Abstract
Aging-induced changes in the immune system are associated with a higher incidence of infection and vaccination failure. Lymph nodes, which filter the lymph to identify and fight infections, play a central role in this process. However, careful characterization of the impact of aging on lymph nodes and associated autoimmune diseases is lacking. We combined single-cell RNA sequencing (scRNA-seq) with flow cytometry to delineate the immune cell atlas of cervical draining lymph nodes (CDLNs) of both young and old mice with or without experimental autoimmune uveitis (EAU). We found extensive and complicated changes in the cellular constituents of CDLNs during aging. When confronted with autoimmune challenges, old mice developed milder EAU compared to young mice. Within this EAU process, we highlighted that the pathogenicity of T helper 17 cells (Th17) was dampened, as shown by reduced GM-CSF secretion in old mice. The mitigated secretion of GM-CSF contributed to alleviation of IL-23 secretion by antigen-presenting cells (APCs) and may, in turn, weaken APCs’ effects on facilitating the pathogenicity of Th17 cells. Meanwhile, our study further unveiled that aging downregulated GM-CSF secretion through reducing both the transcript and protein levels of IL-23R in Th17 cells from CDLNs. Overall, aging altered immune cell responses, especially through toning down Th17 cells, counteracting EAU challenge in old mice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
With a rapidly expanding elderly population, insights into aging-induced physiological changes will assist in healthy aging. The immune system shows marked changes during aging, which are often manifested as compositional and functional alterations in diverse immune cells (Goronzy and Weyand, 2013; Nikolich-Žugich, 2018). Both T and B cell compartments, main components of the adaptive immunity, show a significant shift from naïve to memory cell types with the expression of a relatively restricted antigen-receptor repertoire during aging (Cambier, 2005; Kogut et al., 2012; Nikolich-Žugich, 2018). With respect to innate immunity, the impact of aging on them often varies across cell types, cell states, and tissue contexts (Montgomery and Shaw, 2015). Despite steady progress in understanding the influence of aging on the immunological system, the underlying mechanisms still require further investigation (Nikolich-Žugich, 2018).
Age-related deterioration of the immune system is associated with increased susceptibility to infections and vaccination failure in older people (Gavazzi and Krause, 2002; Pinti et al., 2016; Goronzy and Weyand, 2017). However, there have been insufficient studies about the impact of aging on autoimmune diseases and their results are inconsistent. Although several studies have proposed that older people display greater autoimmunity characterized by increased serum levels of autoantibodies and accumulation of autoreactive T cells due to checkpoint failures during aging (Goronzy and Weyand, 2012; Müller and Pawelec, 2015), the incidence of many autoimmune diseases is lower among the elderly (Cooper and Stroehla, 2003). Indeed, most autoimmune diseases, such as systemic lupus erythematosus and multiple sclerosis, rarely occur in the elderly and, when they occur, may present with milder symptoms than in younger individuals (Cooper and Stroehla, 2003; Rovenský and Tuchynová, 2008).
Autoimmune uveitis (AU) is an autoimmune disease of the central nervous system (CNS) characterized by immune-mediated inflammatory disorders of the eye and is among the leading causes of visual impairments and blindness (de Smet et al., 2011; Thorne et al., 2016). The possible pathogenic mechanisms of AU have been gradually revealed based on studies of experimental autoimmune uveitis (EAU), a widely accepted animal model of AU (Pennesi et al.,2003). However, how aging impacts AU remains to be studied. Clinically, AU predominantly affects people of working age (20 to 50 years of age), and it is hardly observed in the elderly (Papotto et al., 2014). Although a negative relationship between aging and the incidence of AU is commonly found, the underlying mechanism is not clear.
Lymph nodes (LNs) are secondary lymphoid organs that facilitate the interaction among various immune cells (Gasteiger et al., 2016), and they are critical for the regulation of adaptive immune reaction and pathogen clearance (Gasteiger et al., 2016). LNs are also responsible for peripheral tolerance (Samy et al., 2005). However, once the tolerance is broken for some pathological reasons, autoantigens are presented to autoreactive T cells and B cells in LNs (Muñoz et al., 2010; Mochizuki et al., 2013). The interaction with autoantigens results in the proliferation and activation of autoreactive T cells and B cells, as well as the formation of germinal center (GC) in LNs, thus initiating autoimmune diseases (Muñoz et al., 2010; Yu et al., 2015; Theofilopoulos et al., 2017). During aging, LNs undergo structural and functional alterations (Turner and Mabbott, 2017). Specifically, altered structural organization, defective cell trafficking, and delayed germinal center reaction have been observed in aged LNs (Richner et al., 2015a; Turner and Mabbott, 2017). These changes contribute to impaired adaptive immunity, and account for a higher risk of infections and vaccination failure in older people (Richner et al., 2015b; Tabibian-Keissar et al., 2016). However, the impact of aging on autoimmune responses, especially those associated with LNs, has not been defined.
Single-cell RNA sequencing (scRNA-seq) is a powerful tool for analyzing samples with complex cellular constituents (Grün and van Oudenaarden, 2015), and its application has facilitated the understanding of the pathogenesis of many diseases (Rantalainen, 2018; Paik et al., 2020; Luo et al., 2020). scRNA-seq has been applied to profile immune cells of LNs, including axillary, brachial, and inguinal LNs, in various health and disease conditions (Rodda et al., 2018; Blecher-Gonen et al., 2019; Veerman et al., 2019). However, the single-cell transcriptomes of aged LNs and LNs in autoimmune conditions have not been delineated. Cervical draining LNs (CDLNs) are the main draining LNs of the central nervous system (CNS) and contribute to the pathogenesis of autoimmune diseases in CNS (Louveau et al., 2018), which are therefore ideal for exploring immune changes during EAU. Immune cell atlases of aged CDLNs and CDLNs during EAU will facilitate the understanding of the impact of aging on EAU and may provide insights into the interaction between aging and a wider range of autoimmune diseases.
In this study, we used scRNA-seq and flow cytometry to map the immune cell atlas of CDLNs from young and old mice with or without EAU and observed that aging-induced marked changes in the transcriptomes of various immune cell types in CDLNs. In response to EAU challenge, old mice exhibited alterations in immune cell compositions and functions different from those of young mice, which may explain their difference in EAU symptoms. Within this process, our study highlighted that aging weakened the pathogenicity of T helper 17 (Th17) cells, reflected in their reduced GM-CSF secretion. Coculturing antigen-presenting cells (APCs) with these restrained Th17 cells resulted in decreased IL-23 secretion by APCs, which may further impair APCs’ capacity of promoting the pathogenicity of Th17 cells. Meanwhile, aging downregulated GM-CSF secretion of Th17 cells by reducing the mRNA and protein levels of IL-23R in Th17 cells. Overall, aging altered immune responses to EAU challenge, especially through toning down the pathogenic Th17 cells, thus ameliorating EAU development in old mice.
RESULTS
Aging induces complicated and extensive changes in the immune profile of CDLNs
To probe the impact of aging on the immune profile of CDLNs and to fully explore the interaction between aging and autoimmunity, we generated scRNA-seq data from CDLNs of young normal mice (YN), old normal mice (ON), young mice with EAU (YE) and old mice with EAU (OE) (Fig. 1A). EAU was induced by retinal antigen interphotoreceptor retinoid-binding protein 1–20 (IRBP1–20), which is the most common uveitogenic retinal protein in mice (Caspi et al., 2008). Single-cell suspensions were harvested from isolated CDLNs and converted to barcoded scRNA-seq libraries via 10x Genomics for further analysis. We combined the CellRanger software with the Seurat package for initial processing, quality control, and analysis of the sequencing data. After quality control, we obtained a total of 45,857 high-quality cells (Fig. 1A).
Study design and analysis of aging-induced alterations in immune cell profiles of CDLNs. (A) Schematic of the experimental design for single-cell RNA sequencing. CDLNs were harvested from young (Y) and old (O) mice without (N) or with (E) EAU. Each sample included three mice. Samples were processed via scRNA-seq by using the 10x Genomics platform. IRBP1-20: Interphotoreceptor retinoid-binding protein 1-20; PTX: Pertussis toxin. (B) UMAP plot showing clusters of immune cell subsets. (C) Pie charts showing relative cluster abundance of immune cell subset in YN and ON mice. (D) Bar chart showing the cell ratios of immune cell subsets in YN and ON mice derived from scRNA-seq data. (E) Volcano plot showing upregulated and downregulated DEGs of all immune cell types in the ON/YN comparison group. Red and blue dots indicate upregulated and downregulated DEGs in ON group compared to YN group, respectively. (F and G) Representative GO terms and KEGG pathways enriched in upregulated DEGs of total immune cells (F) or immune cell subsets (G) in the ON/YN comparison group
We first examined the general impact of normal aging on the immune profile of CDLNs. We clustered cells from YN and ON mice and identified eight immune cell lineages, including T cells (TC), B cells (BC), conventional dendritic cells (cDC), plasmacytoid dendritic cells (pDC), monocytes (Mono), macrophages (Macro), neutrophils (Neu), and natural killer cells (NK), according to classical markers and other uniquely upregulated genes (Figs. 1B, S1A, and S1B). As shown in Fig. 1C, T cells and B cells are the primary cellular constituents of CDLNs from both YN and ON mice. During aging, the proportion of myeloid cells increased (Fig. 1D). To identify the global gene signatures of CDLN cells associated with normal aging, we identified the differentially expressed genes (DEGs) of all cell types identified in CDLNs from ON and YN mice and conducted Gene Ontology (GO) analysis of these genes (Fig. 1E, 1F; Table S1A). For example, DEG analysis revealed that the naïve phenotype-associated genes Lef1 and Ccr7 were expressed less in ON mice than in YN mice (Fig. 1E; Table S1A). Upregulated DEGs in ON mice were enriched in inflammation-related pathways (annotated as neutrophil degranulation and inflammatory response, the latter harboring DEGs such as Ccl5, S100a11, S100a4) and cytotoxic pathway (annotated as cell killing, associated with DEGs including Ctla2a, Gzmb, and Nkg7) (Patil et al., 2018) (Fig. 1E, 1F; Table S1A). Genes downregulated in ON mice were enriched for pathways such as peptide biosynthetic and amide biosynthetic processes (Fig. S1C). These DEG and GO analyses collectively indicated inflammatory states of aged CDLNs.
We then delineated the aging-induced alterations in the cell type-specific transcriptomes (Table S1B–I). In the context of aging, upregulated DEGs of T cells were enriched in T cell activation, inflammatory response, immunoregulatory interactions, and leukocyte mediated cytotoxicity pathways (Figs. 1G and S1D). In B cells, the immunoglobin production pathway was upregulated (Fig. 1G). Among myeloid cells, cDC and pDC from ON mice highly expressed genes that were enriched in phagocytosis pathways (annotated as phagocytosis or Fc-gamma receptor (FCGR)-dependent phagocytosis) (Fig. 1G), indicating enhanced phagocytosis capacity with diminished antigen-presenting capacity (Savina and Amigorena, 2007). In monocytes and macrophages, we observed aging-upregulated biological pathways related to inflammatory responses, suggesting a bias towards the inflammatory phenotypes (Fig. 1G). In neutrophils, the upregulated neutrophil degranulation pathway suggested that these cells were in active states during aging (Fig. 1G). As for downregulated genes of each major immune cell type in ON mice, pathways such as SRP-dependent cotranslational protein targeting to membrane and amide biosynthetic process were enriched (Fig. S1E). Additionally, downregulated aging DEGs in NK cells were involved in cytotoxicity-related pathways, which was consistent with reduced natural killer cell cytotoxicity during aging as previously reported (Solana et al., 2006) (Fig. S1E). Overall, we have developed a global immune profile of aged CDLNs. Immune cells from aged CDLNs are prone to exhibit inflammatory phenotypes, with each immune cell type showing their respective compositional and functional alterations.
Aging induces compositional and functional changes of T cell and B cell compartments in CDLNs
LNs mainly consist of T cells and B cells, where they are activated by encountering antigen (Gasteiger et al., 2016). T cells and B cells are the main components of adaptive immunity; therefore, they have been the focus of studies on aging and immunity (Goronzy and Weyand, 2019; Elyahu and Monsonego, 2021). Thus, we next examined aging-induced alterations in T cells and B cells.
We identified ten subtypes of T cells, including naïve CD4+ T cells (naïve CD4), naïve CD8+ T cells (naïve CD8), CD4+ T cells with cytotoxic activity (CD4-CTL), CD8+ T cells with cytotoxic activity (CD8-CTL), T helper 17 cells (CD4-Th17), regulatory T cells (CD4-Treg), T follicular helper cells (CD4-Tfh), T helper 1 cells (CD4-Th1), proliferative T cells (Pro-T), and exhausted T cells (CD4-Tex) (Figs. 2A, S2A, and S2B). Naïve T cells dominated in T cell compartments in YN mice, whereas CD4-CTL and CD8-CTL dominated in T cell compartments in ON mice (Fig. 2B). The proportions of most T cell subsets, except for naïve T cells, increased in ON mice compared to YN mice (Fig. 2C and 2D). The proportion of naïve T cells decreased in ON mice, possibly due to their ongoing differentiation into effector and memory T cells during aging as previously reported (Frasca and Blomberg, 2011). To clarify this hypothesis, we conducted pseudotime analysis and observed a pseudotemporal path started from naïve CD4 to effector T cells. Compared to YN mice, ON mice showed more effector T cells, including CD4-Th17, CD4-Th1, CD4-Treg, CD4-Tfh, CD4-CTL, and CD4-Tex in state 1, as well as more CD4-CTL in state 3, but less naïve CD4 T in state 2 (Fig. 2E). This result indicated enhanced differentiation of naïve CD4 into effector T cells in ON mice (Figs. 2F, S2C, and S2D). Similarly, among CD8+ T cells, less naïve CD8 in the initiation and more CD8-CTL in the end of the pseudotemporal path were in ON mice than in YN mice, indicating enhanced differentiation of naïve CD8 into CD8-CTL in ON mice (Fig. S3A–D).
Aging-induced alterations in T cell subsets. (A) UMAP plot showing subclusters of T cell subsets. (B) Pie charts showing relative cluster abundance of T cell subsets in the YN and ON mice. (C and D) Bar charts showing the cell ratios of CD4+ T cell subsets (C) or CD8+ T cell subsets (D) in YN and ON mice derived from scRNA-seq data. (E) Pseudotime trajectory analysis corresponding to the differentiation of effector CD4+ T cells from naïve CD4+ T cells. Cells are colored by pseudotime (top) or cell type (bottom), as indicated. (F) Percentages of CD4+ T cells along the pseudotime for YN and ON mice. (G and H) Representative GO terms and KEGG pathways enriched in upregulated DEGs of naïve CD4+ T cells (G) or Th17 cells (H) in the ON/YN comparison group. (I) Violin plots of expression of Il17a and Il1r1 in Th17 cells of YN and ON mice. (J) Heatmap of relative regulon activity of Bhlhe40 and Junb from Th17 cells in the YN and ON mice. (K) Network plots showing the changes in ligand-receptor interaction events between different cell types in the YN/ON comparison group. Cell-cell communication is indicated by the connected line. The thickness of the lines is positively correlated with the number of ligand-receptor interaction events
GO analysis revealed that the upregulated DEGs of naïve CD4 from ON mice were enriched in T cell activation pathway and regulatory T cell differentiation pathway, including Maf, Ikzf2, and Tnfrsf4 (Figs. 2G and S3E; Table S1J). Cytotoxic genes (Ctla2a and Nkg7) were upregulated in naïve CD8 from ON mice (Fig. S3F; Table S1Q). Upregulated DEGs in naïve CD8 from ON mice were also enriched in the lymphocyte differentiation pathway (Fig. S3G). These results indicated that naïve T cells in aged CDLNs differentiated actively and tended to exhibit a mix of inflammatory, regulatory, and cytotoxic phenotypes. Since Th17 cells are a T cell subset actively involved in AU and EAU (Harrington et al., 2005), we next analyzed their aging-induced transcriptional changes. GO analysis revealed downregulated IL-17 signaling pathway in CD4-Th17 from ON mice compared to YN mice (Fig. 2H). Correspondingly, the expression of Il17a was lower in ON mice compared to YN mice (Figs. 2I and S3H; Table S1K). Moreover, the expression of Il1r1, a pathogenic Th17 cell-related gene (Yasuda et al., 2019), as well as the regulon activity of Bhlhe40 and Junb, two pathogenic transcriptional factors (TFs) of Th17 cells (Lin et al., 2014; Hasan et al., 2017), were all lower in ON mice than in YN mice (Figs. 2I, 2J, and S3I; Table S2). These results may indicate that aging induces functional impairment in Th17 cells. Among other T cell subsets, T cell activation and inflammation-related pathways were generally upregulated in ON mice (Fig. S4A; Table S1J–S). We also observed downregulated pathways such as peptide biosynthetic process and cytoplasmic translation, in most T cell subsets (Fig. S4B; Table S1J–S). Thus, aging induced various alterations in different T cell subsets. Specifically, active differentiation of naïve T cells, impaired function of Th17 cells were observed in aged CDLNs.
As for the B cell compartments, we identified four B cell subsets, i.e., naïve B cells (NBC), germinal B cells (GBC), plasma B cells (PBC), and Fcrl5-positive B cells (Fcrl5-BC) (Fig. S5A–C). Increased proportion of Fcrl5-B cells, germinal B cells, and plasma B cells were observed in ON mice compared to YN mice (Fig. S5D). Pseudotime analysis showed that a pseudotemporal path started from naïve B cells to effector B cells. We observed that more plasma B cells and Fcrl5-B cells were in state 1 and more germinal B cells were in state 2 in ON mice compared to YN mice, indicating enhanced differentiation from naïve B cells to effector B cells (Fig. S5E–H). The relatively large proportion of naïve B cells may explain that although their proportion just slightly decreased in ON mice, active differentiation from naïve subsets to effector subsets still existed in ON mice. We then conducted GO analysis to explore functional changes in B cell subsets. Aging upregulated the antigen processing and presentation pathway in naïve B cells and the immunoglobulin production pathway in plasma B cells, as well as downregulated pathways such as peptide biosynthetic process in all B cell subsets (Fig. S6A and S6B; Table S1T–W). Thus, the two main functions of B cells, namely, antigen processing and presentation, as well as immunoglobulin production, might be preserved and even enhanced in B cells of the elderly.
We further explored the cell-cell interaction among T cell and B cell subsets, along with other major immune cell types (Table S5). The cell-cell interaction events were integrally increased among different cell types during aging (Fig. 2K). Activation and differentiation of T cells require co-stimulation from DC (driven by CD28 and its ligand CD86) (Levine et al., 1995), whereas activation and differentiation of B cells require co-stimulation provided by T cells (driven by CD40 and its ligand CD40LG) (Armitage et al., 1992). We observed enhanced CD28-CD86 interaction between naïve T cells and cDC, accompanied by enhanced CD40-CD40LG interaction between germinal or naïve B cells and Tfh cells in ON compared to YN (Fig. S6C). APRIL, a proliferation-inducing ligand, plays an important role in B cell differentiation (Shabgah et al., 2019; Yeh et al., 2020). We found that the APRIL signaling network was more potent in ON mice than in YN mice (Fig. S6D). These results partially explained the active differentiation from naïve to effector cells and the proportional change of T cell and B cell subsets in old mice. In addition, the interaction of TGF-β (encoded by Tgfb1) and its receptors between Tregs and several other immune cells was among the increased interaction events led by aging (Fig. S6E). TGF-β is the main molecule that mediates the immunosuppressive ability of Tregs (Liu et al., 2018). Thus, Tregs in ON mice might have enhanced immune-repressing capacity compared to those of YN mice as previously reported (Elyahu et al., 2019).
In summary, our single-cell data revealed reduced naïve T cells, and increased effector cells in both T cell and B cell compartments including Th1, Th17, and plasma cells in ON mice. These results indicated that old mice may behave differently when experiencing autoimmune diseases compared to young individuals. Functionally, GO and DEG analyses of T cells indicated that naïve T cells differentiate actively and prefer to exhibit a mixture of inflammatory, regulatory, and cytotoxic phenotypes whereas the function of Th17 cells might be impaired in old mice. B cells from aging CDLNs showed preserved antigen processing and presentation as well as immunoglobin production capacity. In addition, cell-cell communications, including ligand-receptor pairs related to T and B cell activation and immune repressing ability of Tregs, are generally enhanced in old mice.
Aging mitigates EAU symptoms and alters immune cell response to EAU challenge
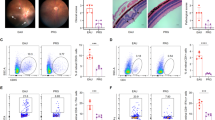
AU was hardly observed in elderly people (Papotto et al., 2014). To elucidate this phenomenon and further analyze the relationship between aging and AU, we next evaluated aging-associated alterations in autoimmune responses during EAU. We first evaluated EAU symptoms in young and old mice by photographing the eye with a fundus camera on day 14 and grading the disease severity according to the published EAU clinical grading scale (Chen and Caspi, 2019a). EAU in the fundus manifested as multiple chorioretinal lesions and/or infiltrations (Fig. 3A and 3B; Table S6). Hemotoxylin and eosin (H&E) staining of sectioned eyeballs showed inflammatory infiltration and retinal folding in EAU lesions (Fig. 3C and 3D; Table S6). EAU challenge enlarged the CDLNs from both young and old mice (Fig. S7A). Intriguingly, OE mice exhibited milder EAU symptoms, defined by lower clinical scores and pathological scores, compared to YE mice (Fig. 3B and 3D; Table S6). This divergence in the EAU severity between young and old mice was in line with the low incidence rate of AU in the elderly population (Papotto et al., 2014).
Milder EAU symptoms and altered immune cell responses to EAU challenge induced by aging. (A) Representative fundus imaging of YN, YE, ON, and OE obtained 14 days after immunization. White arrowheads indicate inflammatory exudation and vascular deformation. (B) Bar plot showing clinical scores of the four groups. Each group contains six mice. The values represent the mean ± SD. Significance was determined using two-way ANOVA. **P < 0.01, ***P < 0.001. (C) Histopathological images of hematoxylin and eosin-stained eye sections of the four groups. Black arrowheads indicate magnified area with infiltration of inflammatory cells and retinal folding. Scale bars, 20 mm. (D) Bar plot showing pathogenic scores of the four groups. Each group contains six mice. The values represent the mean ± SD. Significance was determined using two-way ANOVA. *P < 0.05, ****P < 0.0001. (E) UMAP plot showing clusters of immune cell subsets. (F) Bar chart showing the cell ratios of immune cell subset in the YE/YN and OE/ON comparison groups derived from scRNA-seq data. (G–I) Representative GO terms and KEGG pathways enriched in upregulated DEGs of total immune cells (G), T cells (H), or B cells (I) in the YE/YN and OE/ON comparison groups
To delineate the aging-induced alterations in immune cell response during EAU, CDLN cells from YN, YE, ON, and OE mice were subjected to the single-cell analysis, and major immune cell clusters were identified according to classical markers (Figs. 3E, S7B, and S7C). To demonstrate aging-induced changes in cell composition dynamics in response to EAU, we compared the proportion of each immune cell type between YE and YN mice, in comparison to the change between OE and ON mice (Fig. 3F). Most myeloid cells were increased after EAU development in young mice, but decreased in old mice (Fig. 3F). In contrast, we observed a slight increase in the T cell proportion in OE mice (Fig. 3F). These results showed divergent cell composition changes in response to EAU challenge between young and old mice.
GO analysis showed that total immune cells upregulated the leukocyte differentiation pathway and downregulated the negative regulation of immune system process in response to EAU challenge in both young and old mice (Figs. 3G and S7D; Tables S3A and S4A). When each type of immune cell was analyzed individually, those involved in innate immunity showed comparable upregulated pathways in young and old mice in response to EAU challenge, and most of them were related to enhanced inflammation (Fig. S8A; Tables S3B–I and S4B–I). In contrast, in response to EAU challenge, the IL-17 signaling pathway and the antigen processing and presentation pathway were upregulated in young mice only, whereas T cell-mediated cytotoxicity was upregulated in old mice only (Fig. 3G). The antigen processing and presentation pathway was upregulated in response to EAU challenge in DC from young mice but was barely upregulated in DC from old mice (Fig. S8A). In T cells, the IL-17 signaling pathway was upregulated in YE mice, but not in OE mice (Fig. 3H). This result indicated impaired function of Th17 cells during aging. In B cells, the antigen processing and presentation pathway was enriched in YE mice, whereas the immunoglobulin production pathway was upregulated in OE mice (Fig. 3I). GO analysis of downregulated DEGs showed that negative regulation of immune system process was downregulated in T cells from old mice alone, whereas negative regulation of B cell activation pathway was specifically downregulated in B cells from young mice during EAU (Fig. S8B). In summary, although similar responses in total immune cells are presented in young and old mice in response to EAU challenge, aging clearly induces additional alterations different in T cells and B cells in the EAU model.
Aging induces altered responses of T cell and B cell subsets during EAU
Our data above showed that aging-induced distinct T cell and B cell responses to the EAU challenge. Thus, we next investigated the impact of aging on the response of T cell and B cell subsets to EAU challenge. T cells from YE and OE mice were also clustered into ten subsets, similar to the classification of T cells from YN and ON mice (Figs. 4A and S9A). We then explored different compositional changes of T cell subsets in response to the EAU challenge with respect to age (Fig. 4B). Most CD4+ T cell subsets increased after EAU development in young mice, including CD4-Th1 and CD4-Th17 that could promote EAU pathogenesis (Luger et al., 2008), CD4-Tfh with B cell regulating function (Crotty, 2011) (Fig. 4B). However, this expansion was refrained in old mice (Fig. 4B). Resulted from divergent baseline proportions, although aging refrained most CD4+ effector T cell subsets from extensive expansion in response to EAU challenge, the proportion of most CD4+ effector T cell subsets, including CD4-Th17, CD4-Treg, and CD4-Tfh, was still higher in OE mice than in YE mice (Fig. 4B). The proportion of CD8+ T cell subsets did not increase in response to EAU challenge in both young and old mice (Fig. 4B), suggesting their relatively weaker role in EAU development compared to CD4+ T cells. Thus, we then focused on the aging-induced gene expression alterations in the CD4+ T cell subset in response to EAU challenge.
Altered T cell subset responses to EAU challenge. (A) UMAP plot showing clusters of T cell subsets. (B) The fold change (FC) of cell ratios in different T cell subsets across the four groups (YN, YE, ON, and OE). The numbers on the right indicate the Log2FC values of the cell ratios (YE/YN, OE/ON, and OE/YE). (C) Wind rose diagrams showing the numbers of upregulated and downregulated DEGs of CD4+ T cells in the YE/YN and OE/ON comparison groups. (D) Representative GO terms and KEGG pathways enriched in upregulated DEGs of T cell subsets in the YE/YN and OE/ON comparison groups
After EAU development, 2,242 and 1,611 DEGs were detected in CD4+ T cells of young mice and old mice, respectively (Fig. 4C; Tables S3J–P and S4J–P). In young mice, DEGs in CD4-Th1, CD4-Th17 and CD4-Tex accounted for most DEGs, whereas DEGs in CD4-Tex account for most DEGs in old mice (Fig. 4C). This result suggested that Th1 and Th17 cells played an important role in EAU development in young mice, but their effects were compromised in old mice. We also conducted GO analysis on all T cell subsets (Figs. 4D and S9B; Tables S3J–S and S4J–S). Naïve T cells, including naïve CD4 and naïve CD8, showed upregulated leukocyte differentiation pathway in both young and old mice during EAU (Fig. 4D). In addition, pathways related to inflammation response were only upregulated in effector T cells (including CD4-Th1 and CD4-Tfh) of YE mice, while response to interferon-beta was enriched in CD8-CTL, exclusively in old mice (Fig. 4D). We also observed downregulated DEGs enriched in “amide biosynthetic process” and “SRP-dependent cotranslational protein targeting to membrane” in both young and old mice (Fig. S9B). Thus, aging may weaken the role of Th1 and Th17 cells in EAU development.
In response to EAU challenge, the proportion of the naïve B cell subset reduced, and the proportion of other B cell subsets increased in both young and old mice (Fig. S10A–C). We validated the proportional change of plasma cells using flow cytometry. We found higher proportions of plasma cells in old mice both in baseline level and after EAU development (Fig. S10D and S10E). Tfh cells regulate B cell differentiation into plasma cells and memory B cells (Crotty, 2011). The proportion of Tfh cells was higher in ON mice compared to YN mice (Fig. 2C), which increased after EAU development in both young and old mice but in different ratios (Fig. S10F and S10G), and consistent with the proportional change of plasma cells. At the functional level, GO analysis showed that the antigen processing and presentation-related pathway was upregulated only in naïve B cells in young mice while the immunoglobulin production pathway was upregulated only in plasma cells in old mice after EAU development (Fig. S10H and S10I; Tables S3T–W and S4T–W). Plasma cells are the main antibody-producing cells (Lam and Bhattacharya, 2018). The above results thus suggested that plasma cells from old mice had enhanced immunoglobin production capacity, consistent with previous reports of higher titers of autoantibodies in both older people and aged mice (Bovbjerg et al., 1991; Aprahamian et al., 2008). However, these increased serum levels of autoantibodies in old individuals are due to tissue damage and apoptosis (non-specific autoantibody) instead of specific pathologies (specific autoantibody) (Stacy et al., 2002; Aprahamian et al., 2008; Larbi et al., 2008). These increased non-specific autoantibodies including rheumatoid factor and antinuclear antibodies are not necessarily related to diseases (Moulias et al., 1984; Ruffatti et al., 1990). To identify how aging impacted the function of B cells to produce autoantibodies targeting disease-specific autoantigens, we then measured the level of serum immunoglobulins G (IgG) specific to IRBP1–20 (anti-IRBP1–20 antibody) using ELISA. IRBP1–20 is the classical autoantigen that can induce the immune system to attack the retina and accounts for EAU induction (Avichezer et al., 2003). The anti-IRBP1–20 IgG was undetectable in YN and ON mice and was markedly lower in OE mice compared to YE mice (Fig. S10J). We also quantified the level of total serum IgG, which was higher in ON mice than in YN mice; whereas after EAU development, YE mice showed higher levels of total IgG than OE mice (Fig. S10K). Thus, aging might lead to enhanced non-specific antibody production of B cells but impaired their capacity to produce specific autoantibodies during autoimmune conditions. Retinal autoantibodies have been proven to augment the pathogenic function of autoreactive T cells in EAU development during adoptive transfer experiments (Pennesi et al., 2003). Thus, the impaired ability of B cells to generate specific autoantibodies may also contribute to the milder EAU symptoms in old mice.
Collectively, our above results delineated alterations in T cell and B cell subsets of CDLNs in response to EAU challenge in both young and old mice. Aging leads to refrained expansion of several CD4+ T cell subsets while reserving the proportion dynamics of B cells in response to EAU challenge. In addition, aging reduces specific autoantibody production of B cells during EAU.
EAU challenge induces divergent cell-cell interaction changes between young and old mice
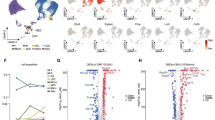
We also explored the cell-cell interaction among T cells, B cells, and other immune cells of the mice with EAU (Table S5). After EAU development, interaction events were evidently increased among cell types in young mice but were not consistent in old mice (Fig. 5A). The increased interaction events after EAU development were mainly enriched in inflammatory and immune cell activation pathways in both young and old mice, but more evident in young mice (Fig. 5B). In addition, the CD28-CD80 interaction related to T cell activation was increased in both young and old mice after EAU development (Fig. 5C). In old mice, the decreased interaction events after EAU development were enriched in chemokine-related pathway (Fig. 5D), whereas the decreased interaction events were rarely presented in young mice. Additionally, PTPRC-CD22 and FAM3C-CLEC2D interactions related to B cell activation (Llibre et al., 2016; Alborzian Deh Sheikh et al., 2021) were enhanced between B cells and Tfh cells in both young and old mice (Fig. 5E; Table S5) but the CD40-CD40LG interaction, another B cell activation pair, was enhanced only in young mice during EAU (Fig. 5F; Table S5). We further compared the interactions among four groups of mice and identified 6 and 9 pairs of ligand-receptors uniquely increased for YE mice and OE mice, respectively (Fig. 5G). Notably, among these unique pairs, GM-CSF and its receptors have been reported to be closely related to CNS autoimmunity (Lotfi et al., 2019; Chong et al., 2020). We found that the interaction of GM-CSF and its receptors mainly occurred between Th17 cells and APCs (Fig. 5H). These results indicated the interaction of GM-CSF and its receptors between Th17 cells and APCs might contribute to distinct EAU symptoms between young and old mice.
EAU challenge induces divergent cell-cell interaction changes between young and old mice. (A) Network plots showing the changes in ligand-receptor interaction events between different cell types in the YE/YN and OE/ON comparison groups. Cell-cell communication is indicated by the connected line. The thickness of the lines is positively correlated with the number of ligand-receptor interaction events. (B) Functional enrichment analysis showing the GO terms and pathways involving increased ligand-receptor interactions in YE/YN and OE/ON comparison groups. (C) Increased ligand-receptor interactions about T cell activation in YE/YN and OE/ON comparison groups. (D) Functional enrichment analysis showing the GO terms and pathways involving decreased ligand-receptor interactions in OE/ON comparison group. (E) Heatmap of the relative expression in interaction pairs: PTPRC_CD22 between CD4-Tfh and germinal B cells, and FAM3C_CLEC2D between naïve B and CD4-Tfh cells. (F) Heatmap of the relative expression in interaction pairs: CD40_CD40LG between germinal B and CD4-Tfh cells, and between naïve B and CD4-Tfh cells. (G) Venn diagrams showing the numbers of interaction pairs across four groups. The unique interaction pairs of YE are labeled in the diagrams. (H) Expression distribution of GM-CSF and its receptors across the four groups
Aging reduces Th17 pathogenicity by weakening the GM-CSF/IL-23/IL-23R positive feedback loop
Th17 cells are essential in the pathogenicity of AU, and adoptive transfer of Th17 can induce EAU development (Luger et al., 2008). To deeply identify aging-associated changes in Th17 cells’ function during EAU, we analyzed DEGs of Th17 cells across four groups of mice. The expression of Csf2 and IL23r in Th17 cells was higher in young mice compared to old mice after EAU development (Fig. S11A). GM-CSF, encoded by Csf2, is dominantly secreted by Th17 cells, which is induced by IL-23 from APCs via IL-23R in Th17 cells. GM-CSF is a hallmark that distinguishes pathogenic Th17 cells from non-pathogenic Th17 cells (Lee et al., 2012; Yasuda et al., 2019), and Th17 cells deficient of GM-CSF lose their pathogenic ability (El-Behi et al., 2011a). Thus, we evaluated GM-CSF and IL-23R expression in Th17 cells by flow cytometry and found that aging increased Th17 cell proportion but reduced their expression of IL-23R and GM-CSF (Figs. 6A–F and S11B). In addition, aging reduced the number of GM-CSF-expressing cells in CDLNs (Fig. 6G and 6H). Correspondingly, the retina-infiltrating T cells in OE mice also exhibited decreased expression of GM-CSF and IL-23R compared to those in YE mice (Fig. 6I–L). These results collectively reflected impaired Th17 pathogenicity led by aging during EAU.
Aging weakened Th17 pathogenicity. (A and B) Representative flow charts (A) and quantification (B) of the proportion of Th17 cells from CDLNs of the four groups. Each group contains six mice. The values represent the mean ± SD from three independent experiments. Significance was determined using two-way ANOVA. **P < 0.01, ***P < 0.001, ****P < 0.0001. (C–F) The proportion of Th17 cells from CDLNs of the four groups expressing GM-CSF (C and D) or IL23R (E and F) was measured by flow cytometry (C and E) and quantified (D and F). Each group contains six mice. The values represent the mean ± SD from three independent experiments. Significance was determined using two-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (G and H) Representative images of immunostaining of cross-sections of CDLNs of the four groups for GM-CSF (green) and nuclei (4’,6-diamidino-2-phenylindole (DAPI)-staining; blue) (G) and quantification of GM-CSF+ cells (H). Each group contains six mice. The values represent the mean ± SD. Significance was determined using two-way ANOVA. **P < 0.01, ****P < 0.0001. Scale bars, 20 mm. (I–L) The proportion of Th17 cells from retina of YE and OE expressing GM-CSF (I and J) or IL-23R (K and L) was measured by flow cytometry (I and K) and quantified (J and L). Each group contains six mice. The values represent the mean ± SD from three independent experiments. Significance was determined using unpaired student’s t test. ****P < 0.0001
Interactions of GM-CSF and its receptors between Th17 cells and APCs only occurred in young mice but not in old mice after EAU development (Fig. 5H). GM-CSF secreted by Th17 cells can augment the secretion of IL-23 from APCs and vice versa, thus constituting a positive feedback loop to maintain Th17 pathogenicity via IL23R expressed in Th17 cells (El-Behi et al., 2011a). The reduced GM-CSF and IL23R expression in Th17 cells may therefore account for abolished interactions between Th17 cells and APCs and may influence IL-23 secretion by APCs. Indeed, we detected decreased serum levels of GM-CSF in old mice by ELISA (Fig. 7A), which was consistent with low GM-CSF levels in elder people (Kim et al., 2011). Additionally, the concentration of IL-23 in serum from old mice was also reduced (Fig. 7B).
Aging weakened the GM-CSF/IL-23/IL-23R positive feedback loop. (A and B) Serum levels of GM-CSF (A) or IL-23 (B) of the four groups. Each group contains six mice. The values represent the mean ± SD. Significance was determined using two-way ANOVA. *P < 0.05, ***P < 0.001, ****P < 0.0001. (C) CD11C+ APCs isolated from YE mice were cocultured with CD4+CCR6+CXCR3− T cells (Th17 cells) isolated from YE mice or OE mice with or without anti-GM-CSF antibody for 72 h. IL-23 level in the culture supernatant was measured by ELISA. The values represent the mean ± SD from six independent experiments. Significance was determined using two-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001. (D and E) CDLN cells from YE and OE mice were collected after immunization on day 14 and cultured with IRBP1–20 alone or with IRBP1–20 plus IL–23. The proportion of GM-CSF-expressing Th17 cells was measured by flow cytometry (D) and quantified (E). The values represent the mean ± SD from six independent experiments. Significance was determined using two-way ANOVA. **P < 0.01, ****P < 0.0001. (F and G) The representative fundus images (F) and clinical scores (G) after induction by CD4+CCR6+CXCR3− T cells (Th17 cells) from YE (YE-AT) or OE (OE-AT) groups. Each group contains six mice. Significance was determined using unpaired student’s t-test. ***P < 0.001. White arrowheads indicate inflammatory exudation and vascular deformation. (H and I) The representative HE staining images (H) and pathological scores (I) after induction by CD4+CCR6+CXCR3− T cells (Th17 cells) from YE (YE-AT) or OE (OE-AT) groups. Black arrowheads indicate magnified area with infiltration of inflammatory cells and retinal folding. Scale bars, 20 mm. Each group contains six mice. Significance was determined using unpaired student’s t-test. ****P < 0.0001. (J–M) The proportion frequency of Th17 cells from CDLN of YE (YE-AT) or OE (OE-AT) groups expressing GM-CSF (J and K) or IL-23R (L and M) was measured by flow cytometry (J and L) and quantified (K and M). Each group contains six mice. The values represent the mean ± SD from three independent experiments. Significance was determined using unpaired student’s t-test. ****P < 0.0001
To explore the above hypothesis, we isolated CD11C+ APCs from YE mice and cocultured them with CD4+CCR6+CXCR3− T cells (Th17 cells) (Zhao et al., 2020) from CDLNs of YE or OE mice. Coculture promoted IL-23 secretion by APCs, and Th17 cells from YE mice have a more potent promoting function on APCs than those from OE mice (Fig. 7C). Additionally, IL-23 secretion was decreased after GM-CSF neutralization (Fig. 7C). This result demonstrated that GM-CSF secreted by Th17 cells was involved in the induced expression of IL-23 in APCs. Moreover, low GM-CSF secretion by Th17 cells and consequently low IL-23 secretion by APCs may in turn, further impair Th17’s pathogenicity due to the compromised IL23-IL23R interaction between APC and Th17 cells. To confirm this in IRBP1–20-specific response, IRBP1–20 was used as a stimulator. We collected CDLN cells from YE and OE mice, cultured these cells with IRBP1–20 alone or with IRBP1–20 plus IL-23, and measured the GM-CSF secretion by Th17 cells. IL-23 enhanced the secretion of GM-CSF by Th17 cells from YE mice more than that by Th17 cells from OE mice, consistent with aging-induced downregulation of IL-23R in OE mice (Fig. 7D and 7E). Thus, our study demonstrated that aging weakened Th17 pathogenicity, which was to some extent, attributed to the downregulated expression of IL-23R. We further conducted an adoptive transfer experiment. Th17 cells isolated from CDLNs of YE mice induced uveitis in mice whereas these cells isolated from OE mice failed to induced EAU in naïve mice (Fig. 7F–I). Correspondingly, a significantly less amount of GM-CSF+ and IL-23R+ Th17 cells were detected in mice that received Th17 cells from OE mice (Fig. 7J–M). This result further confirmed the impaired Th17 cell pathogenicity induced by aging. These results collectively indicated that aging weakened Th17 pathogenicity by weakening the GM-CSF/IL-23/IL-23R positive feedback loop (Fig. S11C).
DISCUSSION
Here, using scRNA-seq, we are the first to map the immune cell atlas of aging LNs. Combining with flow cytometry, we deeply investigate the impact of aging on EAU. The result that old mice developed milder EAU can be attributed to the extensive alterations induced by aging in immune cell response to EAU challenge. Importantly, within this process, our study unveils reduced pathogenicity of Th17 cells from aging mice, reflected as low GM-CSF secretion by Th17 cells resulting from aging-induced downregulation of IL-23R in these cells. These defective Th17 cells contribute to low IL-23 secretion of APCs, which may further impair APCs’ promoting effects on pathogenic Th17 cells.
The impact of aging on immune system has tissue specificity and stage specificity (Consortium., 2020). Although alterations of the immune system during aging have been extensively studied in human peripheral blood and mouse spleen (Elyahu et al., 2019; Zheng et al., 2020), a detailed immune cell atlas in aged lymph nodes, the essential constituents of the immune system, was still missing before our study, let alone a detailed study of cellular alterations in aged CDLNs and associated autoimmune diseases. Here, we for the first time delineate the immune cell atlas of CDLNs of both young and old mice with or without EAU through a combination of scRNA-seq and flow cytometry. It is widely known that aging is commonly associated with the reduction of naïve cells and the expansion of antigen-experienced subsets (Nikolich-Žugich, 2018). We also detected a decrease of naïve subsets and an increase of effector subsets in T cells of aged CDLNs, which might be associated with enhanced differentiation of naïve subsets in old mice compared to young mice as indicated by our GO and pseudotime analyses. In the functional perspective, consistent with previous reports that aging of the immune system was usually accompanied by chronic, low-grade inflammation (Franceschi et al., 2017), DEG and GO analyses of total CDLN immune cells also indicated an inflammatory state of aged CDLNs. Additionally, GO analysis of T cells showed multiple effector states (a mix of inflammatory, cytotoxic, and regulatory phenotypes) of T cells from aging mice, similar to the results of the previous report (Elyahu et al., 2019). Notably, we also observed impaired Th17 cells, which have not gained enough attention before our study, and now we have accentuated its importance in autoimmune responses, especially with respect to age. Regarding B cells, preserved and even enhanced immunoglobin production capacity of plasma cells is indicated by our study. With regard to changes of innate immune cells during aging, varied conclusions had been presented (Shaw et al., 2010; Solana et al., 2012). The proportions of DC, macrophages, NK cells, and neutrophils were reported to be maintained or even increased during aging (Tesar et al., 2006; Shaw et al., 2010; Solana et al., 2012). Our study provided additional evidence showing an increased proportion of innate immune cells except for NK cells during aging (Fig. 1D). At the functional level, GO analysis indicated decreased antigen-presentation capacity of DC and reduced cytotoxicity of NK cells as previously reported (Fig. 1G) (Uyemura et al., 2002; Solana et al., 2006). In addition, it has been proposed that activation of the innate immune system is one of the features of chronic inflammations in the elderly (Franceschi et al., 2018). Consistently, GO analysis of DEGs for innate immune cells provides such evidence, such as the activation of the neutrophils and the shift towards pro-inflammatory phenotype of monocytes and macrophages in the old mice. In summary, aging induces complicated changes in the immune cellular constitutes of CDLNs. These changes are generally consistent with changes in the general immune system induced by aging that have been widely studied (Solana et al., 2012; Nikolich-Žugich, 2018).
The extensive alterations of immune cells indicate an altered autoimmune process in old individuals. However, little attention has been paid to the relationship between aging and autoimmunity before. Our study deliberately compared the autoimmune response to EAU challenge between young and old mice, which may provide references to a wider range of autoimmune diseases. In our study, old mice show much milder EAU symptoms compared to young mice. In addition to being the main orchestrators of adaptive immunity, T cells and B cells significantly contribute to autoimmune diseases (Khan and Ghazanfar, 2018; Meffre and O'Connor, 2019). In AU, T cells are the initiators and play essential roles in its pathogenesis (Weaver et al., 2006; Dittel, 2008), whereas the role of B cells is still uncertain (Smith et al., 2016; Zhu et al., 2021). Our GO analyses showed that T and B cells from young and old mice exhibited distinct upregulated pathways, suggesting their important role in divergent EAU symptoms between young and old mice. The IL-17 signaling was enhanced in young mice but not in old mice after EAU development within both total immune cells and the T cell compartment. This result is in line with the impaired function of Th17 cells in old mice and implicates its role in the aging-induced divergent autoimmune response to EAU challenge. About B cell subsets, although our scRNA-seq data indicated increased immunoglobin production by plasma cells in old mice, ELISA detected decreased IRBP1–20-specific autoantibody production by plasma B cells in old mice. This divergence may result from age-associated defects in the induction of key molecules responsible for class switching and somatic hypermutation (Frasca et al., 2004). In addition, age-related defects in CD4+ T cell cognate helper function also contribute to lower levels of antigen-specific IgG (Eaton et al., 2004). The function of retinal autoantibodies secreted by B cells in EAU has been reported. Although the serum of EAU mice (containing autoantibodies targeting the retina) alone failed to transfer diseases to recipients, transferring autoreactive T cells with that serum can exacerbate EAU symptoms compared to transferring T cells alone, which indicated that retinal autoantibodies may play an assisting role in EAU development (Pennesi et al., 2003). Thus, aging-related decline of antigen-specific autoantibodies in plasma cells may contribute to the milder EAU symptoms. As for innate immune cells, DC from old mice show impaired upregulation in antigen-presentation pathways, whereas no difference was evident in the other innate immune cell types between young and old mice in response to EAU challenge. Overall, aging induces altered immune cell response to EAU challenge, which may counteract EAU challenge in old mice.
Th17 cells are critical pathogenic components of AU and EAU (Amadi-Obi et al., 2007; Wilson et al., 2007). The pathogenicity of Th17 cells can be modulated by environmental factors, such as a high-salt diet (Kleinewietfeld et al., 2013) or environmental toxins (Veldhoen et al., 2008). However, few studies have reported altered pathogenesis of Th17 cells as a consequence of aging and the underlying mechanisms. As discussed above, the function of Th17 cells in EAU development might be weaker in old mice compared to that in young mice. Among the unique ligand-receptor interactions of YE mice that may account for divergent EAU processes between young and old mice, the interaction between GM-CSF (encoded by Csf2) and its receptors has a close relationship to CNS autoimmunity (Ifergan et al., 2017). Expression of GM-CSF is a mark of pathogenic Th17 cells (Codarri et al., 2011; El-Behi et al., 2011b). Flow cytometry showed that Th17 cells of old mice lost their pathogenicity, which was reflected as reduced GM-CSF secretion, during EAU. In addition, Th17 cells defective in GM-CSF expression reduced the IL-23 secretion by cocultured APCs. This effect further impaired the promoting effect of APCs on pathogenic Th17 cells (El-Behi et al., 2011a). GM-CSF is a downstream molecule of IL-23R and the stimulation of IL-23 can induce GM-CSF secretion (El-Behi et al., 2011b; Komuczki et al., 2019). The necessity of IL-23 in Th17 pathogenicity has been demonstrated (El-Behi et al., 2011b; Komuczki et al., 2019). Our scRNA-seq data showed that the expression of IL-23R was lower in old mice compared to that in young mice after EAU development, which was confirmed by flow cytometry. Our in vitro study further supported the correlation of lower IL-23R levels with reduced GM-CSF secretion by Th17 cells from old mice. Taken together, our study is the first to demonstrate that aging weakened Th17 pathogenicity by dampening the GM-CSF/IL-23/IL-23R positive feedback loop.
As for the stage specific contribution of aging to uveitis, this disease is less common in children, with enhanced symptoms and increased incidence with age in people of working age (20 to 50 years of age), but much less severe in the elderly people (>65-year-old) (Papotto et al., 2014; Tsirouki et al., 2016). However, the proportion of Th17 cells, the main pathogenic cell type in uveitis, gradually and continuously increases during aging (ranging from newborn to 60–79-year-old) (Botafogo et al., 2020). Wang et al. demonstrated that, in middle-aged mice (10-month-old), the accumulated DNA and the regulatory subunit of the DNA-dependent protein kinase augment the activation of CD4+ T cells and aging-related auto-inflammation (Wang et al., 2021). This study explained the mechanism of higher incidence of autoimmune uveitis in young adults. However, in the elderly people, it waits to be explained for the lower incidence of autoimmune uveitis and the seemingly contradicting, higher proportion of Th17 cells. And our study filled this gap. We demonstrated that dampening the GM-CSF/IL-23/IL-23R positive feedback loop is key to the weakened pathogenicity of Th17 cells in the old individuals, even though the proportion of Th17 cells is higher in old mice than young mice. Thus, the results of the above study in middle-aged mice and our study in old mice reflect the divergent characteristics of autoimmune inflammation in people of different age groups and actually complement each other.
In summary, our study delineates an immune cell atlas of aging CDLNs and explores the impact of aging on autoimmune response based on EAU model. We demonstrate enormous changes in immune cells in response to EAU challenges between young and old mice that may account for milder EAU symptoms in old mice. Importantly, we are the first to report that aging weakens Th17 pathogenic function and its underlying mechanisms. Our study provides novel insights into the interplay between aging and autoimmunity and serves as a reference for a large community of scientists interested in this subject.
MATERIALS AND METHODS
Mice
Young (2 months old) and old (20 months old) C57BL/6J mice were purchased from SPF Biotechnology Co., Ltd. (Beijing, China). The animal experiments were approved by the Institutional Animal Care Committee (Zhongshan Ophthalmic Center, Sun Yat-Sen University).
EAU model induction and EAU clinical score
For active induction of EAU, mice were injected subcutaneously with an emulsion consisted of 2 mg/mL of retinal antigen interphotoreceptor retinoid-binding protein 1–20 (IRBP1–20) (GPTHLFQPSLVLDMAKVLLD, GiL Biochem, Shanghai, China) and complete Freund’s adjuvant (BD Difco, San Jose, CA, USA) containing 2.5 mg of Mycobacterium tuberculosis strain H37Ra (BD Difco, San Jose, CA, USA) in a 1:1 volume ratio. Additionally, 0.25 µg pertussis toxin (PTX) (List Biological Laboratories, Campbell, California, USA) dissolved in PBS was injected on the same day and 2 days after immunization (Chan et al., 1990; Agarwal et al., 2012; Chen and Caspi, 2019b).
Funduscopic examination of EAU progress with the Micron IV fundus camera (Phoenix Co., Campbell, CA, USA) was performed and the clinical findings were graded from 0 to 4 based on observable infiltration and vasculitis in the retina (Details in Table S6). The clinical score was assessed in a blinded manner.
Flow cytometric analysis
After staining with live/dead dye (Thermo Fisher Scientific, Waltham, MA, USA) for 10 min at 4 °C and washing once with PBS, the cells were stained with antibodies detecting surface markers: CD4 (GK1.5, catalog 100434, BioLegend), IL-23R (12B2B64, catalog 150907, BioLegend), CXCR3 (CXCR3-173, catalog 126506, BioLegend), CCR6 (29-IL17, catalog 129819, BioLegend) and analyzed via flow cytometry (BD LSRFortessa). For intracellular cytokine staining, the cells were stimulated with 5 ng/mL of phorbol myristate acetate (Sigma), 1 μg/mL brefeldin A (Sigma), and 500 ng/mL ionomycin (Sigma) at 37 °C for 4 h. After fixation and permeabilization, harvested cells were stained with antibodies detecting: IL-17A (TC11-18H10.1, catalog 506930, BioLegend), GM-CSF (MP1-22E9, catalog 2131415, Invitrogen). Finally, the cells were measured by flow cytometry. The results were evaluated with FlowJo software (version 10.0.7, Tree Star, Ashland, OR, USA).
Treatment of Draining Lymph node Cells
CDLNs cells were stimulated by IRBP1–20 (20 ng/mL) with or without IL-23 (20 ng/mL, PeproTech, Rocky Hill, NJ, USA) at 37 °C for 72 h in a humidified incubator with 5% CO2. The cells were analyzed by flow cytometry.
Co-culture of CD4+CCR6+CXCR3− T cells with CD11C+ APCs
CD4+CCR6+CXCR3− T cells (Th17 cells) were isolated by cell sorting and CD11C+ APCs were isolated by CD11C positive selection kit (STEMCELL, Vancouver, Canada). Then, sorted CD4+CCR6+CXCR3− T cells and CD11C+ APCs were co-cultured at a ratio of 2:1 with or without anti-GM-CSF (10 g/mL, R&D Systems) for 24 h. The supernatants were used to measure IL-23 by ELISA kit (Invitrogen, Carlsbad, CA, USA).
EAU induction by the adoptive transfer experiment
CD4+CCR6+CXCR3− T cells (Th17 cells) sorted from CDLNs of old and young EAU mice were stimulated with IRBP1–20 (20 μg/mL) for 72 h, washed with PBS three times, then injected through the tail vein into young or old C57BL/6j mice (2 × 107 living cells/ mice).
Pathological examination of eyes
Different groups of mice were euthanized humanely, their eyes were extracted and placed in 10% neutral-buffered formalin for at least 24 h. The fixed eyes were dehydrated in alcohol and embedded in paraffin. Finally, paraffin blocks were sectioned at a thickness of 4 μm. The sections were photographed by a microscope system after staining with hematoxylin and eosin. For each eye, four sections from four different layers were graded. The pathological scores were graded with a score between 0 to 4 as previously described (Chen and Caspi, 2019a). Scoring criteria in details was already in the Table S6.
IRBP antibody assay
Serum levels of mice antibody against IRBP1–20 (IgG) from different groups of mice after immunization were quantified by ELISA. Briefly, the wells were coated with 1 µg/mL IRBP1–20 at 4 °C overnight then blocked with 3% bovine serum albumin in PBS at 4 °C for 2 h. Diluted serum samples at 1:100 in 3% bovine serum albumin (BSA) were added to the wells. After incubation at 37 °C for 1.5 h to allow the antibody to bind the antigen, the sera were removed, and the wells were washed. Bound antibody was detected with goat anti-mouse IgG diluted at 1:2,000 for 1.5 h (Southern Biotechnology, Birmingham, AL, USA). Absorbance was read at 450 nm and mean optical density (OD ± SEM) was calculated for each group after adding stop solution.
Immunofluorescence staining
CDLNs were removed and fixed with 4% paraformaldehyde overnight at 4 °C, dehydrated using sucrose with different concentration gradients (10%, overnight; 20%, 6–8 h; 30%, overnight) and embedded in Tissue-Tek O.C.T Compound (Sakura Finetek USA, Inc). Sagittal sections (4 µm) were cut and blocked with 3% bovine serum albumin in PBS for 30 min at 23 °C ± 2 °C. The sections were incubated with anti-GM-CSF antibodies (1:100, MP1-22E9, catalog 2131415, Invitrogen) overnight at 4 °C and further incubated with the corresponding fluorochrome-conjugated secondary antibody (1:1,000, Cell Signaling Technology, Danvers, MA, USA) for 50 min. After counterstaining with DAPI (catalog ab104139, Abcam), the sections were examined with a fluorescence microscope (Nikon, Tokyo, Japan). The number of immunoreactive cells or percentage of immunoreactive area (20× objective) was analyzed via ImageJ software (v1.46r, NIH, Bethesda, MD, USA).
Enzyme-linked immunosorbent assay (ELISA)
Mouse sera or supernatants form cell coculture medium were analyzed with the following ELISA kits according to the manufacturer’s instructions: IL-23 Mouse ELISA Kit (BMS6017, Thermo Fisher Scientific) and GM-CSF Mouse ELISA Kit (BMS612, Thermo Fisher Scientific).
Single-cell collection
CDLNs were transferred to DMEM (Thermo Fisher Scientific) containing 2% FBS (Thermo Fisher Scientific), Pen/Strep antibiotics (Thermo Fisher Scientific), 3 mg/mL Collagenase IV (Sigma-Aldrich) and 40 mg/mL DNase I (Sigma-Aldrich), and incubated at 37 °C for 15 min. Digested cells were collected and filtered through a 70 um cell strainer. The single cell suspension was generated at 1 × 107 cells/mL (viability ≥ 85%) as determined using the Countess® II Automated Cell Counter.
scRNA-seq library preparation
scRNA-seq libraries were generated utilizing the Chromium Single Cell 5’ Library and Gel Bead Kit (10× Genomics, 120237) according to the manufacturer’s instruction with some modifications. In detail, after washing with 0.04% BSA buffer (0.02 g BSA dissolved in 50 mL deionized PBS), cells were captured in droplets. Then, reverse transcription, emulsion breaking, barcoded-cDNA purification with Dynabeads, and PCR amplification were conducted step by step. The amplified cDNA was then used for 5’ gene expression library construction. Specifically, fragmenting and end-repair, double-size selection with SPRIselect beads, and sequencing were conducted on 50 ng of amplified cDNA using NovaSeq platform (Illumina NovaSeq6000) to yield 150 bp paired-end reads.
Quality control
Raw data (Raw reads) of fastq files were assembled from the Raw BCL files using Illumina’s bcl2fastq converter. For primary quality control with FastQC software (S Andrews, 2010), the following parameters were assessed, (1) contained N was no more than 3; (2) the proportion of base with quality value below 5 was no more than 20%; (3) adapter sequence was removed. All the downstream analyses were based on the cleaned data with high quality.
Analysis of scRNA-seq data
The command “cellranger count” in CellRanger Software Suite (version 3.1.0) was used to demultiplex and barcode the sequences from NovaSeq system. Calculation of the single-cell expression matrix was performed via the Seurat software (version 2.3.4) for filtering, data normalization, dimensionality reduction, clustering, and differentially expressed gene analysis (Butler et al., 2018).
Quality control
Before proceeding further, cells with a number of genes <300 or >4,000, or with a ratio of mitochondrial genes >15% were excluded. Most cells with high expression of Hbb-a1 and Hbb-bs that were recognized as red blood cells were filtered. The batch effect across different samples was removed with the Harmony package (version 1.0) (Korsunsky et al., 2019).
Dimensionality reduction and clustering analysis
For analysis of scRNA-seq data with Seurat, the data were normalized via “NormalizeData’’ function, followed by PCA on top 2,000 variable genes via ‘‘FindVariableGenes’’ function with the default parameters. ‘‘FindClusters’’ function was used to cluster cells and ‘‘RunUMAP’’ function was used to visualize with a 2-dimensional UMAP algorithm. In addition, ‘‘FindAllMarkers’’ function with default parameters was used to generate marker genes of different clusters.
Cell type composition variation analysis
The number of cells from different cell types across YN, YE, ON, and OE mice were divided by the total number of cells in the same group to generate the cell type ratio. The Log2fold-change (Log2FC) in YE/YN, OE/ON, OE/YE comparison groups were used to recognize the alteration of cell types during aging and EAU (|Log2FC| > 0.5).
DEG analysis
DEG analysis from different kinds of cells between different groups (ON/YN, YE/YN, OE/ON) was performed by ‘‘FindMarkers’’ function (adjusted P value < 0.05, |LogFC| > 0.25). Before DEG analysis, we excluded the cell types that were missing or had fewer than three cells in the comparison groups.
Gene Ontology enrichment analysis
The Metascape webtool (www.metascape.org) (Zhou et al., 2019) was used to perform DEGs gene ontology (GO) and pathway enrichment analysis. Among the top 50 enriched GO terms or pathways across different kinds of cells, 5–10 GO terms or pathways that were associated with aging or EAU were visualized with ggplot2 (Ginestet, 2011).
Cell-cell communication
The intercellular communication was performed with CellPhoneDB (version 1.1.0) (Vento-Tormo et al., 2018). We selected and analyzed the ligand-receptor pairs that existed in more than 10% of a given cell type. We compared the mean of expression from ligand-receptor pairs in different cell types then selected pairs with P < 0.05 for further computerization of intercellular communication.
Cell–cell signaling pathways
Cell–cell signaling pathways were analyzed using the CellChat package (version 1.1.0) (Jin et al., 2021) to predicts major signaling inputs and outputs for cells and how those cells and signals coordinate for functions by network analysis and pattern recognition approaches.
Transcriptional factor (TF)
According to the workflow (http://scenic.aertslab.org/), TF-binding motifs were performed with the GENIE3 packages (version 1.6.0) (Huynh-Thu et al., 2010) and the RcisTarget database (version 1.4.0) (Verfaillie et al., 2015) of the SCENIC (version 1.1.2.2) (Aibar et al., 2017). TFs were identified by SCENIC only if they were found to be active in more than 1% of all cells and correlated with at least one other regulon (|r| > 0.3).
Pseudotime analysis
Pseudotime analysis was performed with the Monocle2 package (Qiu et al., 2017). Gene ordering was performed by using a cutoff of expression in at least 10 cells and a combination of intercluster differential expression and dispersion with a q-value cutoff of <0.1. The structure of the trajectory was plotted in 2-dimensional space using the DDRTree dimensionality reduction algorithm, and the cells were ordered in pseudotime.
Statistical analysis
GraphPad Prism Software was used for data analysis and presentation. The values are represented as the mean ± SD. Statistical analysis was performed with an unpaired, two-tailed Student’s t-test or one-way ANOVA. P values above 0.05 were considered as not significant, ns; *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Data availability statement
The single-cell sequencing data have been deposited at Genome Sequence Archive (GSA) with the project number PRJCA006064 and GSA accession number CRA004687.
ABBREVIATIONS
APCs, antigen-presenting cells; AU, autoimmune uveitis; BC, B cells; BSA, bovine serum albumin; CD4-CTL, CD4+ T cells with cytotoxic activity; CD8-CTL, CD8+ T cells with cytotoxic activity; CNS, central nervous system; CDLNs, cervical draining lymph nodes; cDC, conventional dendritic cells; DEGs, differentially expressed genes; EAU, experimental autoimmune uveitis; FCGR, Fc-gamma receptor; Fcrl5-BC, Fcrl5-positive B cells; GO, Gene Ontology; GBC, germinal B cells; GC, germinal center; H&E, Hemotoxylin and eosin; IgG, immunoglobulins G; IRBP1-20, interphotoreceptor retinoid-binding protein 1-20; LNs, Lymph nodes; Macro, macrophages; Mono, monocytes; NBC, naïve B cells; naïve CD4, naïve CD4+ T cells; naïve CD8, naïve CD8+ T cells; NK, natural killer cell; Neu, neutrophils; OE, old mice with EAU; ON, old normal mice; PTX, pertussis toxin; PBC, plasma B cells; pDC, plasmacytoid dendritic cells; Pro-T, proliferative T cells; scRNA-seq, single-cell RNA sequencing; TC, T cells; Tex, exhausted T cells; Tfh, T follicular helper cells; TFs, transcriptional factors; Th1, T helper 1 cells; Th17, T helper 17 cells; Treg, regulatory T cells; YE, young mice with EAU; YN, young normal mice.
DECLARATIONS
This work was supported by the National Key Research and Development Program of China (2017YFA0105804), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16010000), the National Key Research and Development Program of China (2020YFA0804000, 2018YFC2000100, 2017YFA0103304, 2020YFA0803401, 2019YFA0802202), the National Natural Science Foundation of China (Grant Nos. 81921006, 81625009, 91749202, 91949209, 81822018, 82125011, 82122024, 31970597), the Key Research Program of the Chinese Academy of Sciences (KFZD-SW-221), and the 14th Five-year Network Security and Informatization Plan of Chinese Academy of Sciences (WX145XQ07-18). He Li, Lei Zhu, Rong Wang, Lihui Xie, Jie Ren, Shuai Ma, Weiqi Zhang, Xiuxing Liu, Zhaohao Huang, Binyao Chen, Zhaohuai Li, Huyi Feng, Guang-Hui Liu, Si Wang, Jing Qu, and Wenru Su declare that they have no conflict of interest. All procedures followed were in accordance with the ethical standards of the Institutional Animal Care Committee (Zhongshan Ophthalmic Center, Sun Yat-Sen University). He Li, Lei Zhu, Rong Wang, Lihui Xie, Jie Ren, Shuai Ma, Weiqi Zhang, Xiuxing Liu, Zhaohao Huang, Binyao Chen, Zhaohuai Li, Huyi Feng, Guang-Hui Liu, Si Wang, Jing Qu, and Wenru Su declare that they agreed to participate in the project. He Li, Lei Zhu, Rong Wang, Lihui Xie, Jie Ren, Shuai Ma, Weiqi Zhang, Xiuxing Liu, Zhaohao Huang, Binyao Chen, Zhaohuai Li, Huyi Feng, Guang-Hui Liu, Si Wang, Jing Qu, and Wenru Su declare that they agreed to publish the project. The single-cell sequencing data is deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG, https://bigd.big.ac.cn/gsa/), Chinese Academy of Sciences, with the project number PRJCA006064 and GSA accession number CRA004687.
References
Agarwa RK, Silver PB, Caspi RR (2012) Rodent models of experimental autoimmune uveitis. Methods in Molecular Biology (clifton, N.J.) 900:443–469
Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine J, Geurts P, Aerts J et al (2017) SCENIC: single-cell regulatory network inference and clustering. Nat Methods 14(11):1083–1086
Amadi-Obi A, Yu C, Liu X, Mahdi RM, Clarke GL, Nussenblatt RB, Gery I, Lee YS, Egwuagu CE (2007) TH17 cells contribute to uveitis and scleritis and are expanded by IL-2 and inhibited by IL-27/STAT1. Comp Study 13(6):711–718
Andrews SA (2010) FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Aprahamian T, Takemura Y, Goukassian D, Walsh K (2008) Ageing is associated with diminished apoptotic cell clearance in vivo. Clin Exp Immunol 152(3):448–455
Armitage RJ, Fanslow WC, Strockbine L, Sato TA, Clifford KN, Macduff BM, Anderson DM, Gimpel SD, Davis-Smith T, Maliszewski CR (1992) Molecular and biological characterization of a murine ligand for CD40. Nature 357(6373):80–82
Avichezer D, Liou GI, Chan C, Lewis GM, Wiggert B, Donoso LA, Nickerson JM, Crawford MA, Caspi RR (2003) Interphotoreceptor retinoid-binding protein (IRBP)-deficient C57BL/6 mice have enhanced immunological and immunopathogenic responses to IRBP and an altered recognition of IRBP epitopes. J Autoimmun 21(3):185–194
Blecher-Gonen R, Bost P, Hilligan KL, David E, Salame TM, Roussel E, Connor LM, Mayer JU, Bahar Halpern K, Tóth B et al (2019) Single-cell analysis of diverse pathogen responses defines a molecular roadmap for generating antigen-specific immunity. Cell Syst 8(2):109-121.e6
Botafogo V, Pérez-Andres M, Jara-Acevedo M, Bárcena P, Grigore G, Hernández-Delgado A, Damasceno D, Comans S, Blanco E, Romero A et al (2020) Age Distribution of Multiple Functionally Relevant Subsets of CD4+ T Cells in Human Blood Using a Standardized and Validated 14-Color EuroFlow Immune Monitoring Tube. Front Immunol 11166:14
Bovbjerg DH, Kim YT, Schwab R, Schmitt K, DeBlasio T, Weksler ME (1991) “Cross-wiring” of the immune response in old mice: increased autoantibody response despite reduced antibody response to nominal antigen. Cell Immunol 135(2):519–525
Butler A, Hoffman P, Smibert P, Papalexi E, Satija R (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36(5):411–420
Cambier J (2005) Immunosenescence: a problem of lymphopoiesis, homeostasis, microenvironment, and signaling. Immunol Rev 2055:6
Caspi RR, Silver PB, Luger D, Tang J, Cortes LM, Pennesi G, Mattapallil MJ, Chan C (2008) Mouse models of experimental autoimmune uveitis. Ophthalmic Res 40(3–4):169–174
Chan CC, Caspi RR, Ni M, Leake WC, Wiggert B, Chader GJ, Nussenblatt RB (1990) Pathology of experimental autoimmune uveoretinitis in mice. J Autoimmun 3(3):247–255
Chen J, Caspi RR (2019a) Clinical and functional evaluation of ocular inflammatory disease using the model of experimental autoimmune uveitis, Springer, New York, pp 211–227
Chen J, Caspi RR (2019b) Clinical and functional evaluation of ocular inflammatory disease using the model of experimental autoimmune uveitis. Methods in Molecular Biology (clifton, N.J.) 1899211–227:44–469
Chong WP, Mattapallil MJ, Raychaudhuri K, Bing SJ, Wu S, Zhong Y, Wang W, Chen Z, Silver PB, Jittayasothorn Y et al (2020) The cytokine IL-17A Limits Th17 pathogenicity via a negative feedback loop driven by autocrine induction of IL-24. Immunity 53(2):384-397.e5
Codarri L, Gyülvészi G, Tosevski V, Hesske L, Fontana A, Magnenat L, Suter T, Becher B (2011) RORγt drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol 12(6):560–567
Consortium., T. M (2020) A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583(7817):590–595
Cooper GS, Stroehla BC (2003) The epidemiology of autoimmune diseases. Autoimmun Rev 2(3):119–125
Crotty S (2011) Follicular helper CD4 T cells (TFH). Ann Rev Immunol 29621:63
de Smet MD, Taylor SRJ, Bodaghi B, Miserocchi E, Murray PI, Pleyer U, Zierhut M, Barisani-Asenbauer T, LeHoang P, Lightman S (2011) Understanding uveitis: the impact of research on visual outcomes. Prog Retin Eye Res 30(6):452–470
Dittel BN (2008) CD4 T cells: balancing the coming and going of autoimmune-mediated inflammation in the CNS. Brain Behav Immun 22(4):421–430
Eaton SM, Burns EM, Kusser K, Randall TD, Haynes L (2004) Age-related defects in CD4 T cell cognate helper function lead to reductions in humoral responses. J Exp Med 200(12):1613–1622
El-Behi M, Ciric B, Dai H, Yan Y, Cullimore M, Safavi F, Zhang G, Dittel BN, Rostami A (2011a) The encephalitogenicity of TH17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol 12(6):568–575
El-Behi M, Ciric B, Dai H, Yan Y, Cullimore M, Safavi F, Zhang G, Dittel BN, Rostami A (2011b) The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol 12(6):568–575
Elyahu Y, Monsonego A (2021) Thymus involution sets the clock of the aging T-cell landscape: Implications for declined immunity and tissue repair. Ageing Res Rev 65101:231
Elyahu Y, Hekselman I, Eizenberg-Magar I, Berner O, Strominger I, Schiller M, Mittal K, Nemirovsky A, Eremenko E, Vital A et al (2019) Aging promotes reorganization of the CD4 T cell landscape toward extreme regulatory and effector phenotypes. Sci Adv 5(8):8330–8330
Franceschi C, Garagnani P, Parini P, Giuliani C, Santoro A (2018) Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat Rev Endocrinol 14(10):576–590
Franceschi C, Garagnani P, Vitale G, Capri M, Salvioli S (2017) Inflammaging and “Garb-aging.” Trends Endocrinol Metab 28(3):199–212
Frasca D, Blomberg BB (2011) Aging affects human B cell responses. J Clin Immunol 31(3):430–435
Frasca D, Van der Put E, Riley RL, Blomberg BB (2004) Reduced Ig class switch in aged mice correlates with decreased E47 and activation-induced cytidine deaminase. J Immunol 172(4):2155–2162
Gasteiger G, Ataide M, Kastenmüller W (2016) Lymph node - an organ for T-cell activation and pathogen defense. Immunol Rev 271(1):200–220
Gavazzi G, Krause K (2002) Ageing and infection. Lancet Infect Dis 2(11):659–666
Ginestet C (2011) ggplot2: elegant graphics for data analysis. J R Stat Soc Ser A 174(1):245–245
Goronzy JJ, Weyand CM (2012) Immune aging and autoimmunity. Cell Mol Life Sci 69(10):1615–1623
Goronzy JJ, Weyand CM (2013) Understanding immunosenescence to improve responses to vaccines. Nat Immunol 14(5):428–436
Goronzy JJ, Weyand CM (2017) Successful and maladaptive T cell aging. Immunity 46(3):364–378
Goronzy JJ, Weyand CM (2019) Mechanisms underlying T cell ageing. Nat Rev Immunol 19(9):573–583
Grün D, van Oudenaarden A (2015) Design and analysis of single-cell sequencing experiments. Cell 163(4):799–810
Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT (2005) Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol 6(11):1123–1132
Hasan Z, Koizumi S, Sasaki D, Yamada H, Arakaki N, Fujihara Y, Okitsu S, Shirahata H, Ishikawa H (2017) JunB is essential for IL-23-dependent pathogenicity of Th17 cells. Nat Commun 8(1):1–12
Huynh-Thu VA, Irrthum A, Wehenkel L, Geurts P (2010) Inferring regulatory networks from expression data using tree-based methods. Nat Commun 8(1):1–12
Ifergan I, Davidson TS, Kebir H, Xu D, Palacios-Macapagal D, Cann J, Rodgers JM, Hunter ZN, Pittet CL, Beddow S et al (2017) Targeting the GM-CSF receptor for the treatment of CNS autoimmunity. J Autoimmun 84:1–11
Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan C, Myung P, Plikus MV, Nie Q (2021) Inference and analysis of cell-cell communication using Cell Chat. Nat Commun 12(1):1088
Khan U, Ghazanfar H (2018) T lymphocytes and autoimmunity. Int Rev Cell Mol Biol 44:341125–341168
Kim HO, Kim H, Youn J, Shin E, Park S (2011) Serum cytokine profiles in healthy young and elderly population assessed using multiplexed bead-based immunoassays. J Transl Med 9113:3892
Kleinewietfeld M, Manzel A, Titze J, Kvakan H, Yosef N, Linker RA, Muller DN, Hafler DA (2013) Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature 496(7446):518–522
Kogut I, Scholz JL, Cancro MP, Cambier JC (2012) B cell maintenance and function in aging. Semin Immunol 24(5):342–349
Komuczki J, Tuzlak S, Friebel E, Hartwig T, Spath S, Rosenstiel P, Waisman A, Opitz L, Oukka M, Schreiner B et al (2019) Fate-mapping of GM-CSF expression identifies a discrete subset of inflammation-driving t helper cells regulated by cytokines IL-23 and IL-1β. Immunity 50(5):1289-1304.e6
Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh P, Raychaudhuri S (2019) Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16(12):1289–1296
Lam WY, Bhattacharya D (2018) Metabolic links between plasma cell survival, secretion, and stress. Trends Immunol 39(1):19–27
Larbi A, Fülöp T, Pawelec G (2008) Immune receptor signaling, aging and autoimmunity. Adv Exp Med Biol 640312:24
Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA et al (2012) Induction and molecular signature of pathogenic TH17 cells. Nat Immunol 13(10):991–999
Levine BL, Ueda Y, Craighead N, Huang ML, June CH (1995) CD28 ligands CD80 (B7–1) and CD86 (B7–2) induce long-term autocrine growth of CD4+ T cells and induce similar patterns of cytokine secretion in vitro. Int Immunol 7(6):891–904
Lin C, Bradstreet TR, Schwarzkopf EA, Sim J, Carrero JA, Chou C, Cook LE, Egawa T, Taneja R, Murphy TL et al (2014) "Bhlhe40 controls cytokine production by T cells and is essential for pathogenicity in autoimmune neuroinflammation. Nat Commun 5:1
Liu M, Li S, Li MO (2018) TGF-β control of adaptive immune tolerance: a break from Treg cells. BioEssays 40(11):1800063
Llibre A, López-Macías C, Marafioti T, Mehta H, Partridge A, Kanzig C, Rivellese F, Galson JD, Walker LJ, Milne P et al (2016) LLT1 and CD161 expression in human germinal centers promotes B cell activation and CXCR4 downregulation. J Immunol 196(5):2085–2094
Lotfi N, Thome R, Rezaei N, Zhang G, Rezaei A, Rostami A, Esmaeil N (2019) Roles of GM-CSF in the pathogenesis of autoimmune diseases: an update. Front Immunol 101265:158
Louveau A, Herz J, Alme MN, Salvador AF, Dong MQ, Viar KE, Herod SG, Knopp J, Setliff JC, Lupi AL et al (2018) CNS lymphatic drainage and neuroinflammation are regulated by meningeal lymphatic vasculature. Nat Neurosci 21(10):1380–1391
Luger D, Silver PB, Tang J, Cua D, Chen Z, Iwakura Y, Bowman EP, Sgambellone NM, Chan C, Caspi RR (2008) Either a Th17 or a Th1 effector response can drive autoimmunity: conditions of disease induction affect dominant effector category. J Exp Med 205(4):799–810
Luo G, Gao Q, Zhang S, Yan B (2020) Probing infectious disease by single-cell RNA sequencing: progresses and. Comput Struct Biotechnol J 182962:2971
Meffre E, O’Connor KC (2019) Impaired B-cell tolerance checkpoints promote the development of autoimmune diseases and pathogenic autoantibodies. Immunol Rev 292(1):90–101
Mochizuki M, Sugita S, Kamoi K (2013) Immunological homeostasis of the eye. Progress Retinal Eye Res 3310:27
Montgomery RR, Shaw AC (2015) Paradoxical changes in innate immunity in aging: recent progress and new directions. J Leukoc Biol 98(6):937–943
Moulias R, Proust J, Wang A, Congy F, Marescot MR, Deville Chabrolle A, Paris Hamelin A, Lesourd B (1984) Age-related increase in autoantibodies. Lancet 323(8386):1128–1129
Müller L, Pawelec G (2015) As we age: Does slippage of quality control in the immune system lead to collateral damage? Ageing Res Rev 23:116–123
Muñoz LE, Janko C, Schulze C, Schorn C, Sarter K, Schett G, Herrmann M (2010) Autoimmunity and chronic inflammation - two clearance-related steps in the etiopathogenesis of SLE. Autoimmun Rev 10(1):38–42
Nikolich-Žugich J (2018) The twilight of immunity: emerging concepts in aging of the immune system. Nat Immunol 19(1):10–19
Paik DT, Cho S, Tian L, Chang HY, Wu JC (2020) Single-cell RNA sequencing in cardiovascular development, disease and medicine. Nat Rev Cardiol 17(8):457–473
Papotto PH, Marengo EB, Sardinha LR, Goldberg AC, Rizzo LV (2014) Immunotherapeutic strategies in autoimmune uveitis. Autoimmun Rev 13(9):909–916
Patil VS, Madrigal A, Schmiedel BJ, Clarke J, O’Rourke P, de Silva AD, Harris E, Peters B, Seumois G, Weiskopf D et al (2018) Precursors of human CD4(+) cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci Immunol 3(19):8664
Pennesi G, Mattapallil MJ, Sun S, Avichezer D, Silver PB, Karabekian Z, David CS, Hargrave PA, McDowell JH, Smith WC et al (2003) A humanized model of experimental autoimmune uveitis in HLA class II transgenic mice. J Clin Investig 111(8):1171–1180
Pinti M, Appay V, Campisi J, Frasca D, Fülöp T, Sauce D, Larbi A, Weinberger B, Cossarizza A (2016) Aging of the immune system: focus on inflammation and vaccination. Eur J Immunol 46(10):2286–2301
Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C (2017) Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14(10):979–982
Rantalainen M (2018) Application of single-cell sequencing in human cancer. Brief Funct Genomics 17(4):273–282
Richner JM, Gmyrek GB, Govero J, Tu Y, van der Windt GJW, Metcalf TU, Haddad EK, Textor J, Miller MJ, Diamond MS (2015a) Age-dependent cell trafficking defects in draining lymph nodes impair adaptive immunity and control of west Nile virus infection. PLoS Pathog 11(7):e1005027
Richner JM, Gmyrek GB, Govero J, Tu Y, van der Windt GJW, Metcalf TU, Haddad EK, Textor J, Miller MJ, Diamond MS (2015b) Age-Dependent Cell Trafficking Defects in Draining Lymph Nodes Impair Adaptive Immunity and Control of West Nile Virus Infection. PLoS Pathog 11(7):1005027
Rodda LB, Lu E, Bennett ML, Sokol CL, Wang X, Luther SA, Barres BA, Luster AD, Ye CJ, Cyster JG (2018) Single-cell RNA sequencing of lymph node stromal cells reveals niche-associated heterogeneity. Immunity 48(5):1014-1028.e6
Rovenský J, Tuchynová A (2008) Systemic lupus erythematosus in the elderly. Autoimmun Rev 7(3):235–239
Ruffatti A, Rossi L, Calligaro A, Del Ross T, Lagni M, Marson P, Todesco S (1990) Autoantibodies of systemic rheumatic diseases in the healthy elderly. Gerontology 36(2):104–111
Samy ET, Parker LA, Sharp CP, Tung KSK (2005) Continuous control of autoimmune disease by antigen-dependent polyclonal CD4+CD25+ regulatory T cells in the regional lymph node. J Exp Med 202(6):771–781
Savina A, Amigorena S (2007) Phagocytosis and antigen presentation in dendritic cells. Immunol Rev 219(1):143–156
Shabgah AG, Shariati-Sarabi Z, Tavakkol-Afshari J, Mohammadi M (2019) The role of BAFF and APRIL in rheumatoid arthritis. J Cell Physiol 234(10):17050–17063
Shaw AC, Joshi S, Greenwood H, Panda A, Lord JM (2010) Aging of the innate immune system. Curr Opin Immunol 22(4):507–513
Sheikh AAD, Akatsu C, Abdu-Allah HHM, Suganuma Y, Imamura A, Ando H, Takematsu H, Ishida H, Tsubata T (2021) The Protein Tyrosine Phosphatase SHP-1 (PTPN6) but Not CD45 (PTPRC) Is Essential for the Ligand-Mediated Regulation of CD22 in BCR-Ligated B Cells. J Immunol 206(11):2544–2551
Smith JR, Stempel AJ, Bharadwaj A, Appukuttan B (2016) Involvement of B cells in non-infectious uveitis. Clin Trans Immunol 5(2):63
Solana R, Pawelec G, Tarazona R (2006) Aging and Innate Immunity. Immunity (cambridge, Mass) 24(5):491–494
Solana R, Tarazona R, Gayoso I, Lesur O, Dupuis G, Fulop T (2012) Innate immunosenescence: effect of aging on cells and receptors of the innate immune system in humans. Semin Immunol 24(5):331–341
Stacy S, Krolick KA, Infante AJ, Kraig E (2002) Immunological memory and late onset autoimmunity. Mech Ageing Dev 123(8):975–985
Tabibian-Keissar H, Hazanov L, Schiby G, Rosenthal N, Rakovsky A, Michaeli M, Shahaf GL, Pickman Y, Rosenblatt K, Melamed D et al (2016) Aging affects B-cell antigen receptor repertoire diversity in primary and secondary lymphoid tissues. Eur J Immunol 46(2):480–492
Tesar BM, Walker WE, Unternaehrer J, Joshi NS, Chandele A, Haynes L, Kaech S, Goldstein DR (2006) Murine [corrected] myeloid dendritic cell-dependent toll-like receptor immunity is preserved with aging. Aging Cell 5(6):473–486
Theofilopoulos AN, Kono DH, Baccala R (2017) The multiple pathways to autoimmunity. Nat Immunol 18(7):716–724
Thorne JE, Suhler E, Skup M, Tari S, Macaulay D, Chao J, Ganguli A (2016) Prevalence of Noninfectious Uveitis in the United States: A Claims-Based Analysis. JAMA Ophthalmol 134(11):1237–1245
Tsirouki T, Dastiridou A, Symeonidis C, Tounakaki O, Brazitikou I, Kalogeropoulos C, Androudi S (2016) A focus on the epidemiology of uveitis. Ocul Immunol Inflamm 26(1):2–16
Turner VM, Mabbott NA (2017) Structural and functional changes to lymph nodes in ageing mice. Immunology 151(2):239–247
Uyemura K, Castle SC, Makinodan T (2002) The frail elderly: role of dendritic cells in the susceptibility of infection. Mech Ageing Dev 123(8):955–962
Veerman K, Tardiveau C, Martins F, Coudert J, Girard J (2019) Single-cell analysis reveals heterogeneity of high endothelial venules and different regulation of genes controlling lymphocyte entry to lymph nodes. Cell Rep 26(11):3116-3131.e5
Veldhoen M, Hirota K, Westendorf AM, Buer J, Dumoutier L, Renauld J, Stockinger B (2008) The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 453(7191):106–109
Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, Park J, Stephenson E, Polański K, Goncalves A et al (2018) Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563(7731):347–353
Verfaillie A, Imrichova H, Janky R, Aerts S (2015) iRegulon and i-cisTarget: reconstructing regulatory networks using motif and track enrichment. Curr Protoc Bioinform 52(1):2–16
Wang Y, Fu Z, Li X, Liang Y, Pei S, Hao S, Zhu Q, Yu T, Pei Y, Yuan J et al (2021) Cytoplasmic DNA sensing by KU complex in aged CD4+ T cell potentiates T cell activation and aging-related autoimmune inflammation. Immunity 54(4):632-647.e9
Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM (2006) Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity 24(6):677–688
Wilson NJ, Boniface K, Chan JR, McKenzie BS, Blumenschein WM, Mattson JD, Basham B, Smith K, Chen T, Morel F et al (2007) Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol 8(9):950–957
Yasuda K, Takeuchi Y, Hirota K (2019) The pathogenicity of Th17 cells in autoimmune diseases. Seminars Immunopathol 41(3):283–297
Yeh T, Okano T, Naruto T, Yamashita M, Okamura M, Tanita K, Du L, Pan-Hammarström Q, Mitsuiki N, Okada S et al (2020) APRIL-dependent lifelong plasmacyte maintenance and immunoglobulin production in humans. J Allergy Clin Immunol 146(5):1109-1120.e4
Yu J, Zhou X, Chang M, Nakaya M, Chang J, Xiao Y, Lindsey JW, Dorta-Estremera S, Cao W, Zal A et al (2015) Regulation of T-cell activation and migration by the kinase TBK1 during neuroinflammation. Nat Commun 6(1):1–13
Zhao Y, Yamasaki R, Yamaguchi H, Nagata S, Une H, Cui Y, Masaki K, Nakamuta Y, Iinuma K, Watanabe M et al (2020) Oligodendroglial connexin 47 regulates neuroinflammation upon autoimmune demyelination in a novel mouse model of multiple sclerosis. Proc Natl Acad Sci USA 117(4):2160–2169
Zheng Y, Liu X, Le W, Xie L, Li H, Wen W, Wang S, Ma S, Huang Z, Ye J et al (2020) A human circulating immune cell landscape in aging and COVID-19. Protein Cell 11(10):740–770
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK (2019) Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10(1):1523
Zhu L, Chen B, Su W (2021) A review of the various roles and participation levels of B-cells in non-infectious uveitis. Front Immunol 12:676046
Author information
Authors and Affiliations
Corresponding authors
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, H., Zhu, L., Wang, R. et al. Aging weakens Th17 cell pathogenicity and ameliorates experimental autoimmune uveitis in mice. Protein Cell 13, 422–445 (2022). https://doi.org/10.1007/s13238-021-00882-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13238-021-00882-3