang G et al (2020) Deep learning system to screen coronavirus disease 2019 pneumonia. arXiv preprint arXiv:2002.09334
Chen J, Wu L, Zhang J, Zhang L, Gong D, Zhao Y, Hu S, Wang Y, Hu X, Zheng B et al (2020) Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. medRxiv
Sethy PK, Behera SK (2020) Detection of coronavirus disease (covid-19) based on deep features. Preprints
Wang L, Wong A (2020) Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest radiography images. arxiv:2003.09871
Narin A, Kaya C, Pamuk Z (2020) Automatic detection of coronavirus disease (covid-19) using X-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.10849
Apostolopoulos Ioannis D, Mpesiana Tzani A (2020) Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med 1:1
Santosh KC, Vajda S, Antani SK, Thoma GR (2016) Edge map analysis in chest X-rays for automatic pulmonary abnormality screening. Int J Comput Assist Radiol Surg 11(9):1637–1646
Karargyris A, Siegelman J, Tzortzis D, Jaeger S, Candemir S, Xue Z, Santosh KC, Vajda S, Antani SK, Folio LR, Thoma GR (2016) Combination of texture and shape features to detect pulmonary abnormalities in digital chest X-rays. Int J Comput Assist Radiol Surg 11(1):99–106
Vajda S, Karargyris A, Jäger S, Santosh KC, Candemir S, Xue Z, Antani Sameer K, Thoma George R (2018) Feature selection for automatic tuberculosis screening in frontal chest radiographs. J Med Syst 42(8):1–11
Santosh KC, Antani S (2017) Automated chest X-ray screening: can lung region symmetry help detect pulmonary abnormalities? IEEE Trans Med Imaging 37(5):1168–1177
Christian S, Vincent V, Sergey I, Jon S, Zbigniew W (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826
Paul CJ (2020) Covid-19 image data collection. https://github.com/ieee8023/covid-chestxray-dataset
Paul M (2020) Kaggle chest X-ray images (pneumonia) dataset. https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia/
U.S. National Library of Medicine. Tuberculosis chest X-ray image data sets. https://ceb.nlm.nih.gov/tuberculosis-chest-X-ray-image-data-sets/ (2020)
Pranav R, Jeremy I, Kaylie Z, Brandon Y, Hershel M, Tony D, Daisy D, Aarti B, Curtis L, Katie S et al (2017) Chexnet: radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv preprintarXiv:1711.05225
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declared no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Note that the study has no clinical implications. Instead, we solely aimed to check whether the proposed Truncated Inception Net could possibly be used in detecting COVID-19 positive cases using CXRs.
Rights and permissions
About this article
Cite this article
Das, D., Santosh, K.C. & Pal, U. Truncated inception net: COVID-19 outbreak screening using chest X-rays. Phys Eng Sci Med 43, 915–925 (2020). https://doi.org/10.1007/s13246-020-00888-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-020-00888-x